Physiological Fitness and the Pathophysiology of Chronic Lymphocytic Leukemia (CLL)
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Characteristics
2.2. Clinical Characteristics
2.3. Physical Performance and Fitness
2.4. Blood Sampling
2.5. Autologous Plasma Incubation with Cell Lines
2.6. Exosomal miRNA
miRNA Analyses
2.7. Flow Cytometry
2.8. Nuclear Magnetic Resonance (NMR) Spectroscopy
2.9. Statistical Analyses
3. Results
3.1. Group Demographics, Clinical Measures, Physical Fitness, and Function
3.2. Cell Line Growth with Autologous Plasma
3.3. Exosomal microRNA Profiles
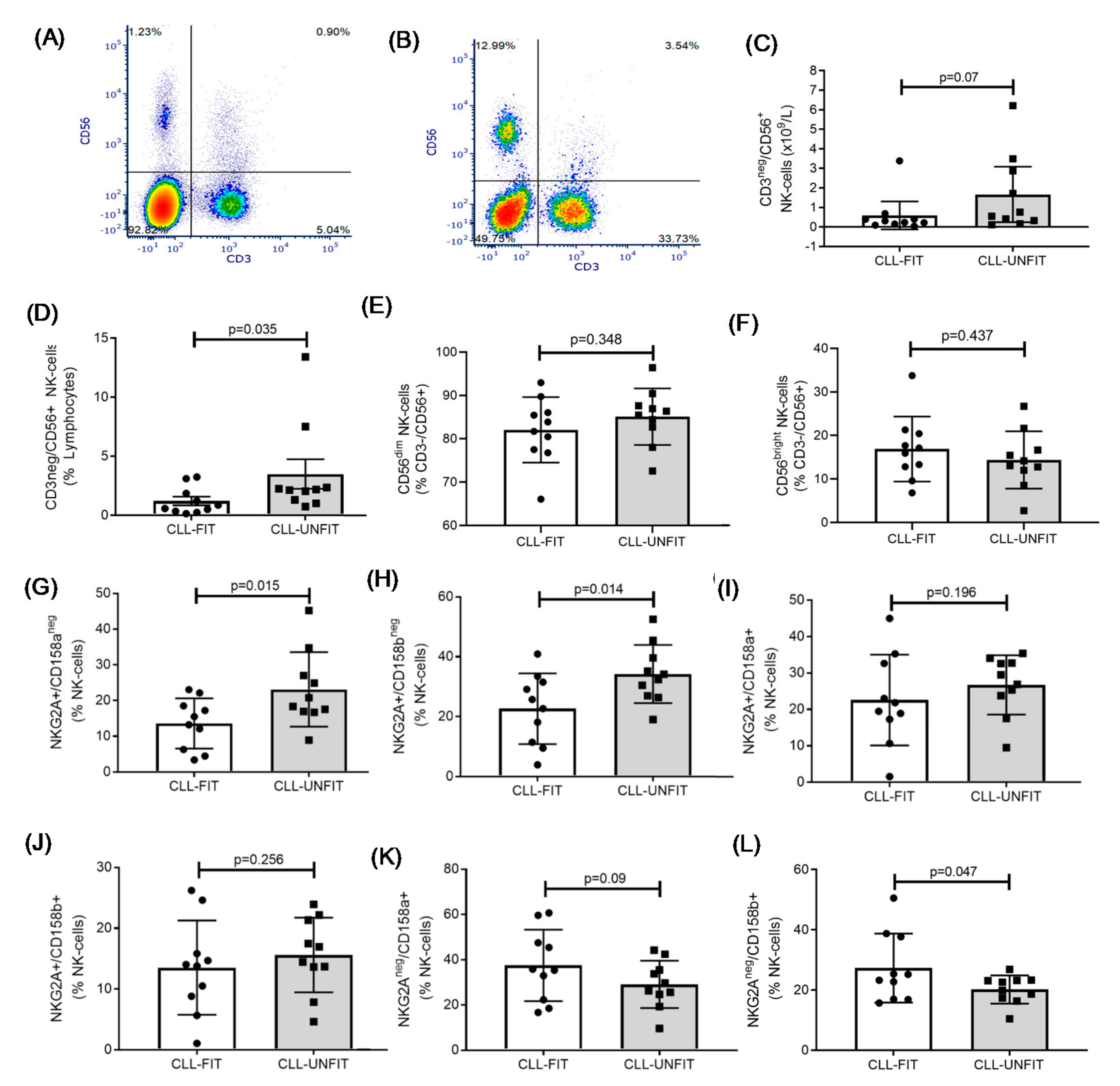
3.4. NK Cell Immunophenotype
3.5. NMR Measured Lipids, Lipoproteins (LipoProfile®), and Inflammatory Profiles
3.6. Correlations between miRNAs, Immune Cells, and Lipids
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rai, K.R.; Jain, P. Chronic lymphocytic leukemia (CLL)-Then and now. Am. J. Hematol. 2016, 91, 330–340. [Google Scholar] [CrossRef] [PubMed]
- Washburn, L. CME Chronic lymphocytic leukemia Chronic lymphocytic leukemia: The most common leukemia in adults. J. Am. Acad. Physician Assist. 2011, 24, 54–58. [Google Scholar] [CrossRef]
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R. SEER Cancer Statistics Review, 1975–2017; National Cancer Institute: Bethesda, MD, USA, 2020. Available online: https://seer.cancer.gov/csr/1975_2017/ (accessed on 1 November 2020).
