Modeling Gastrointestinal Diseases Using Organoids to Understand Healing and Regenerative Processes
Abstract
1. Introduction
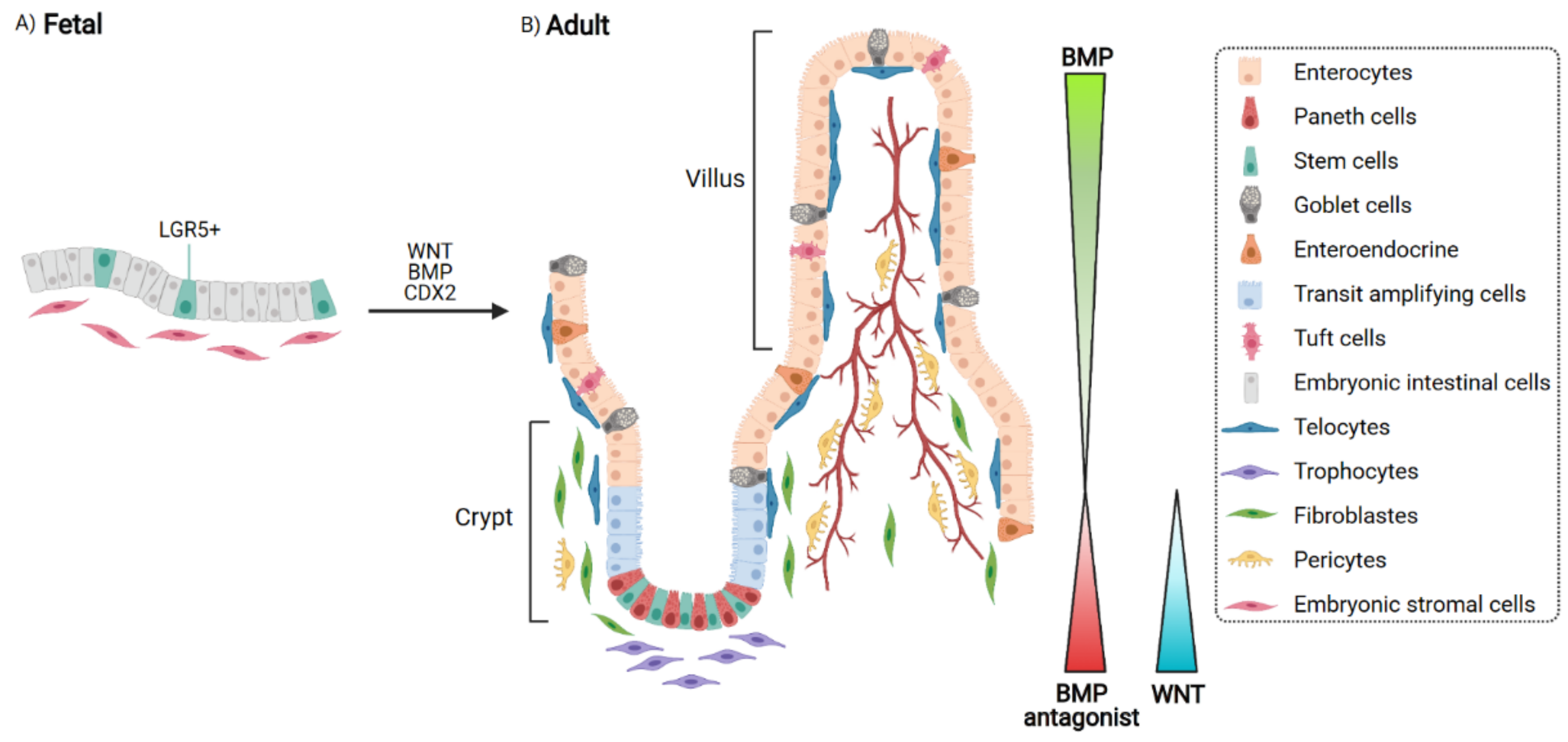
2. Epithelial Cells and Their Niche: When Homeostasis Takes Place
3. Deregulated Niche and Epithelial Barrier Loss
Using Organoid to Model Damaged Epithelium and Wound-Healing Processes
4. Organoid-Based Regenerative Therapy: Repairing the Epithelial Barrier
4.1. The Process of Regeneration Is A Healing Phase
4.2. Role of the Stroma in the Regenerative Process
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fuchs, E.; Tumbar, T.; Guasch, G. Socializing with the neighbors: Stem cells and their niche. Cell 2004, 116, 769–778. [Google Scholar] [CrossRef]
- Hsu, Y.-C.; Fuchs, E. A family business: Stem cell progeny join the niche to regulate homeostasis. Nat. Rev. Mol. Cell Biol. 2012, 13, 103–114. [Google Scholar] [CrossRef]
- Hsu, Y.-C.; Li, L.; Fuchs, E. Transit-amplifying cells orchestrate stem cell activity and tissue regeneration. Cell 2014, 157, 935–949. [Google Scholar] [CrossRef]
- Thoo, L.; Noti, M.; Krebs, P. Keep calm: The intestinal barrier at the interface of peace and war. Cell Death Dis. 2019, 10, 849. [Google Scholar] [CrossRef] [PubMed]
- McCauley, H.A.; Wells, J. Pluripotent stem cell-derived organoids: Using principles of developmental biology to grow human tissues in a dish. Development 2017, 144, 958–962. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Matano, M.; Toshimitsu, K.; Takano, A.; Mikami, Y.; Nishikori, S.; Sugimoto, S.; Sato, T. Human intestinal organoids maintain self-renewal capacity and cellular diversity in niche-inspired culture condition. Cell Stem Cell 2018, 23, 787–793. [Google Scholar] [CrossRef]
- Taeko, K.N.; Donahue, B.; Shroyer, N.F. Intestinal development and differentiation. Exp. Cell Res. 2011, 317, 2702–2710. [Google Scholar]
- Zorn, A.M.; Wells, J.M. Vertebrate Endoderm Development and Organ Formation. Annu. Rev. Cell Dev. Biol 2009, 25, 221–251. [Google Scholar] [CrossRef] [PubMed]
- Spence, J.R.; Lauf, R.; Shroyer, N.F. Vertebrate intestinal endoderm development. Dev. Dyn. 2011, 240, 501–520. [Google Scholar] [CrossRef]
- Guiu, J.; Hannezo, E.; Yui, S.; Demharter, S.; Ulyanchenko, S.; Maimets, M.; Jørgensen, A.; Perlman, S.; Lundvall, L.; Mamsen, L.S.; et al. Tracing the origin of adult intestinal stem cells. Nature 2019, 570, 107–111. [Google Scholar] [CrossRef]
- Gehart, H.; Clevers, H. Tales from the crypt: New insights into intestinal stem cells. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 19–34. [Google Scholar] [CrossRef] [PubMed]
