The Role of HSP90 in Preserving the Integrity of Genomes Against Transposons Is Evolutionarily Conserved
Abstract
:1. HSP90 Is an Evolutionarily Conserved Molecular Chaperone
2. HSP90 Acts to Maintain the Integrity of the Genomes
2.1. Transposable Elements and the piRNA Pathway
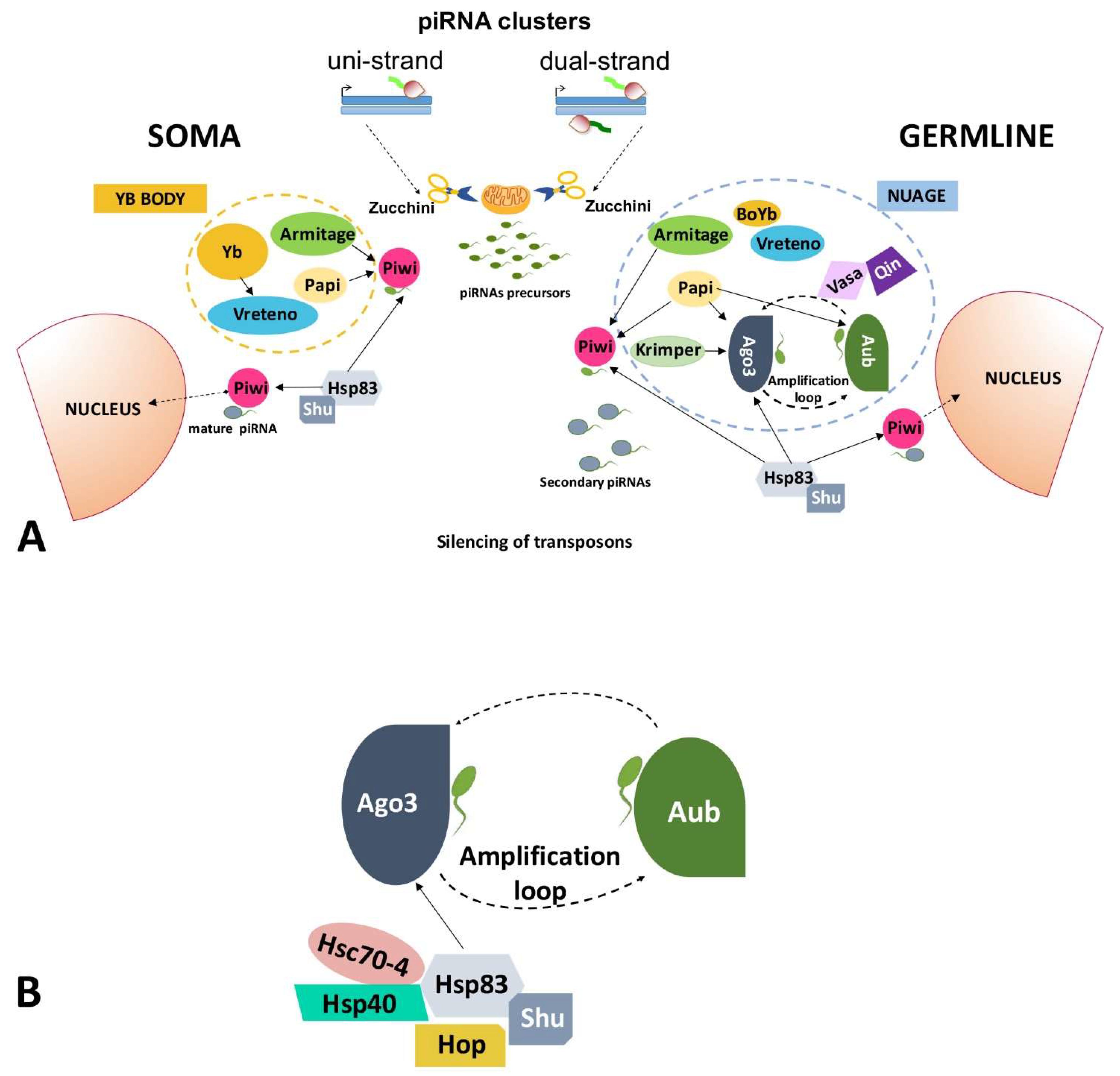
2.1.1. piRNA Pathway in Germ Cells of the Ovary
2.1.2. piRNA Pathway in Somatic Cells of the Ovary
2.2. HSP90 Has a Role in the Regulation of Transposable Elements
3. HSP90, Transposable Elements, and Neurological Diseases
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Taipale, M.; Jarosz, D.F.; Lindquist, S. HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 2010, 11, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Prodromou, C. Mechanisms of Hsp90 regulation. Biochem. J. 2016, 473, 2439–2452. [Google Scholar] [CrossRef] [Green Version]
- Schopf, F.H.; Biebl, M.M.; Buchner, J. The HSP90 chaperone machinery. Nat. Rev. Mol. Cell Biol. 2017, 18, 345–360. [Google Scholar] [CrossRef]
- Mimnaugh, E.G.; Chavany, C.; Neckers, L. Polyubiquitination and proteasomal degradation of the p185c–erbB-2 receptor protein-tyrosine kinase induced by geldanamycin. J. Biol. Chem. 1996, 271, 22796–22801. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Zhou, L.; Prodromou, C.; Savic, V.; Pearl, L.H. HECTD3 Mediates an HSP90-Dependent Degradation Pathway for Protein Kinase Clients. Cell Rep. 2017, 19, 2515–2528. [Google Scholar] [CrossRef] [Green Version]
- Ziegler, A.N.; Levison, S.W.; Wood, T.L. Insulin and IGF receptor signaling in neural-stem-cell homeostasis. Nat. Rev. Endocrinol. 2015, 11, 161–170. [Google Scholar] [CrossRef] [Green Version]
- Quintà, H.R.; Galigniana, M.D. The neuroregenerative mechanism mediated by the Hsp90-binding immunophilin FKBP52 resenbles the early steps of neuronal differentiation. Br. J. Pharmacol. 2012, 166, 637–649. [Google Scholar]
- Trepel, J.; Mollapour, M.; Giaccone, G.; Neckers, L. Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer 2010, 10, 537–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Csermely, P.; Schnaider, T.; Soti, C.; Prohaszka, Z.; Nardai, G. The 90-kDa molecular chaperone family: Structure, function, and clinical applications. A comprehensive review. Pharmacol. Ther. 1998, 79, 129–168. [Google Scholar] [CrossRef]
- Chen, B.; Zhong, D.; Monteiro, A. Comparative genomics and evolution of the HSP90 family of genes across all kingdoms of organisms. BMC Genom. 2006, 7, 156. [Google Scholar] [CrossRef] [Green Version]
- Sreedhar, A.S.; Kalmar, E.; Csermely, P.; Shen, Y.F. Hsp90 isoforms: Functions, expression and clinical importance. FEBS Lett. 2004, 562, 11–15. [Google Scholar] [CrossRef]
- Chen, B.; Piel, W.H.; Bruford, E.; Monteiro, A. The HSP90 family of genes in the human genome: Insights into their divergence and evolution. Genomics 2005, 86, 627–637. [Google Scholar] [CrossRef] [PubMed]
- Cutforth, T.; Rubin, G.M. Mutations in Hsp83 and cdc37 impair signaling by the sevenless receptor tyrosine kinase in Drosophila. Cell 1994, 77, 1027–1036. [Google Scholar] [CrossRef]