- Goldin, L.R.; Björkholm, M.; Kristinsson, S.Y.; Turesson, I.; Landgren, O. Elevated risk of chronic lymphocytic leukemia and other indolent non-Hodgkin’s lymphomas among relatives of patients with chronic lymphocytic leukemia. Haematologica 2009, 94, 647–653. [Google Scholar] [CrossRef]
- Scarfò, L.; Ferreri, A.J.; Ghia, P. Chronic lymphocytic leukaemia. Crit. Rev. Oncol. 2016, 104, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Shanafelt, T.D.; Rabe, K.G.; Kay, N.E.; Zent, C.S.; Jelinek, D.F.; Reinalda, M.S.; Schwager, S.M.; Bowen, D.A.; Slager, S.L.; Hanson, C.A.; et al. Age at diagnosis and the utility of prognostic testing in patients with chronic lymphocytic leukemia. Cancer 2010, 116, 4777–4787. [Google Scholar] [CrossRef]
- Solomon, B.M.; Rabe, K.G.; Slager, S.L.; Brewer, J.D.; Cerhan, J.R.; Shanafelt, T.D. Overall and Cancer-Specific Survival of Patients With Breast, Colon, Kidney, and Lung Cancers with and without Chronic Lymphocytic Leukemia: A SEER Population-Based Study. J. Clin. Oncol. 2013, 31, 930–937. [Google Scholar] [CrossRef]
- Rossi, D.; De Paoli, L.; Rossi, F.M.; Cerri, M.; Deambrogi, C.; Rasi, S.; Zucchetto, A.; Capello, D.; Gattei, V.; Gaïdano, G. Early stage chronic lymphocytic leukaemia carrying unmutated IGHV genes is at risk of recurrent infections during watch and wait. Br. J. Haematol. 2008, 141, 734–736. [Google Scholar] [CrossRef]
- Riches, J.C.; Gribben, J.G. Immunomodulation and Immune Reconstitution in Chronic Lymphocytic Leukemia. Semin. Hematol. 2014, 51, 228–234. [Google Scholar] [CrossRef]
- Dighiero, G.; Maloum, K.; Desablens, B.; Cazin, B.; Navarro, M.; Leblay, R.; Leporrier, M.; Jaubert, J.; Lepeu, G.; Dreyfus, B.; et al. Chlorambucil in indolent chronic lymphocytic leukemia. French Cooperative Group on Chronic Lymphocytic Leukemia. N. Engl. J. Med. 1998, 338, 1506–1514. [Google Scholar] [CrossRef] [PubMed]
- Shustik, C.; Mick, R.; Silver, R.; Sawitsky, A.; Rai, K.; Shapiro, L. Treatment of early chronic lymphocytic leukemia: Intermittent chlorambucil versus observation. Hematol. Oncol. 1988, 6, 7–12. [Google Scholar] [CrossRef]
- Goede, V.; Bahlo, J.; Chataline, V.; Eichhorst, B.; Dürig, J.; Stilgenbauer, S.; Kolb, G.; Honecker, F.; Wedding, U.; Hallek, M. Evaluation of geriatric assessment in patients with chronic lymphocytic leukemia: Results of the CLL9 trial of the German CLL study group. Leuk. Lymphoma 2015, 57, 789–796. [Google Scholar] [CrossRef]
- Sitlinger, A.; Brander, D.M.; Bartlett, D.B. Impact of exercise on the immune system and outcomes in hematologic malignancies. Blood Adv. 2020, 4, 1801–1811. [Google Scholar] [CrossRef]
- Streckmann, F.; Kneis, S.; Leifert, J.A.; Baumann, F.T.; Kleber, M.; Ihorst, G.; Herich, L.; Grüssinger, V.; Gollhofer, A.; Bertz, H. Exercise program improves therapy-related side-effects and quality of life in lymphoma patients undergoing therapy. Ann. Oncol. 2014, 25, 493–499. [Google Scholar] [CrossRef]
- Courneya, K.S.; Sellar, C.M.; Stevinson, C.; McNeely, M.L.; Peddle-McIntyre, C.J.; Friedenreich, C.M.; Tankel, K.; Basi, S.; Chua, N.; Mazurek, A.; et al. Randomized Controlled Trial of the Effects of Aerobic Exercise on Physical Functioning and Quality of Life in Lymphoma Patients. J. Clin. Oncol. 2009, 27, 4605–4612. [Google Scholar] [CrossRef] [PubMed]
- Koelwyn, G.J.; Quail, D.F.; Zhang, X.; White, R.M.; Jones, L.W. Exercise-dependent regulation of the tumour microenvironment. Nat. Rev. Cancer 2017, 17, 620–632. [Google Scholar] [CrossRef] [PubMed]
- Ngo, T.H.; Barnard, R.J.; Tymchuk, C.N.; Cohen, P.; Aronson, W.J. Effect of diet and exercise on serum insulin, IGF-I, and IGFBP-1 levels and growth of LNCaP cells in vitro (United States). Cancer Causes Control. 2002, 13, 929–935. [Google Scholar] [CrossRef] [PubMed]