- Holloway, E.M.; Czerwinski, M.; Tsai, Y.H.; Wu, J.H.; Wu, A.; Childs, C.J.; Walton, K.D.; Sweet, C.W.; Yu, Q.; Glass, I.; et al. Mapping Development of the Human Intestinal Niche at Single-Cell Resolution. Cell Stem Cell 2021, 28, 568–580. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, N.; Kraiczy, J.; Shivdasani, R.A. Cellular and molecular architecture of the intestinal stem cell niche. Nat. Cell Biol. 2020, 22, 1033–1041. [Google Scholar] [CrossRef] [PubMed]
- Kondo, A.; Kaestner, K.H. Emerging diverse roles of telocytes. Development 2019, 146. [Google Scholar] [CrossRef] [PubMed]
- Shoshkes-Carmel, M.; Wang, Y.J.; Wangensteen, K.J.; Toth, B.; Kondo, A.; Itzkovitz, S.; Kaestner, K.H. Subepithelial telocytes are the source of Wnts that support intestinal crypts. Nature 2018, 557, 242–246. [Google Scholar] [CrossRef] [PubMed]
- McCarthy, N.; Manieri, E.; Storm, E.E.; Saadatpour, A.; Luoma, A.M.; Kapoor, V.N.; Madha, S.; Gaynor, L.T.; Cox, C.; Keerthivasan, S.; et al. Distinct Mesenchymal Cell Populations Generate the Essential Intestinal BMP Signaling Gradient. Cell Stem Cell 2020, 26, 391–402. [Google Scholar] [CrossRef]
- Sanne, L.; Rosekrans, B.B.; Muncan, V.; van den Brink, G.R. Esophageal development and epithelial homeostasis. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 309, G216–G228. [Google Scholar]
- Reid, B.J.; Li, X.; Galipeau, P.C.; Vaughan, T.L. Barrett’s oesophagus and oesophageal adenocarcinoma: Time for a new synthesis. Nat. Rev. Cancer 2020, 10, 87–101. [Google Scholar] [CrossRef]
- Jiang, M.; Li, H.; Zhang, Y.; Yang, Y.; Lu, R.; Liu, K.; Lin, S.; Lan, X.; Wang, H.; Wu, H.; et al. Transitional basal cells at the squamous-columnar junction generate Barrett’s oesophagus. Nature 2017, 550, 529–533. [Google Scholar] [CrossRef]
- Souza, R.F.; Krishnan, K.; Spechler, S.J. Acid, bile, and CDX: The ABCs of making Barrett’s metaplasia. Am. J. Physiol. Gastrointest. Liver Physiol. 2008, 295, G211–G218. [Google Scholar] [CrossRef]
- Wang, D.H.; Clemons, N.J.; Miyashita, T.; Dupuy, A.J.; Zhang, W.; Szczepny, A.; Corcoran-Schwartz, I.M.; Wilburn, D.L.; Montgomery, E.A.; Wang, J.S.; et al. Aberrant epithelial-mesenchymal Hedgehog signaling characterizes Barrett’s metaplasia. Gastroenterology 2010, 138, 1810–1822. [Google Scholar] [CrossRef]
- Willet, S.G.; Mills, J.C. Stomach Organ and Cell Lineage Differentiation: From Embryogenesis to Adult Homeostasis. Cell. Mol. Gastroenterol. Hepatol. 2016, 2, 546–559. [Google Scholar] [CrossRef]
- Hunt, R.H.; Camilleri, M.; Crowe, S.E.; El-Omar, E.M.; Fox, J.G.; Kuipers, E.J.; Malfertheiner, P.; McColl, K.E.L.; Pritchard, D.M.; Rugge, M.; et al. The stomach in health and disease. Gut 2015, 64, 1650–1668. [Google Scholar] [CrossRef]