- Birnby, D.A.; Link, E.M.; Vowels, J.J.; Tian, H.; Colacurcio, P.L.; Thomas, J.H. A transmembrane guanylylcyclase (DAF-11) and Hsp90 (DAF-21) regulate a com-mon set of chemosensory behaviors in Caenorhabditis elegans. Genetics 2000, 155, 85–104. [Google Scholar] [CrossRef]
- Zhao, R.; Davey, M.; Hzu, Y.-C.; Kaplanet, P.; Tong, A.; Parsons, A.B.; Krogan, N.; Cagney, G.; Mai, D.; Greenblatt, J.; et al. Navigating the chaperone network: An integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 2005, 120, 715–727. [Google Scholar] [CrossRef] [Green Version]
- Kaziales, A.; Barkovits, K.; Marcus, K.; Richter, K. Glucocorticoid receptor complexes form cooperatively with the Hsp90 co-chaperones Pp5 and FKBPs. Sci. Rep. 2020, 10, 10733. [Google Scholar] [CrossRef] [PubMed]
- Shelton, L.B.; Baker, J.D.; Zheng, D.; Sullivan, L.E.; Solanki, P.K.; Webster, J.M.; Sun, Z.; Sabbagh, J.J.; Nordhues, B.A.; Koren, J.; et al. Hsp90 activator Aha1 drives production of pathological tau aggregates. Proc. Natl. Acad. Sci. USA. 2017, 114, 9707–9712. [Google Scholar] [CrossRef] [Green Version]
- Blair, L.J.; Sabbagh, J.J.; Dickey, C.A. Targeting Hsp90 and its co-chaperones to treat Alzheimer’s disease. Expert Opin. Ther. Targets 2014, 18, 1219–1232. [Google Scholar] [CrossRef] [Green Version]
- Sawarkar Sawarkar, R.; Sievers, C.; Paro, R. Hsp90 globally targets paused RNA polymerase to regulate gene expression in response to environmental stimuli. Cell 2012, 149, 807–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawarkar, R.; Paro, R. Hsp90@ chromatin. nucleus: An emerging hub of a networker. Trends Cell Biol. 2013, 23, 193–201. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, R.; Furukawa, Y.; Morita, M.; Iimura, Y.; Silva, F.P.; Li, M.; Yagyu, R.; Nakamura, Y. SMYD3 encodes a histone methyltransferase involved in the proliferation of cancer cells. Nat. Cell Biol. 2004, 6, 731–740. [Google Scholar] [CrossRef] [PubMed]
- Tariq, M.; Nussbaumer, U.; Chen, Y.; Beisel, C.; Paro, R. Trithorax requires Hsp90 for maintenance of active chromatin at sites of gene expression. Proc. Natl. Acad. Sci. USA 2009, 106, 1157–1162. [Google Scholar] [CrossRef] [Green Version]
- Pennisi, R.; Ascenzi, P.; Di Masi, A. Hsp90: A new player in DNA repair? Biomolecules 2015, 5, 2589–2618. [Google Scholar] [CrossRef] [Green Version]
- Specchia, V.; Piacentini, L.; Tritto, P.; Fanti, L.; D’Alessandro, R.; Palumbo, G.; Pimpinelli, S.; Bozzetti, M.P. Hsp90 prevents phenotypic variation by suppressing the mutagenic activity of transposons. Nature 2010, 463, 662–665. [Google Scholar] [CrossRef] [PubMed]
- Gangaraju, V.K.; Yin, H.; Weiner, M.M.; Wang, J.; Huang, X.A.; Lin, H. Drosophila Piwi functions in Hsp90-mediated suppression of phenotypic variation. Nat. Genet. 2011, 43, 153. [Google Scholar] [CrossRef] [Green Version]
- Ryan, C.P.; Broenlie, J.C.; Whyard, S. Hsp90 and physiological stress are linked to autonomous transposon mobility and heritable genetic change in nematodes. Genome Biol. Evol. 2016, 8, 3794–3805. [Google Scholar] [CrossRef] [Green Version]