- Barnard, R.J.; Gonzalez, J.H.; Liva, M.E.; Ngo, T.H. Effects of a Low-Fat, High-Fiber Diet and Exercise Program on Breast Cancer Risk Factors In Vivo and Tumor Cell Growth and Apoptosis In Vitro. Nutr. Cancer 2006, 55, 28–34. [Google Scholar] [CrossRef]
- Dethlefsen, C.; Lillelund, C.; Midtgaard, J.; Andersen, C.; Pedersen, B.K.; Christensen, J.F.; Hojman, P. Exercise regulates breast cancer cell viability: Systemic training adaptations versus acute exercise responses. Breast Cancer Res. Treat. 2016, 159, 469–479. [Google Scholar] [CrossRef]
- Devin, J.L.; Hill, M.M.; Mourtzakis, M.; Quadrilatero, J.; Jenkins, D.G.; Skinner, T.L. Acute high intensity interval exercise reduces colon cancer cell growth. J. Physiol. 2019, 597, 2177–2184. [Google Scholar] [CrossRef] [PubMed]
- Rozovski, U.; Keating, M.J.; Estrov, Z. Targeting inflammatory pathways in chronic lymphocytic leukemia. Crit. Rev. Oncol. 2013, 88, 655–666. [Google Scholar] [CrossRef]
- Caligaris-Cappio, F. Inflammation, the microenvironment and chronic lymphocytic leukemia. Haematology 2011, 96, 353–355. [Google Scholar] [CrossRef][Green Version]
- Schulz, A.; Toedt, G.; Zenz, T.; Stilgenbauer, S.; Lichter, P.; Seiffert, M. Inflammatory cytokines and signaling pathways are associated with survival of primary chronic lymphocytic leukemia cells in vitro: A dominant role of CCL2. Haematologica 2011, 96, 408–416. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, C.; Pedersen, K.S.; Hojman, P. Every exercise bout matters: Linking systemic exercise responses to breast cancer control. Breast Cancer Res. Treat. 2017, 162, 399–408. [Google Scholar] [CrossRef] [PubMed]
- Hertlein, E.; Beckwith, K.A.; Lozanski, G.; Chen, T.L.; Towns, W.H.; Johnson, A.J.; Lehman, A.; Ruppert, A.S.; Bolon, B.; Andritsos, L.; et al. Characterization of a New Chronic Lymphocytic Leukemia Cell Line for Mechanistic In Vitro and In Vivo Studies Relevant to Disease. PLoS ONE 2013, 8, e76607. [Google Scholar] [CrossRef]
- International CLL-IPI Working Group. An international prognostic index for patients with chronic lymphocytic leukaemia (CLL-IPI): A meta-analysis of individual patient data. Lancet Oncol. 2016, 17, 779–790. [Google Scholar] [CrossRef]
- Burr, J.F.; Bredin, S.S.D.; Faktor, M.D.; Warburton, D.E.R. The 6-Minute Walk Test as a Predictor of Objectively Measured Aerobic Fitness in Healthy Working-Aged Adults. Physician Sportsmed. 2011, 39, 133–139. [Google Scholar] [CrossRef]
- Bartlett, D.B.; Willis, L.H.; Slentz, C.A.; Hoselton, A.; Kelly, L.; Huebner, J.L.; Kraus, V.B.; Moss, J.; Muehlbauer, M.J.; Spielmann, G.; et al. Ten weeks of high-intensity interval walk training is associated with reduced disease activity and improved innate immune function in older adults with rheumatoid arthritis: A pilot study. Arthritis Res. 2018, 20, 1–15. [Google Scholar] [CrossRef]
- Herbert, Z.T.; Thimmapuram, J.; Xie, S.; Kershner, J.P.; Kolling, F.W.; Ringelberg, C.S.; Leclerc, A.; Alekseyev, Y.O.; Fan, J.; Podnar, J.W.; et al. Multisite Evaluation of Next-Generation Methods for Small RNA Quantification. J. Biomol. Tech. JBT 2020, 31, 47–56. [Google Scholar] [CrossRef]
- Mannerström, B.; Paananen, R.O.; Abu-Shahba, A.G.; Moilanen, J.; Seppänen-Kaijansinkko, R.; Kaur, S. Extracellular small non-coding RNA contaminants in fetal bovine serum and serum-free media. Sci. Rep. 2019, 9, 5538. [Google Scholar] [CrossRef] [PubMed]
- Karlsen, T.A.; Aae, T.F.; Brinchmann, J.E. Robust profiling of microRNAs and isomiRs in human plasma exosomes across 46 individuals. Sci. Rep. 2019, 9, 1–9. [Google Scholar] [CrossRef]
- Andrews, S.; Krueger, F.; Segonds-Pichon, A.; Biggins, L.; Krueger, C.; Wingett, S.; Montgomery, J. Available online: https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/ (accessed on 1 April 2018).