- Wright, N.A.; Pike, C.; Elia, G. Induction of a novel epidermal growth factor-secreting cell lineage by mucosal ulceration in human gastrointestinal stem cells. Nature 1990, 343, 82–85. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, M.; Reddy, K.M.; Marsicano, E. Peptic Ulcer Disease and Helicobacter pylori infection. Mo. Med. 2018, 115, 219–224. [Google Scholar] [PubMed]
- Moon, H.; Zhu, J.; Donahue, L.R.; Choi, E.; White, A.C. Krt5 +/Krt15 + foregut basal progenitors give rise to cyclooxygenase-2-dependent tumours in response to gastric acid stress. Nat. Commun. 2019, 10, 2225. [Google Scholar] [CrossRef] [PubMed]
- Barbara, P.D.; van den Brink, G.R.; Roberts, D.J. Development and differentiation of the intestinal epithelium. Cell. Mol. Life Sci. 2003, 60, 1322–1332. [Google Scholar] [CrossRef] [PubMed]
- Kinchen, J.; Chen, H.; Parikh, K.; Antanaviciute, A.; Jagielowicz, M.; Fawkner-Corbett, D.; Ashley, N.; Cubitt, L.; Mellado-Gomez, E.; Attar, M.; et al. Structural Remodeling of the Human ColonicMesenchyme in Inflammatory Bowel Disease. Cell 2018, 175, 372–386. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q. A Comprehensive Review and Update on the Pathogenesis of Inflammatory Bowel Disease. J. Immunol Res. 2019, 2019, 7247238. [Google Scholar] [CrossRef]
- Pleguezuelos-Manzano, C.; Puschhof, J.; van den Brink, S.; Geurts, V.; Beumer, J.; Clevers, H. Establishment and culture of human intestinal organoids derived from adult stem cells. Curr. Protoc. Immunol. 2020, 130, e106. [Google Scholar] [CrossRef]
- Holloway, E.M.; Capeling, M.M.; Spence, J.R. Biologically inspired approaches to enhance human organoid complexity. Development 2019, 146, dev166173. [Google Scholar] [CrossRef]
- Sato, T.; Vries, R.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Spence, J.R.; Mayhew, C.; Rankin, S.A.; Kuhar, M.F.; Vallance, J.E.; Tolle, K.; Hoskins, E.E.; Kalinichenko, V.V.; Wells, S.I.; Zorn, A.M.; et al. Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 2011, 470, 105–109. [Google Scholar] [CrossRef]
- Uchida, H.; Machida, M.; Miura, T.; Kawasaki, T.; Okazaki, T.; Sasaki, K.; Sakamoto, S.; Ohuchi, N.; Kasahara, M.; Umezawa, A.; et al. A xenogeneic-free system generating functional human gut organoids from pluripotent stem cells. JCI Insight 2017, 2, e86492. [Google Scholar] [CrossRef] [PubMed]
- Gerbe, F.; Sidot, E.; Smyth, D.J.; Ohmoto, M.; Matsumoto, I.; Dardalhon, V.; Cesses, P.; Garnier, L.; Pouzolles, M.; Brulin, B.; et al. Intestinal epithelial tuft cells initiate type 2 mucosal immunity to helminth parasites. Nature 2016, 529, 226–230. [Google Scholar]
- Nicholas, W.; Bellono, J.R.; Bayrer, D.B.; Castro, L.J.; Zhang, C.; O’Donnell, T.; Brierley, S.M.; Ingraham, H.A.; Julius, D. Enterochromaffin cells are gut chemosensors that couple to sensory neural pathways. Cell 2017, 170, 185–198. [Google Scholar]