- McClintock, B. The origin and behavior of mutable loci in maize. Proc. Natl. Acad. Sci. USA 1950, 36, 344–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pimpinelli, S.; Berloco, M.; Fanti, L.; Dimitri, P.; Bonaccorsi, S.; Marchetti, E.; Caizzi, R.; Caggese, C.; Gatti, M. Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc. Natl. Acad. Sci. USA 1995, 92, 3804–3808. [Google Scholar] [CrossRef] [Green Version]
- Lander, E.S. Initial impact of the sequencing of the human genome. Nature 2011, 470, 187–197. [Google Scholar] [CrossRef] [PubMed]
- Van Luenen, H.G.; Colloms, S.D.; Plasterk, R.H. The mechanism of transposition of Tc3 in C. elegans. Cell 1994, 79, 293–301. [Google Scholar] [CrossRef]
- Palazzo, A.; Marconi, S.; Specchia, V.; Bozzetti, M.P.; Ivics, Z.; Caizzi, R.; Marsano, R.M. Functional characterization of the Bari1 transposition system. PLoS ONE 2013, 8, e79385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCullers, T.J.; Steiniger, M. Transposable elements in Drosophila. Mob. Genet. Elem. 2017, 7, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chénais, B.; Caruso, A.; Hiard, S.; Casse, N. The impact of transposable elements on eukaryotic genomes: From genome size increase to genetic adaptation to stressful environments. Gene 2012, 509, 7–15. [Google Scholar] [CrossRef]
- Bourque, G.; Leong, B.; Vega, V.B.; Chen, X.; Lee, Y.L.; Srinivasan, K.G.; Chew, J.; Ruan, Y.; Wei, C.; Ng, H.H.; et al. Evolution of the mammalian transcription factor binding repertoire via transposable elements. Genome Res. 2008, 18, 1752–1762. [Google Scholar] [CrossRef] [Green Version]
- Cohen, C.J.; Lock, W.M.; Mager, D.L. Endogenous retroviral LTRs as promoters for human genes: A critical assessment. Gene 2009, 448, 105–114. [Google Scholar] [CrossRef]
- Teng, L.; Firpi, H.A.; Tan, K. Enhancers in embryonic stem cells are enriched for transposable elements and genetic variations associated with cancers. Nucleic Acids Res. 2011, 39, 7371–7379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Keightley, P.D.; Halligan, D.L. Effect of divergence time and recombination rate on molecular evolution of Drosophila INE-1 transposable elements and other candidates for neutrally evolving sites. J. Mol. Evol. 2007, 65, 627. [Google Scholar] [CrossRef]
- Czech, B.; Munafò, M.; Ciabrelli, F.; Eastwood, E.L.; Fabry, M.H.; Kneuss, E.; Hannon, G.J. piRNA-guided genome defense: From biogenesis to silencing. Annu. Rev. Genet. 2018, 52, 131–157. [Google Scholar] [CrossRef] [PubMed]
- Aravin, A.; Gaidatzis, D.; Pfeffer, S.; Lagos-Quintana, M.; Landgraf, P.; Iovino, N.; Morris, P.; Brownstein, M.J.; Kuramochi-Miyagawa, S.; Nakano, T.; et al. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 2006, 442, 203–207. [Google Scholar] [CrossRef]
- Vagin, V.V.; Sigova, A.; Li, C.; Seitz, H.; Gvozdev, V.; Zamore, P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 2006, 313, 320–324. [Google Scholar] [CrossRef] [PubMed]
- Specchia, V.; Benna, C.; Mazzotta, G.M.; Piccin, A.; Zordan, M.A.; Costa, R.; Bozzetti, M.P. aubergine gene overexpression in somatic tissues of auberginesting mutants interferes with the RNAi pathway of a yellow hairpin dsRNA in Drosophila melanogaster. Genetics 2008, 178, 1271–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malone, C.D.; Hannon, G.J. Small RNAs as guardians of the genome. Cell 2009, 136, 656–668. [Google Scholar] [CrossRef] [Green Version]