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, 25. [Google Scholar] [CrossRef]
- Kozomara, A.; Griffiths-Jones, S. miRBase: Annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014, 42, D68–D73. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Huber, W.; Carey, V.J.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, B.S.; Bravo, H.C.; Davis, S.; Gatto, L.; Girke, T.; et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 2015, 12, 115–121. [Google Scholar] [CrossRef] [PubMed]
- Flores-Guerrero, J.L.; Connelly, M.A.; Shalaurova, I.; Gruppen, E.G.; Kieneker, L.M.; Dullaart, R.P.; Bakker, S.J. Lipoprotein insulin resistance index, a high-throughput measure of insulin resistance, is associated with incident type II diabetes mellitus in the Prevention of Renal and Vascular End-Stage Disease study. J. Clin. Lipidol. 2019, 13, 129–137.e1. [Google Scholar] [CrossRef] [PubMed]
- Otvos, J.D.; Shalaurova, I.; Wolak-Dinsmore, J.; Connelly, M.A.; Mackey, R.H.; Stein, J.H.; Tracy, R.P. GlycA: A Composite Nuclear Magnetic Resonance Biomarker of Systemic Inflammation. Clin. Chem. 2015, 61, 714–723. [Google Scholar] [CrossRef]
- Gruppen, E.G.; Kunutsor, S.K.; Kieneker, L.M.; Van Der Vegt, B.; Connelly, M.A.; De Bock, G.H.; Gansevoort, R.T.; Bakker, S.J.; Dullaart, R.P.; Vegt, B.; et al. GlycA, a novel pro-inflammatory glycoprotein biomarker is associated with mortality: Results from the PREVEND study and meta-analysis. J. Intern. Med. 2019, 286, 596–609. [Google Scholar] [CrossRef]
- Shalaurova, I.; Connelly, M.A.; Garvey, W.T.; Otvos, J.D. Lipoprotein Insulin Resistance Index: A Lipoprotein Particle–Derived Measure of Insulin Resistance. Metab. Syndr. Relat. Disord. 2014, 12, 422–429. [Google Scholar] [CrossRef]
- Otvos, J.D.; Jeyarajah, E.J.; Bennett, D.W. Quantification of plasma lipoproteins by proton nuclear magnetic resonance spectroscopy. Clin. Chem. 1991, 37, 377–386. [Google Scholar] [CrossRef]
- Wolak-Dinsmore, J.; Gruppen, E.G.; Shalaurova, I.; Matyus, S.P.; Grant, R.P.; Gegen, R.; Bakker, S.J.; Otvos, J.D.; Connelly, M.A.; Dullaart, R.P. A novel NMR-based assay to measure circulating concentrations of branched-chain amino acids: Elevation in subjects with type 2 diabetes mellitus and association with carotid intima media thickness. Clin. Biochem. 2018, 54, 92–99. [Google Scholar] [CrossRef] [PubMed]
- Balatti, V.; Pekarky, Y.; Croce, C.M. Role of microRNA in chronic lymphocytic leukemia onset and progression. J. Hematol. Oncol. 2015, 8, 12. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.-H.; Moles, R.; Nicot, C. Clinical significance of microRNAs in chronic and acute human leukemia. Mol. Cancer 2016, 15, 1–16. [Google Scholar] [CrossRef] [PubMed]
- McCaw, L.; Shi, Y.; Wang, G.; Li, Y.-J.; Spaner, D.E. Low Density Lipoproteins Amplify Cytokine-signaling in Chronic Lymphocytic Leukemia Cells. EBioMedicine 2017, 15, 24–35. [Google Scholar] [CrossRef] [PubMed]
- Friedman, D.R. Lipids and Their Effects in Chronic Lymphocytic Leukemia. EBioMedicine 2017, 15, 2–3. [Google Scholar] [CrossRef] [PubMed]
- Huergo-Zapico, L.; Acebes-Huerta, A.; Gonzalez-Rodriguez, A.P.; Contesti, J.; Gonzalez-García, E.; Payer, A.R.; Villa-Alvarez, M.; Fernández-Guizán, A.; López-Soto, A.; Gonzalez, S. Expansion of NK Cells and Reduction of NKG2D Expression in Chronic Lymphocytic Leukemia. Correlation with Progressive Disease. PLoS ONE 2014, 9, e108326. [Google Scholar] [CrossRef]
- Forconi, F.; Moss, P. Perturbation of the normal immune system in patients with CLL. Blood 2015, 126, 573–581. [Google Scholar] [CrossRef]
- Rundqvist, H.; Augsten, M.; Stromberg, A.; Rullman, E.; Mijwel, S.; Kharaziha, P.; Panaretakis, T.; Gustafsson, T.; Östman, A. Effect of Acute Exercise on Prostate Cancer Cell Growth. PLoS ONE 2013, 8, e67579. [Google Scholar] [CrossRef]
- Pedersen, L.; Idorn, M.; Olofsson, G.H.; Lauenborg, B.; Nookaew, I.; Hansen, R.H.; Johannesen, H.H.; Becker, J.C.; Pedersen, K.S.; Dethlefsen, C.; et al. Voluntary Running Suppresses Tumor Growth through Epinephrine- and IL-6-Dependent NK Cell Mobilization and Redistribution. Cell Metab. 2016, 23, 554–562. [Google Scholar] [CrossRef]