- Min, S.; Kim, S.; Cho, S.-W. Gastrointestinal tract modeling using organoids engineered with cellular and microbiota niches. Exp. Mol. Med. 2020, 52, 227–237. [Google Scholar] [CrossRef] [PubMed]
- Kasagi, Y.; Chandramouleeswaran, P.M.; Whelan, K.A.; Tanaka, K.; Giroux, V.; Sharma, M.; Wang, J.; Benitez, A.J.; DeMarshall, M.; Tobias, J.W.; et al. The Esophageal Organoid System Reveals Functional Interplay Between Notch and Cytokines in Reactive Epithelial Changes. Cell. Mol. Gastroenterol. Hepatol. 2018, 5, 333–352. [Google Scholar]
- Trisno, S.L.; Philo, K.E.D.; McCracken, K.W.; Catá, E.M.; Ruiz-Torres, S.; Rankin, S.A.; Han, L.; Nasr, T.; Chaturvedi, P.; Rothenberg, M.E.; et al. Esophageal Organoids from Human Pluripotent Stem Cells Delineate Sox2 Functions during Esophageal Specification. Cell Stem Cell 2018, 23, 501–515. [Google Scholar] [CrossRef]
- Nakagawa, H.; Kasagi, Y.; Karakasheva, T.A.; Hara, T.; Aaron, B.; Shimonosono, M.; Kijima, T.; Giroux, V.; Bailey, D.; Wilkins, B.; et al. Modeling Epithelial Homeostasis and Reactive Epithelial Changes in Human and Murine Three-Dimensional Esophageal Organoids. Curr. Protoc. Stem Cell Biol. 2020, 52, e106. [Google Scholar]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.J.; van Es, J.H.; van den Brink, S.; van Houdt, W.J.; Pronk, A.; van Gorp, J.; Siersema, P.D.; et al. Long-term Expansion of Epithelial Organoids from Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef]
- Liu, X.; Cheng, Y.; Abraham, J.M.; Wang, Z.; Wang, Z.; Ke, X.; Yan, R.; Shin, E.J.; Ngamruengphong, S.; Khashab, M.; et al. Modeling Wnt signaling by CRISPR-Cas9 genome editing facilitates neoplasia inhuman Barrett epithelial organoids. Cancer Lett. 2018, 436, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Simmini, S.; Bialecka, M.; Huch, M.; Kester, L.; van de Wetering, M.; Sato, T.; Beck, F.; van Oudenaarden, A.; Clevers, H.; Deschamps, J. Transformation of intestinal stem cells into gastric stem cells on loss of transcription factor Cdx2. Nat. Comm. 2014, 11, 5728. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ouyang, H.; Yamamoto, Y.; Kumar, P.A.; Wei, T.S.; Dagher, R.; Vincent, M.; Lu, X.; Bellizzi, A.M.; Ho, K.Y.; et al. Residual embryonic cells as precursors of a Barrett’s-like metaplasia. Cell 2011, 145, 1023–1035. [Google Scholar] [CrossRef] [PubMed]
- Barker, N.; Huch, M.; Kujala, P.; van de Wetering, M.; Snippert, H.J.; van Es, J.H.; Sato, T.; Stange, D.E.; Begthel, H.; van den Born, M.; et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell 2010, 6, 25–36. [Google Scholar] [CrossRef]