- Senti, K.A.; Brennecke, J. The piRNa pathway: A fly’s perspective on the guardian of the genome. Trens Gent. 2010, 26, 499–509. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, T.; Tomizawa, S.I.; Mitsuya, K.; Totoki, Y.; Yamamoto, Y.; Kuramochi-Miyagawa, S.; Iida, N.; Hoki, Y.; Murphy, P.J.; Toyoda, A.; et al. Role for piRNAs and noncoding RNA in de novo DNA methylation of the imprinted mouse Rasgrf1 locus. Science 2011, 332, 848–852. [Google Scholar] [CrossRef] [Green Version]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef] [Green Version]
- Nishida, K.M.; Saito, K.; Mori, T.; Kawamura, Y.; Nagami-Okada, T.; Inagaki, S.; Siomi, H.; Siomi, M.C. Gene silencing mechanisms mediated by Aubergine–piRNA complexes in Drosophila male gonad. RNA 2007, 13, 1911–1922. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gunawardane, L.S.; Saito, K.; Nishida, K.M.; Miyoshi, K.; Kawamura, Y.; Nagami, T.; Siomi, H.; Siomi, M.C. A slicer-mediated mechanism for repeat-associated siRNA 5’end formation in Drosophila. Science 2007, 315, 1587–1590. [Google Scholar] [CrossRef] [Green Version]
- Klattenhoff, C.; Theurkauf, W. Biogenesis and germline functions of piRNAs. Development 2008, 135, 3–9. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.Z.; Xu, J.; Koppetsch, B.S.; Wang, J.; Tipping, C.; Ma, S.; Weng, Z.; Theurkauf, W.E.; Zamore, P.D. Heterotypic piRNA Ping-Pong Requies Qin, a Protein with Both E3 Ligase and Tudor Domains. Mol. Cell 2011, 44, 572–584. [Google Scholar] [CrossRef] [Green Version]
- Ge, D.T.; Wang, W.; Tipping, C.; Gainetdinov, I.; Zhiping, W.; Zamore., P.D. The RNA-Binding ATPase, Armitage, Couples piRNAAmplification in Nuage to Phased piRNA Production on Mitochondria. Mol. Cell 2019, 74, 982–995. [Google Scholar] [CrossRef] [PubMed]
- Xiol, J.; Spinelli, P.; Laussmann, M.A.; Homolka, D.; Yang, Z.; Cora, E.; Couté, Y.; Conn, S.; Jan, K.; Sachidanandam, R.; et al. RNA clamping by Vasa assembles a piRNA amplifier complex on transposon transcripts. Cell 2014, 157, 1698–1711. [Google Scholar] [CrossRef] [Green Version]
- Bozzetti, M.P.; Specchia, V.; Cattenoz, P.B.; Laneve, P.; Geusa, A.; Sahin, H.B.; DiTommaso, S.; Friscini, A.; Massari, S.; Diebold, C.; et al. The Drosophila fragile X mental retardation protein participates in the piRNA pathway. J. Cell Sci. 2015, 128, 2070–2084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, K.; Iwasaki, Y.W.; Shibuya, A.; Carninci, P.; Tsuchizawa, Y.; Ishizu, H.; Siomi, M.; Siomi, H. Krimper enforces an antisense bias on piRNA pools by binding AGO3 in the Drosophila germline. Mol. Cell 2015, 59, 553–563. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Ishizu, H.; Komai, M.; Kotani, H.; Kawamura, Y.; Nishida, K.M.; Siomi, H.; Siomi, C. Roles for the Yb body components Armitage and Yb in primary piRNA biogenesis in Drosophila. Genes Dev. 2010, 24, 2493–2498. [Google Scholar] [CrossRef] [Green Version]