- Bigley, A.B.; Rezvani, K.; Chew, C.; Sekine, T.; Pistillo, M.; Crucian, B.; Bollard, C.M.; Simpson, R.J. Acute exercise preferentially redeploys NK-cells with a highly-differentiated phenotype and augments cytotoxicity against lymphoma and multiple myeloma target cells. Brain Behav. Immun. 2014, 39, 160–171. [Google Scholar] [CrossRef]
- Barber, J.L.; Kraus, W.E.; Church, T.S.; Hagberg, J.M.; Thompson, P.D.; Bartlett, D.B.; Beets, M.W.; Earnest, C.P.; Huffman, K.M.; Landers-Ramos, R.Q.; et al. Effects of regular endurance exercise on GlycA: Combined analysis of 14 exercise interventions. Atherosclerosis 2018, 277, 1–6. [Google Scholar] [CrossRef]
- Bartlett, D.B.; Slentz, C.A.; Connelly, M.A.; Piner, L.W.; Willis, L.H.; Bateman, L.A.; Granville, E.O.; Bales, C.W.; Huffman, K.M.; Kraus, W.E. Association of the Composite Inflammatory Biomarker GlycA, with Exercise-Induced Changes in Body Habitus in Men and Women with Prediabetes. Oxidative Med. Cell. Longev. 2017, 2017, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Braga, T.V.; Evangelista, F.C.G.; Gomes, L.C.; Araújo, S.S.D.S.; Carvalho, M.D.G.; Sabino, A.D.P. Evaluation of MiR-15a and MiR-16-1 as prognostic biomarkers in chronic lymphocytic leukemia. Biomed. Pharmacother. 2017, 92, 864–869. [Google Scholar] [CrossRef] [PubMed]
- Pekarsky, Y.; Croce, C.M. Role of miR-15/16 in CLL. Cell Death Differ. 2015, 22, 6–11. [Google Scholar] [CrossRef]
- Kaur, G.; Ruhela, V.; Rani, L.; Gupta, A.; Sriram, K.; Gogia, A.; Sharma, A.; Kumar, L.; Gupta, R. RNA-Seq profiling of deregulated miRs in CLL and their impact on clinical outcome. Blood Cancer J. 2020, 10, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Balatti, V.; Acunzo, M.; Pekarky, Y.; Croce, C.M. Novel Mechanisms of Regulation of miRNAs in CLL. Trends Cancer 2016, 2, 134–143. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Calin, G.A.; Ferracin, M.; Cimmino, A.; Di Leva, G.; Shimizu, M.; Wojcik, S.E.; Iorio, M.V.; Visone, R.; Sever, N.I.; Fabbri, M.; et al. A MicroRNA Signature Associated with Prognosis and Progression in Chronic Lymphocytic Leukemia. N. Engl. J. Med. 2005, 353, 1793–1801. [Google Scholar] [CrossRef]
- Haderk, F.; Hanna, B.; Richter, K.; Schnölzer, M.; Zenz, T.; Stilgenbauer, S.; Lichter, P.; Seiffert, M. Extracellular vesicles in chronic lymphocytic leukemia. Leuk. Lymphoma 2013, 54, 1826–1830. [Google Scholar] [CrossRef]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet. 2011, 12, 99–110. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Whiteside, T.L. Tumor-Derived Exosomes and Their Role in Cancer Progression. Int. Rev. Cytol. 2016, 74, 103–141. [Google Scholar] [CrossRef]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Nisticò, N.; Maisano, D.; Iaccino, E.; Vecchio, E.; Fiume, G.; Rotundo, S.; Quinto, I.; Mimmi, S. Role of Chronic Lymphocytic Leukemia (CLL)-Derived Exosomes in Tumor Progression and Survival. Pharmaceutics 2020, 13, 244. [Google Scholar] [CrossRef] [PubMed]
- Yeh, Y.-Y.; Ozer, H.G.; Lehman, A.M.; Maddocks, K.; Yuh-Ying, Y.; Johnson, A.J.; Byrd, J.C. Characterization of CLL exosomes reveals a distinct microRNA signature and enhanced secretion by activation of BCR signaling. Blood 2015, 125, 3297–3305. [Google Scholar] [CrossRef]
- Bobrie, A.; Colombo, M.; Raposo, G.; Théry, C. Exosome Secretion: Molecular Mechanisms and Roles in Immune Responses. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef] [PubMed]
- Nair, V.D.; Ge, Y.; Li, S.; Pincas, H.; Jain, N.; Seenarine, N.; Amper, M.A.S.; Goodpaster, B.H.; Walsh, M.J.; Coen, P.M.; et al. Sedentary and Trained Older Men Have Distinct Circulating Exosomal microRNA Profiles at Baseline and in Response to Acute Exercise. Front. Physiol. 2020, 11, 605. [Google Scholar] [CrossRef]
- Moussay, E.; Wang, K.; Cho, J.-H.; van Moer, K.; Pierson, S.; Paggetti, J.; Nazarov, P.V.; Palissot, V.; Hood, L.E.; Berchem, G.; et al. MicroRNA as biomarkers and regulators in B-cell chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2011, 108, 6573–6578. [Google Scholar] [CrossRef]