- McCracken, K.W.; Catá, E.M.; Crawford, C.M.; Sinagoga, K.L.; Schumacher, M.; Rockich, B.E.; Tsai, Y.-H.; Mayhew, C.N.; Spence, J.R.; Zavros, Y.; et al. Modelling human development and disease in pluripotent stem-cell-derived gastric organoids. Nature 2014, 516, 400–404. [Google Scholar] [CrossRef] [PubMed]
- Bartfeld, S.; Bayram, T.; van de Wetering, M.; Huch, M.; Begthel, H.; Kujala, P.; Vries, R.; Peters, P.J.; Clevers, H. In Vitro Expansion of Human Gastric Epithelial Stem Cells and Their Responses to Bacterial Infection. Gastroenterology 2015, 148, 126–136. [Google Scholar] [CrossRef]
- Huang, J.Y.; Sweeney, E.G.; Sigal, M.; Zhang, H.C.; Remington, S.J.; Cantrell, M.A.; Kuo, C.J.; Guillemin, K.; Amieva, M.R. Chemodetection and Destruction of Host Urea Allows Helicobacter pylori to Locate the Epithelium. Cell Host Microbe 2015, 18, 147–156. [Google Scholar] [CrossRef]
- Schumacher, M.A.; Aihara, E.; Feng, R.; Engevik, A.; Shroyer, N.F.; Ottemann, K.M.; Worrell, R.T.; Montrose, M.H.; Shivdasani, R.A.; Zavros, Y. The use of murine-derived fundic organoids in studies of gastric physiology. J. Physiol. 2015, 593, 1809–1827. [Google Scholar] [CrossRef]
- Engevik, K.A.; Hanyu, H.; Matthis, A.L.; Zhang, T.; Frey, M.R.; Oshima, Y.; Aihara, E.; Marshall, H. Trefoil factor 2 activation of CXCR4 requires calcium mobilization to drive epithelial repair in gastric organoids. J. Physiol. 2019, 597, 2673–2690. [Google Scholar] [CrossRef]
- Sayed, I.M.; Sahan, A.Z.; Venkova, T.; Chakraborty, A.; Mukhopadhyay, D.; Bimczok, D.; Beswick, E.J.; Reyes, V.E.; Pinchuk, I.; Sahoo, D.; et al. Helicobacter pylori infection downregulates the DNA glycosylase NEIL2, resulting in increased genome damage and inflammation in gastric epithelial cells. J. Biol. Chem. 2020, 295, 11082–11098. [Google Scholar] [CrossRef]
- Dotti, I.; Mora-Buch, R.; Ferrer-Picón, E.; Planell, N.; Jung, P.; Masamunt, M.C.; Leal, R.F.; de Carpi, J.M.; Llach, J.; Ordás, I.; et al. Alterations in the epithelial stem cell compartment could contribute to permanent changes in the mucosa of patients with ulcerative colitis. Gut 2017, 66, 2069–2079. [Google Scholar] [CrossRef] [PubMed]
- D’Aldebert, E.; Quaranta, M.; Sébert, M.; Bonnet, D.; Kirzin, S.; Portier, G.; Duffas, J.; Chabot, S.; Lluel, P.; Allart, S.; et al. Characterization of Human Colon Organoids From Inflammatory Bowel Disease Patients. Front. Cell Dev. Biol. 2020, 8, 363. [Google Scholar] [CrossRef] [PubMed]
- Esterházy, D.; Canesso, M.C.; Mesin, L.; Muller, P.A.; de Castro, T.B.R.; Lockhart, A.; ElJalby, M.; Faria, A.M.C.; Mucida, D. Compartmentalized gut lymph node drainage dictates adaptive immune responses. Nature 2019, 569, 126–130. [Google Scholar] [CrossRef] [PubMed]