- Izumi, N.; Kawaoka, S.; Yasuhara, S.; Suzuki, Y.; Sugano, S.; Katsuma, S.; Tomari, Y. Hsp90 facilitates accurate loading of precursor piRNAs into PIWI proteins. RNA 2013, 19, 896–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olivieri, D.; Senti, K.A.; Subramanian, S.; Sachidanandam, R.; Brennecke, J. The cochaperone shutdown defines a group of biogenesis factors essential for all piRNA populations in Drosophila. Mol. Cell 2012, 47, 954–969. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preall, J.B.; Czech, B.; Guzzardo, P.M.; Muerdter, F.; Hannon, G.J. Shutdown is a component of the Drosophila piRNA biogenesis machinery. RNA 2012, 18, 1446–1457. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Qi, H.; Wang, J.; Lin, H. PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development 2011, 138, 1863–1873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malone, C.D.; Brennecke, J.; Dus, M.; Stark, A.; McCombie, W.R.; Sachinandam, R.; Hannon, G.J. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 2009, 137, 522–535. [Google Scholar] [CrossRef] [Green Version]
- Qi, H.; Watanabe, T.; Ku, H.Y.; Liu, N.; Zhong, M.; Lin, H. The Yb body, a major site for Piwi-associated RNA biogenesis and a gateway for Piwi expression and transport to the nucleus in somatic cells. J. Biol. Chem. 2011, 286, 3789–3797. [Google Scholar] [CrossRef] [Green Version]
- Rozhkov, N.V.; Hammell, M.; Hannon, G.J. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 2013, 27, 400–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brower-Toland, B.; Findley, S.D.; Jiang, L.; Liu, L.; Yin, H.; Dus, M.; Zhou, P.; Elgin, S.C.R.; Lim, H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007, 21, 2300–2311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.A.; Yin, H.; Sweeenev, S.; Raha, D.; Snyder, M.; Lin, H. A major epigenetic programming mechanism guided by piRNAs. Dev. Cell 2013, 24, 502–516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishizu, H.; Kinoshita, T.; Hirakata, S.; Komatsuzaki, C.; Siomi, M.C. Distinct and Collaborative Functions of Yb and Armitage in Transposon-Targeting piRNA Biogenesis. Cell Rep. 2019, 27, 1822–1835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bozzetti, M.P.; Fanti, L.; Di Tommaso, S.; Piacentini, L.; Berloco, M.; Tritto, P.; Specchia, V. The “Special” crystal-stellate system in Drosophila melanogaster reveals mechanisms underlying piRNA pathway-mediated canalization. Genet. Res. Int. 2012, 2012, 324293. [Google Scholar]
- Piacentini, L.; Fanti, L.; Specchia, V.; Bozzetti, M.P.; Berloco, M.; Palumbo, G.; Pimpinelli, S. Transposons, environmental changes, and heritable induced phenotypic variability. Chromosoma 2014, 123, 345–354. [Google Scholar] [CrossRef] [Green Version]
- Rutherford, S.L.; Lindquist, S. Hsp90 as a capacitor for morphological evolution. Nature 1998, 396, 336–342. [Google Scholar] [CrossRef]
- Xiol, J.; Cora, E.; Koglgruber, R.; Chuma, S.; Subramanian, S.; Hosokawa, M.; Reuter, M.; Yang, Z.; Berninger, P.; Palencia, A.; et al. A role for Fkbp6 and the chaperone machinery in piRNA amplification and transposon silencing. Mol. Cell 2012, 47, 970–979. [Google Scholar] [CrossRef] [Green Version]