- Marcucci, G.; Mrózek, K.; Radmacher, M.D.; Bloomfield, C.D.; Croce, C.M. MicroRNA expression profiling in acute myeloid and chronic lymphocytic leukaemias. Best Pract. Res. Clin. Haematol. 2009, 22, 239–248. [Google Scholar] [CrossRef]
- Cimmino, A.; Calin, G.A.; Fabbri, M.; Iorio, M.V.; Ferracin, M.; Shimizu, M.; Wojcik, S.E.; Aqeilan, R.I.; Zupo, S.; Dono, M.; et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc. Natl. Acad. Sci. USA 2005, 102, 13944–13949. [Google Scholar] [CrossRef] [PubMed]
- Mittelbrunn, M.; Gutiérrez-Vázquez, C.; Villarroya-Beltri, C.; González, S.; Sánchez-Cabo, F.; González, M.Á.; Bernad, A.; Sánchez-Madrid, F. Unidirectional transfer of microRNA-loaded exosomes from T cells to antigen-presenting cells. Nat. Commun. 2011, 2, 282. [Google Scholar] [CrossRef]
- Graner, M.W.; Schnell, S.; Olin, M.R. Tumor-derived exosomes, microRNAs, and cancer immune suppression. Semin. Immunopathol. 2018, 40, 505–515. [Google Scholar] [CrossRef]
- Ma, F.; Xu, S.; Liu, X.; Zhang, Q.; Xu, X.; Liu, M.; Hua, M.; Li, N.; Yao, H.; Cao, X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat. Immunol. 2011, 12, 861–869. [Google Scholar] [CrossRef]
- Sullivan, R.P.; Leong, J.W.; Fehniger, T.A. MicroRNA regulation of natural killer cells. Front. Immunol. 2013, 4, 44. [Google Scholar] [CrossRef]
- Wang, P.; Gu, Y.; Zhang, Q.; Han, Y.; Hou, J.; Lin, L.; Wu, C.; Bao, Y.; Su, X.; Jiang, M.; et al. Identification of Resting and Type I IFN-Activated Human NK Cell miRNomes Reveals MicroRNA-378 and MicroRNA-30e as Negative Regulators of NK Cell Cytotoxicity. J. Immunol. 2012, 189, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Donatelli, S.S.; Zhou, J.M.; Gilvary, D.L.; Eksioglu, E.A.; Chen, X.; Cress, W.D.; Haura, E.B.; Schabath, M.B.; Coppola, D.; Wei, S.; et al. TGF-β-inducible microRNA-183 silences tumor-associated natural killer cells. Proc. Natl. Acad. Sci. USA 2014, 111, 4203–4208. [Google Scholar] [CrossRef]
- Sadallah, S.; Schmied, L.; Eken, C.; Charoudeh, H.N.; Amicarella, F.; Schifferli, J.A. Platelet-Derived Ectosomes Reduce NK Cell Function. J. Immunol. 2016, 197, 1663–1671. [Google Scholar] [CrossRef] [PubMed]
- Pesce, S.; Greppi, M.; Ferretti, E.; Obino, V.; Carlomagno, S.; Rutigliani, M.; Thoren, F.B.; Sivori, S.; Castagnola, P.; Candiani, S.; et al. miRNAs in NK Cell-Based Immune Responses and Cancer Immunotherapy. Front. Cell Dev. Biol. 2020, 8. [Google Scholar] [CrossRef]
- Kraus, W.E.; Houmard, J.A.; Duscha, B.D.; Knetzger, K.J.; Wharton, M.B.; McCartney, J.S.; Bales, C.W.; Henes, S.; Samsa, G.P.; Otvos, J.D.; et al. Effects of the Amount and Intensity of Exercise on Plasma Lipoproteins. N. Engl. J. Med. 2002, 347, 1483–1492. [Google Scholar] [CrossRef] [PubMed]
- Kelley, G.; Kelley, K.S.; Tran, Z.V. Aerobic exercise, lipids and lipoproteins in overweight and obese adults: A meta-analysis of randomized controlled trials. Int. J. Obes. 2005, 29, 881–893. [Google Scholar] [CrossRef] [PubMed]
- Chow, S.; Buckstein, R.; Spaner, D.E. A link between hypercholesterolemia and chronic lymphocytic leukemia. Leuk. Lymphoma 2015, 57, 1–6. [Google Scholar] [CrossRef]
- Mozessohn, L.; Earle, C.; Spaner, D.; Cheng, S.Y.; Kumar, M.; Buckstein, R. The Association of Dyslipidemia with Chronic Lymphocytic Leukemia: A Population-Based Study. J. Natl. Cancer Inst. 2016, 109, djw226. [Google Scholar] [CrossRef] [PubMed]
- Yavasoglu, I.; Sargin, G.; Yilmaz, F.; Altındag, S.; Akgun, G.; Tombak, A.; Toka, B.; Dal, S.; Ozbas, H.; Cetin, G.; et al. Cholesterol Levels in Patients with Chronic Lymphocytic Leukemia. J. Natl. Med. Assoc. 2017, 109, 23–27. [Google Scholar] [CrossRef]
- Sankanagoudar, S.; Singh, G.; Mahapatra, M.; Kumar, L.; Chandra, N.C. Cholesterol Homeostasis in Isolated Lymphocytes: A Differential Correlation between Male Control and Chronic Lymphocytic Leukemia Subjects. Asian Pac. J. Cancer Prev. 2017, 18, 23–30. [Google Scholar]
- Friedman, D.R.; Magura, L.A.; Warren, H.A.C.; Harrison, J.D.; Diehl, L.F.; Weinberg, J.B. Statin use and need for therapy in chronic lymphocytic leukemia. Leuk. Lymphoma 2010, 51, 2295–2298. [Google Scholar] [CrossRef]