- Sayed, I.M.; Suarez, K.; Lim, E.; Singh, S.; Pereira, M.; Ibeawuchi, S.R.; Katkar, G.; Dunkel, Y.; Mittal, Y.; Chattopadhyay, R.; et al. Host engulfment pathway controls inflammation in inflammatory bowel disease. FEBS J. 2020, 287, 3967–3988. [Google Scholar] [CrossRef] [PubMed]
- Middendorp, S.; Schneeberger, K.; Wiegerinck, C.L.; Mokry, M.; Akkerman, R.D.L.; Wijngaaeden, S.V.; Clevers, H.; Nieuwenhuis, E.E.S. Adult Stem Cells in the Small Intestine Are Intrinsically Programmed with Their Location-Specific Function. Stem Cells 2014, 32, 1083–1091. [Google Scholar] [CrossRef]
- Yui, S.; Nakamura, T.; Sato, T.; Nemoto, Y.; Mizutani, T.; Zheng, X.; Ichinose, S.; Nagaishi, T.; Okamoto, R.; Tsuchiya, K.; et al. Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5+ stem cell. Nat. Med. 2012, 18, 618–623. [Google Scholar] [CrossRef]
- Sugimoto, S.; Ohta, Y.; Fujii, M.; Matano, M.; Shimokawa, M.; Nanki, K.; Date, S.; Nishikori, S.; Nakazato, Y.; Nakamura, T.; et al. Reconstruction of the Human Colon Epithelium In Vivo. Cell Stem Cell 2018, 22, 171–176.e175. [Google Scholar] [CrossRef]
- Fordham, R.P.; Yui, S.; Hannan, N.R.F.; Soendergaard, C.; Madgwick, A.; Schweiger, P.J.; Nielsen, O.H.; Vallier, L.; Pedersen, R.A.; Nakamura, T.; et al. Transplantation of Expanded Fetal Intestinal Progenitors Contributes to Colon Regeneration after Injury. Cell Stem Cell 2013, 13, 734–744. [Google Scholar] [CrossRef]
- Cruz-Acuña, R.; Quiros, M.; Farkas, A.E.; Dedhia, P.H.; Huang, S.; Siuda, D.; García-Hernández, V.; Miller, A.J.; Spence, J.R.; Nusrat, A.; et al. Synthetic hydrogels for human intestinal organoid generation and colonic wound repair. Nat. Cell Biol. 2017, 119, 1326–1335. [Google Scholar] [CrossRef]
- Sugimoto, S.; Kobayashi, E.; Fujii, M.; Ohta, Y.; Arai, K.; Matano, M.; Ishikawa, K.; Miyamoto, K.; Toshimitsu, K.; Takahashi, S.; et al. An organoid-based organ-repurposing approach to treat short bowel syndrome. Nature 2021, 592, 99–108. [Google Scholar] [PubMed]
- Meran, L.; Massie, I.; Campinoti, S.; Weston, A.E.; Gaifulina, R.; Tullie, L.; Faull, P.; Orford, M.; Kucharska, A.; Baulies, A.; et al. Engineering transplantable jejunal mucosal grafts using patient-derived organoids from children with intestinal failure. Nat. Med. 2020, 26, 1593–1601. [Google Scholar] [CrossRef] [PubMed]
- Engevik, A.C.; Feng, R.; Choi, E.; White, S.; Bertaux-Skeirik, N.; Li, J.; Mahe, M.M.; Aihara, E.; Yang, L.; DiPasquale, B.; et al. The Development of Spasmolytic Polypeptide/TFF2-Expressing Metaplasia (SPEM) During Gastric Repair Is Absent in the Aged Stomach. Cell Mol. Gastroenterol. Hepatol. 2016, 2, 605–624. [Google Scholar] [CrossRef]