- Ohsako, S.; Bunick, D.; Hayashi, Y. Immunocytochemical observation of the 90 KD heat shock protein (HSP90): High expression in primordial and pre-meiotic germ cells of male and female rat gonads. J. Histochem. Cytochem. 1995, 43, 67–76. [Google Scholar] [CrossRef]
- Cappucci, U.; Noro, F.; Casale, A.M.; Fanti, L.; Berloco, M.; Alagia, A.A.; Grassi, L.; Le Pera, L.; Piacentini, L.; Pimpineli, S. The Hsp70 chaperone is a major player in stress-induced transposable eletn activation. Proc. Natl. Acad. Sci. USA 2019, 116, 17943–17950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coufal, N.G.; Garcia-Perez, J.L.; Peng, G.E.; Yeo, G.W.; Mu, Y.; Loyci, M.T.; Morell, M.; O’Sgea, K.S.; Moran., J.V.; Gage, F.H. L1 retrotransposition in human neural progenitor cells. Nature 2009, 460, 1127–1131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baillie, J.K.; Barnett, M.W.; Upton, K.R.; Gerhardt, D.J.; Richmond, T.A.; De SApio, F.; Brennan, P.M.; Rizzu, P.; Smith, S.; Fell, M.; et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature 2011, 479, 534–537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muotri, A.R.; Marchetto, M.C.; Coufal, N.G.; Oefner, R.; Yeo, G.; Nakashima, K.; Gage, F.H. L1 retrotransposition in neurons is modulated by MeCP2. Nature 2010, 468, 443–446. [Google Scholar] [CrossRef] [PubMed]
- Kuwabara, T.; Hsieh, J.; Muotri, A.; Yeo, G.; Warashira, M.; Chichung, L.; Moore, L.; Makashima, K.; Asashima, M.; Gage, F. Wnt-mediated activation of NeuroD1 and retro-elements during adult neurogenesis. Nat. Neurosci. 2009, 12, 1097–1105. [Google Scholar] [CrossRef] [Green Version]
- Dubnau, J.; Grady, L.; Kitamoto, T.; Tully, T. Disruption of neurotransmission in Drosophila mushroom body blocks retrieval but not acquisition of memory. Nature 2001, 411, 476–480. [Google Scholar] [CrossRef]
- Perrat, P.N.; DasGupta, S.; Wang, J.; Theurkauf, W.; Weng, Z.; Rosbash, M.; Waddell, S. Transposition-driven genomic heterogeneity in the Drosophila brain. Science 2013, 340, 91–95. [Google Scholar] [CrossRef] [Green Version]
- Amir, R.E.; Van den Veyver, I.B.; Wan, M.; Tran, C.Q.; Francke, U.; Zonghbi, H.Y. Rett syndrome is cuased by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat. Genet. 1999, 23, 185–188. [Google Scholar] [CrossRef]
- Jiang, F.; Lu, F.; Li, P.; Liu, W.; Zhao, L.; Wang, Q.; Cao, X.; Zhang, L.; Zhang, Y.Q. Drosophila Homolog of FMRP Maintains Genome Integrity by Interacting with Piwi. J. Genet. Genom. 2016, 43, 11–24. [Google Scholar] [CrossRef]
- Specchia, V.; Puricella, A.; D’Attis, S.; Massari, S.; Giangrande, A.; Bozzetti, M.P. Drosophila melanogaster as a Model to Study the Multiple Phenotypes, Related to Genome Stability of the Fragile-X Syndrome. Front. Genet. 2019, 10, 10. [Google Scholar] [CrossRef]
- Drozd, M.; Bardoni, B.; Capovilla, M. Modeling Fragile X Syndrome in Drosophila. Front. Mol. Neurosci. 2018, 11, 124. [Google Scholar] [CrossRef]