- Wang, Y.-C.; Li, Y.; Wang, X.-Y.; Zhang, D.; Zhang, H.; Wu, Q.; He, Y.-Q.; Wang, J.-Y.; Zhang, L.; Xia, H.; et al. Circulating miR-130b mediates metabolic crosstalk between fat and muscle in overweight/obesity. Diabetology 2013, 56, 2275–2285. [Google Scholar] [CrossRef]
- Zhang, J.; Jazii, F.R.; Haghighi, M.M.; Alvares, D.; Liu, L.; Khosraviani, N.; Adeli, K. miR-130b is a potent stimulator of hepatic very-low-density lipoprotein assembly and secretion via marked induction of microsomal triglyceride transfer protein. Am. J. Physiol. Metab. 2020, 318, E262–E275. [Google Scholar] [CrossRef]
- Duggal, N.A.; Niemiro, G.; Harridge, S.D.R.; Simpson, R.J.; Lord, J.M. Can physical activity ameliorate immunosenescence and thereby reduce age-related multi-morbidity? Nat. Rev. Immunol. 2019, 19, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Moro-García, M.A.; Fernández-García, B.; Echeverría, A.; Rodríguez-Alonso, M.; Suárez-García, F.M.; Solano-Jaurrieta, J.J.; López-Larrea, C.; Alonso-Arias, R. Frequent participation in high volume exercise throughout life is associated with a more differentiated adaptive immune response. Brain Behav. Immun. 2014, 39, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Woods, J.; Ceddia, M.; Wolters, B.; Evans, J.; Lu, Q.; McAuley, E. Effects of 6 months of moderate aerobic exercise training on immune function in the elderly. Mech. Ageing Dev. 1999, 109, 1–19. [Google Scholar] [CrossRef]
- Yan, H.; Kuroiwa, A.; Tanaka, H.; Shindo, M.; Kiyonaga, A.; Nagayama, A. Effect of moderate exercise on immune senescence in men. Graefe’s Arch. Clin. Exp. Ophthalmol. 2001, 86, 105–111. [Google Scholar] [CrossRef]
- Wang, J.-S.; Weng, T.-P. Hypoxic exercise training promotes antitumour cytotoxicity of natural killer cells in young men. Clin. Sci. 2011, 121, 343–353. [Google Scholar] [CrossRef]
- Wherry, E.J. T cell exhaustion. Nat. Immunol. 2011, 12, 492–499. [Google Scholar] [CrossRef] [PubMed]
- Wensveen, F.M.; Jelenčić, V.; Polić, B. NKG2D: A Master Regulator of Immune Cell Responsiveness. Front. Immunol. 2018, 9, 441. [Google Scholar] [CrossRef] [PubMed]
- Béziat, V.; Descours, B.; Parizot, C.; Debre, P.; Vieillard, V. NK Cell Terminal Differentiation: Correlated Stepwise Decrease of NKG2A and Acquisition of KIRs. PLoS ONE 2010, 5, e11966. [Google Scholar] [CrossRef] [PubMed]


| CLL-FIT (N = 10) | CLL-UNFIT (N = 10) | p-Value | 95% CI | Effect Size (d) | |
|---|---|---|---|---|---|
| Demographics | |||||
| Age (years) | 65.8 ± 11.1 | 69.8 ± 10.8 | 0.426 | −14.3, 6.3 | 0.37 |
| Sex (M/F) 1 | 5/5 | 5/5 | 1.000 | 0.00 | |
| Height (cm) | 167.8 ± 17.0 | 170.8 ± 11.9 | 0.648 | −16.8, 10.7 | 0.20 |
| Weight (kg) | 66.2 ± 16.8 | 85.8 ± 24.1 | 0.049 | −39.1, −0.1 | 0.94 |
| BMI (kg/m2) | 23 ± 2.9 | 29 ± 6.2 | 0.013 | −10.5, −1.4 | 1.24 |
| Resting Heart Rate (bpm) | 68 ± 12 | 72 ± 12 | 0.438 | −15.3, 6.9 | 0.33 |
| Clinical Characteristics | |||||
| CLL-IPI Score | 2.4 ± 2.2 | 1.3 ± 1.1 | 0.175 | −0.53, 2.74 | 0.63 |
| RAI Stage [N (%)] 1 | |||||
| Stage 0 | 9 (90) | 7 (70) | 0.453 | ||
| Stage I | 1 (10) | 2 (20) | 0.346 | ||
| Unknown | 0 | 1 (10) | |||
| Disease Duration (years) | 5.0 ± 2.9 | 4.9 ± 2.7 | 0.937 | −2.5, 2.7 | 0.04 |
| Cytogenetics [N (%)] 1 | |||||
| 13q Deletion | 7 (70) | 5 (50) | 0.361 | 0.20 | |
| 17p Deletion | 3 (30) | 0 | 0.060 | 0.42 | |
| 11q Deletion | 2 (20) | 0 | 0.136 | 0.33 | |
| Trisomy 12 | 0 | 2 (20) | 0.136 | 0.33 | |
| TP53 Mutated | 4 (40) | 1 (10) | 0.291 | 0.35 | |
| IGHV Mutated1 | 6 (60) | 6 (60) | 0.766 | 0.16 | |
| CD38 Expression >30% 1 | 1 (10) | 0 | 0.119 | 0.46 | |
| WBC Counts (×103/µL) | 65 ± 54 | 51 ± 51 | 0.553 | −35.1, 63.5 | 0.27 |
| Lymphocytes | 59 ± 52 | 44 ± 49 | 0.517 | −32.7, 62.8 | 0.30 |
| CD19+/CD5+ CLL-cells | 48.1 ± 48.6 | 40.8 ± 41.2 | 0.359 | −35.0, 49.7 | 0.16 |
| CD4+ T-cells | 1.6 ± 2.3 | 5.1 ± 7.4 | 0.169 | −8.6, 1.6 | 0.64 |
| CD8+ T-cells | 0.8 ± 0.9 | 2.2 ± 2.3 | 0.088 | −3.0, 0.2 | 0.80 |
| Monocytes | 1.4 ± 1.4 | 1.8 ± 2.3 | 0.694 | −2.1, 1.5 | 0.21 |
| Neutrophils | 3.4 ± 1.6 | 4.9 ± 1.6 | 0.046 | −3.0, −0.3 | 0.94 |
| Neutrophil: T-cell | 3.7 ± 5.7 | 2.4 ± 2.5 | 0.510 | −2.2, 0.8 | 0.30 |