- Bertaux-Skeirik, N.; Wunderlich, M.; Teal, E.; Chakrabarti, J.; Biesiada, J.; Mahe, M.; Sundaram, N.; Gabre, J.; Hawkins, J.; Jian, G.; et al. CD44 variant isoform 9 emerges in response to injury and contributes to the regeneration of the gastric epithelium. J. Pathol. 2017, 242, 463–475. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, M.; Kosaric, N.; Bonham, C.A.; Gurtner, G.C. Wound Healing: A Cellular Perspective. Physiol. Rev. 2019, 99, 665–706. [Google Scholar] [CrossRef]
- Stzepourginski, I.; Nigro, G.; Jacob, J.-M.; Dulauroy, S.; Sansonetti, P.J.; Eberl, G.; Peduto, L. CD34+ mesenchymal cells are a major component of the intestinal stem cells niche at homeostasis and after injury. Proc. Natl. Acad. Sci. USA 2017, 114, E506–E513. [Google Scholar] [CrossRef]
- Nikolaev, M.; Mitrofanova, O.; Broguiere, N.; Geraldo, S.; Dutta, D.; Tabata, Y.; Elci, B.; Brandenberg, N.; Kolotuev, I.; Gjorevski, N.; et al. Homeostatic mini-intestines through scaffold-guided organoid morphogenesis. Nature 2020, 585, 574–578. [Google Scholar] [CrossRef]
- Hinman, S.S.; Wang, Y.; Kim, R.; Allbritton, N.L. In vitro generation of self-renewing human intestinal epithelia over planar and shaped collagen hydrogels. Nat. Protoc. 2021, 16, 352–382. [Google Scholar] [CrossRef]
- Sontheimer-Phelps, A.; Chou, D.B.; Tovaglieri, A.; Ferrante, T.C.; Duckworth, T.; Fadel, C.; Frismantas, V.; Sutherland, A.D.; Jalili-Firoozinezhad, S.; Kasendra, M.; et al. Human Colon-on-a-Chip Enables Continuous In Vitro Analysis of Colon Mucus Layer Accumulation and Physiology. Cell Mol. Gastroenterol. Hepatol. 2020, 9, 507–526. [Google Scholar] [CrossRef]
- Shin, W.; Kim, H.J. Intestinal barrier dysfunction orchestrates the onset of inflammatory host–microbiome cross-talk in a human gut inflammation-on-a-chip. Proc. Natl. Acad. Sci. USA 2018, 115, E10539–E10547. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ollivier, A.; Mahe, M.M.; Guasch, G. Modeling Gastrointestinal Diseases Using Organoids to Understand Healing and Regenerative Processes. Cells 2021, 10, 1331. https://doi.org/10.3390/cells10061331
Ollivier A, Mahe MM, Guasch G. Modeling Gastrointestinal Diseases Using Organoids to Understand Healing and Regenerative Processes. Cells. 2021; 10(6):1331. https://doi.org/10.3390/cells10061331
Chicago/Turabian StyleOllivier, Alexane, Maxime M. Mahe, and Géraldine Guasch. 2021. "Modeling Gastrointestinal Diseases Using Organoids to Understand Healing and Regenerative Processes" Cells 10, no. 6: 1331. https://doi.org/10.3390/cells10061331
APA StyleOllivier, A., Mahe, M. M., & Guasch, G. (2021). Modeling Gastrointestinal Diseases Using Organoids to Understand Healing and Regenerative Processes. Cells, 10(6), 1331. https://doi.org/10.3390/cells10061331