- Caudy, A.A.; Myers, M.; Hammon, G.J.; Hammond, S.M. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002, 16, 2491–2496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishizuka, A.; Siomi, M.C.; Siomi, H. A Drosophila Fragile X protein interacts with component of RNAi and ribosomal proteins. Genes Dev. 2002, 16, 2497–2508. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Xu, S.; Xia, L.; Wang, J.; Wen, S.; Jin, P.; Chen, D. The bantam microRNA is associated with Drosophila fFagile X mental retardation protein and regulates the fate of germline stem cells. PLoS Genet. 2009, 5, e1000444. [Google Scholar] [CrossRef]
- Minakhina, S.; Changela, N.; Steward, R. Zfrp8/PDCD2 is required in ovarian stem cells and interacts with the piRNA pathway machinery. Development 2014, 141, 259–268. [Google Scholar] [CrossRef] [Green Version]
- Tan, W.; Schauder, C.; Naryshkina, T.; Minakhina, S.; Steward, R. Zfrp8 forms a complex with fragile-X mental retardation protein and regulates its localization and function. Dev. Biol. 2016, 410, 202–212. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doll, C.A.; Vita, D.J.; Broadie, K. Fragile X mental retardation protein requirements in activity-dependent critical period neural circuit refinement. Curr. Biol. 2017, 27, 2318–2330. [Google Scholar] [CrossRef]
- Chen-Plotkin, A.S.; Lee, V.M.Y.; Trojanowski, J.Q. TAR DNA-binding protein 43 in neurodegenerative disease. Nat. Rev. Neurol. 2010, 6, 211–220. [Google Scholar] [CrossRef] [Green Version]
- Neumann, M.; Sampathu, D.M.; Kwong, L.K.; Truax, A.C.; Micsenyi, M.C.; Chou, T.T.; Bruce, J.; Schuck, T.; Grossman, M.; Clark, C.M.; et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006, 314, 130–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, E.B.; Lee, V.M.; Trojanowski, J.Q. Gains or losses: Molecular mechanisms of TDP43-mediated neurodegeneration. Nat. Rev. Neurosci. 2012, 13, 38–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krug, L.; Chatterjee, N.; Borges-Monroy, R.; Hearn, S.; Liao, W.W.; Morrill, K.; Rozhkov, N.; Theodorou, D.; Hammell, M.; Dubnau, J. Retrotransposon activation contributes to neurodegeneration in a Drosophila TDP-43 model of ALS. PLoS Genet. 2017, 13, e1006635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romano, G.; Klima, R.; Feiguin, F. TDP-43 prevents retrotransposon activation in the Drosophila motor system through regulation of Dicer-2 activity. BMC Biol. 2020, 18, 32. [Google Scholar] [CrossRef] [PubMed]
- Coyne, A.N.; Yamada, S.B.; Siddegowda, B.B.; Estes, P.S.; Zaepfel, B.L.; Johannesmeyer, J.S.; Lockwood, D.B.; Pham, L.T.; Hart, M.P.; Cassel, J.A.; et al. Fragile X protein mitigates TDP-43 toxicity by remodeling RNA granules and restoring translation. Hum. Mol. Genet. 2015, 24, 6886–6898. [Google Scholar] [CrossRef] [PubMed]
- Majumder, P.; Chu, J.F.; Chatterjee, B.; Swamy, K.B.; Shen, C.J. Co-regulation of mRNA translation by TDP-43 and Fragile X Syndrome protein FMRP. Acta Neuropathol. 2016, 132, 721–738. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jinwal, U.K.; Abisambra, J.F.; Zhang, J.; Dharia, S.; O’Leary, J.C.; Patel, T.; Braswell, K.; Jani, T.; Gestwicki, J.E.; Dickey, C.A. Cdc37/Hsp90 protein complex disruption triggers an autophagic clearance cascade for TDP-43 protein. J. Biol. Chem. 2012, 287, 24814–24820. [Google Scholar] [CrossRef] [Green Version]