| T-cell: Monocyte | 2.1 ± 1.4 | 6.5 ± 7.6 | 0.086 | −9.6, 0.7 | 0.81 |
| Platelets (×103/µL) | 149 ± 43 | 196 ± 62 | 0.065 | −96.4, 3.2 | 0.88 |
| Hemoglobin (g/dL) | 13.0 ± 1.7 | 13.6 ± 1.5 | 0.433 | −2.1, 0.92 | 0.37 |
| β2-microglobulin (mg/dL) | 1.9 ± 0.7 | 2.4 ± 0.9 | 0.228 | −1.3, 0.33 | 0.57 |
| ICAM-1 (ng/mL) | 709.1 ± 171.0 | 685.1 ± 158.6 | 0.755 | −136, 184 | 0.15 |
| sCD20 (pg/mL) | 220.8 ± 306.9 | 118.2 ± 181.3 | 0.417 | −158, 364 | 0.40 |
| Fitness and Performance | |||||
| eVO2peak (mL/kg/min) | 34.2 ± 3.3 | 24.9 ± 3.2 | <0.001 | 6.2, 12.4 | 2.86 |
| 6MWT (m) | 500 ± 77 | 399 ± 107 | 0.027 | 12.6, 188 | 1.08 |
| SPPB Score | 12 ± 0 | 9.9 ± 2.4 | 0.012 | 0.52, 3.68 | 1.24 |
| TUG (sec) | 9.4 ± 1.6 | 11.6 ± 4.5 | 0.170 | −5.6, 1.07 | 0.65 |
| Grip Strength Right (kg) | 33.3 ± 12.0 | 32.7 ± 16.7 | 0.930 | −13.1, 14.3 | 0.04 |
| Grip Strength Left (kg) | 32.4 ± 14.3 | 30.5 ± 16.1 | 0.780 | −12.4, 16.3 | 0.12 |
| Best Grip (BMI normalized) | 1.4 ± 0.4 | 1.1 ± 0.5 | 0.171 | −0.15, 0.77 | 0.66 |
| IPAQ (total hours/week) | 31.9 ± 29.1 | 26.4 ± 16.8 | 0.610 | −10.2, 50.6 | 0.23 |
| SBAS [N (%)] 1 | |||||
| Inactive | 2 (20) | 3 (30) | 0.936 | ||
| Light | 4 (40) | 3 (30) | |||
| Moderate | 2 (20) | 2 (20) | 0.21 | ||
| Hard | 1 (10) | 1 (10) | |||
| Very Hard | 1 (10) | 1 (10) | |||
| CLL-FIT (N = 10) | CLL-UNFIT (N = 10) | p-Value | 95% CI | Effect Size (d) | |
|---|---|---|---|---|---|
| Inflammation | |||||
| GlycA (µmol/L) | 354.7 ± 31.1 | 442.0 ± 97.0 | 0.014 | −155.0, −19.6 | 1.21 |
| Insulin Resistance | |||||
| LP-IR (1-100) | 33.7 ± 18.1 | 65.7 ± 22.2 | 0.007 | −54.3, −9.7 | 1.58 |
| Triglycerides | |||||
| Total Triglycerides (mg/dL) | 101.4 ± 40.6 | 165.2 ± 72.0 | 0.025 | −118.7, 8.9 | 1.09 |
| TRL Triglycerides (mg/dL) | 78.1 ± 38.8 | 143.4 ± 67.7 | 0.016 | −117.1, −13.5 | 1.18 |
| Total TRLP (nmol/L) | 127.6 ± 58.4 | 189.8 ± 70.8 | 0.046 | −123.2, −1.2 | 0.96 |
| Large (nmol/L) | 2.5 ± 3.4 | 8.9 ± 6.2 | 0.011 | −11.1, −1.7 | 1.28 |
| Very Small (nmol/L) | 73.4 ± 38.4 | 124.4 ± 63.7 | 0.044 | −100.4, −1.6 | 0.97 |
| Cholesterol (mg/dL) | |||||
| Total Cholesterol | 194.4 ± 29.1 | 195.4 ± 39.4 | 0.949 | −33.5, 31.5 | 0.03 |
| TRL Cholesterol | 22.5 ± 10.8 | 34.9 ± 13.3 | 0.034 | −23.8, −1.0 | 1.02 |
| LDL Cholesterol | 106.3 ± 25.7 | 105.5 ± 29.7 | 0.949 | −25.3, 26.9 | 0.03 |
| HDL Cholesterol | 65.5 ± 10.0 | 54.8 ± 11.6 | 0.040 | 0.5, 20.9 | 0.99 |
| HDL-P (µmol/L) | |||||
| Total | 24.7 ± 2.4 | 23.8 ± 4.2 | 0.559 | −2.3, 4.1 | 0.26 |
| Large | 3.4 ± 1.4 | 1.9 ± 1.6 | 0.043 | 0.1, 2.9 | 1.00 |
| Lipoprotein Size (nm) | |||||
| TRL | 43.7 ± 4.7 | 50.3 ± 7.2 | 0.027 | −12.3, −0.9 | 1.09 |
| LDL | 20.8 ± 0.4 | 20.4 ± 0.4 | 0.059 | 0.02, 0.8 | 1.00 |
| HDL | 9.2 ± 0.4 | 8.8 ± 0.5 | 0.073 | −0.03, 0.8 | 0.88 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sitlinger, A.; Deal, M.A.; Garcia, E.; Thompson, D.K.; Stewart, T.; MacDonald, G.A.; Devos, N.; Corcoran, D.; Staats, J.S.; Enzor, J.; et al. Physiological Fitness and the Pathophysiology of Chronic Lymphocytic Leukemia (CLL). Cells 2021, 10, 1165. https://doi.org/10.3390/cells10051165
Sitlinger A, Deal MA, Garcia E, Thompson DK, Stewart T, MacDonald GA, Devos N, Corcoran D, Staats JS, Enzor J, et al. Physiological Fitness and the Pathophysiology of Chronic Lymphocytic Leukemia (CLL). Cells. 2021; 10(5):1165. https://doi.org/10.3390/cells10051165
Chicago/Turabian StyleSitlinger, Andrea, Michael A. Deal, Erwin Garcia, Dana K. Thompson, Tiffany Stewart, Grace A. MacDonald, Nicolas Devos, David Corcoran, Janet S. Staats, Jennifer Enzor, and et al. 2021. "Physiological Fitness and the Pathophysiology of Chronic Lymphocytic Leukemia (CLL)" Cells 10, no. 5: 1165. https://doi.org/10.3390/cells10051165
APA StyleSitlinger, A., Deal, M. A., Garcia, E., Thompson, D. K., Stewart, T., MacDonald, G. A., Devos, N., Corcoran, D., Staats, J. S., Enzor, J., Weinhold, K. J., Brander, D. M., Weinberg, J. B., & Bartlett, D. B. (2021). Physiological Fitness and the Pathophysiology of Chronic Lymphocytic Leukemia (CLL). Cells, 10(5), 1165. https://doi.org/10.3390/cells10051165