- Bohush, A.; Bieganowski, P.; Filipek, A. Hsp90 and Its Co-Chaperones in Neurodegenerative Diseases. Int. J. Mol. Sci. 2019, 20, e4976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, C.; Jeong, H.H.; Hsieh, Y.C.; Klein, H.U.; Bennett, D.A.; De Jager, L.; Liu, Z.; Shulman, J.M. Tau activates transposable elements in Alzheimer’s disease. Cell Rep. 2018, 23, 2874–2880. [Google Scholar] [CrossRef] [PubMed]
| Genes | Abbreviation in Drosophila | Ortholog in Humans | Protein Type and Domains | Protein Function in the piRNA Pathway | References |
|---|---|---|---|---|---|
| aubergine | aub | PIWIL1 | Argonaute Protein | primary pathway ping-pong pathway | [45,48,49] |
| ago 3 | ago3 | PIWIL2 | Argonaute Protein | ping-pong pathway | [45,48,49] |
| piwi | piwi | PIWIL3 | Argonaute Protein | primary pathway | [46] |
| armitage | armi | MOV10L1 | RNA helicases (SDE3) | primary pathway phased piRNA pathway | [50] |
| vasa | vasa | DDX4 | DEAD RNA helicase | primary pathway ping-pong pathway; | [51] |
| dFmr1 | dFmr1 | FMR1 | Tudor protein KH domain RGG domain | primary pathway ping-pong pathway | [52] |
| krimper | krimp | TDRD6 | Tudor protein | ping-pong pathway | [53] |
| papi | papi | TDRKH | Tudor protein KH domain | primary pathway ping-pong pathway | [51] |
| qin/kumo | qin/kumo | TDRD1 | Tudor protein RING domain | primary pathway (dual strand cluster) | [49] |
| vreteno | vret | TDRD15 | Tudor protein RRM domain | primary pathway | [54] |
| Yb | Yb | DDX46 | Tudor protein DEAD RNA helicase | primary pathway (single strand cluster) | [42] |
| zucchini | zuc | PLD6 | Tudor protein nuclease domain | primary pathway phased piRNA pathway | [50] |
| hsp83 | hsp83 | HSP90 | Heat shock protein | primary pathway ping-pong pathway | [24] |
| hsp40 | hsp40 | DNAJB5 | Heat shock protein | primary pathway ping-pong pathway | [55] |
| hsp70 cognate-4 | hsc70-4 | HSPA8 | Heat shock protein cognate | primary pathway ping-pong pathway | [55] |
| hsp70/hsp90 organizing protein | hop (Sti1) | HOP1 (STIP1) | Cochaperon | primary pathway ping-pong pathway | [55] |
| shutdown | shu | FKBP6 | Cochaperon | primary pathway ping-pong pathway | [56,57] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Specchia, V.; Bozzetti, M.P. The Role of HSP90 in Preserving the Integrity of Genomes Against Transposons Is Evolutionarily Conserved. Cells 2021, 10, 1096. https://doi.org/10.3390/cells10051096
Specchia V, Bozzetti MP. The Role of HSP90 in Preserving the Integrity of Genomes Against Transposons Is Evolutionarily Conserved. Cells. 2021; 10(5):1096. https://doi.org/10.3390/cells10051096
Chicago/Turabian StyleSpecchia, Valeria, and Maria Pia Bozzetti. 2021. "The Role of HSP90 in Preserving the Integrity of Genomes Against Transposons Is Evolutionarily Conserved" Cells 10, no. 5: 1096. https://doi.org/10.3390/cells10051096
APA StyleSpecchia, V., & Bozzetti, M. P. (2021). The Role of HSP90 in Preserving the Integrity of Genomes Against Transposons Is Evolutionarily Conserved. Cells, 10(5), 1096. https://doi.org/10.3390/cells10051096






