Riluzole Administration to Rats with Levodopa-Induced Dyskinesia Leads to Loss of DNA Methylation in Neuronal Genes
Abstract
1. Introduction
2. Materials and Methods
2.1. Abnormal Involuntary Movements (AIMs) Rat Model
2.2. AIMs Scoring
2.3. Reduced Representation Bisulfite Sequencing (RRBS)
2.4. Bioinformatic Analysis
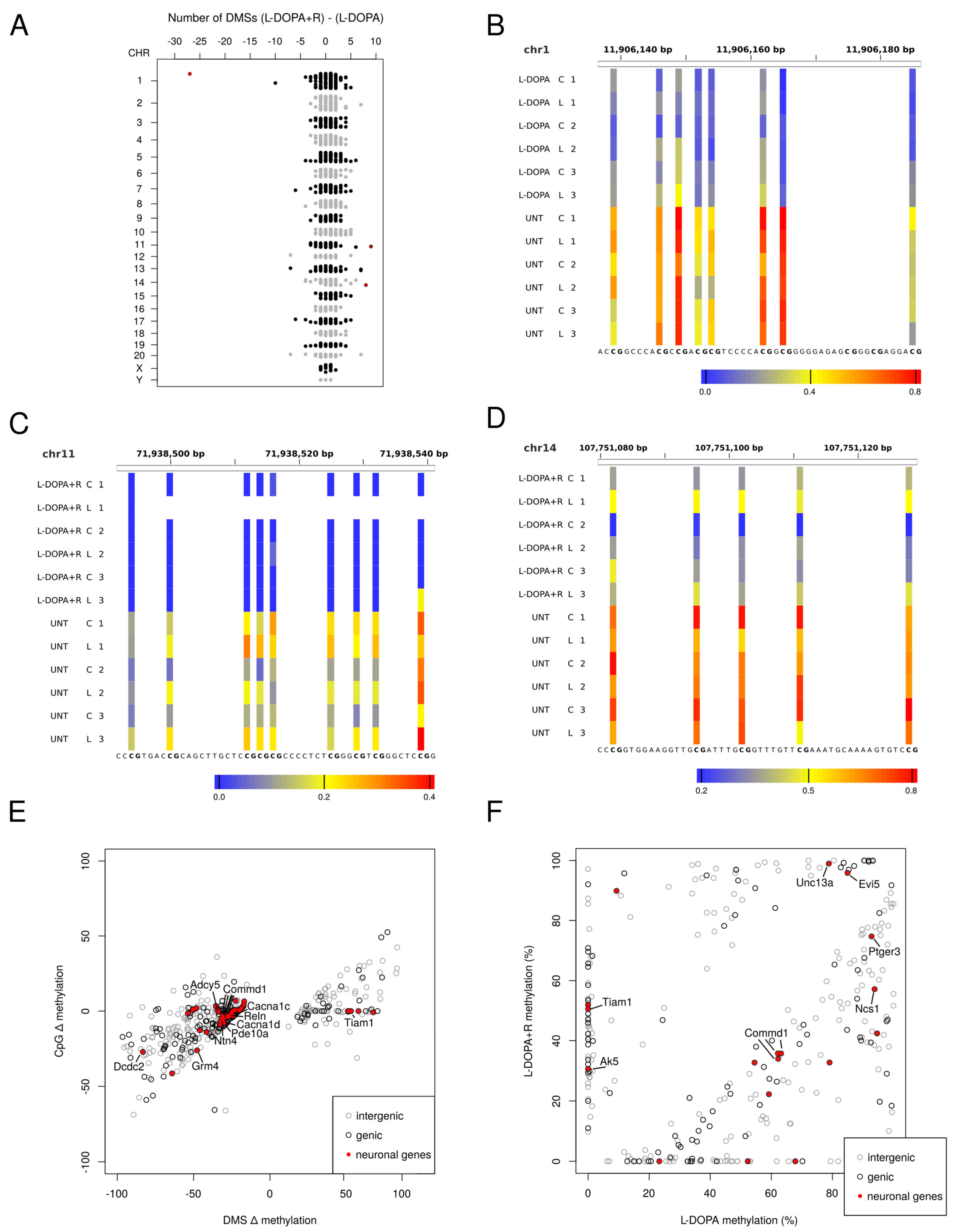
3. Results
3.1. DNA Methylation Analysis
3.1.1. Differentially Methylated Sites (DMS)
3.1.2. Genomic Distribution of DMS
3.1.3. Genes Undergoing DNA Methylation Alteration in the LID Model
3.1.4. Identification of DMS with Differential Methylation between L-DOPA and L-DOPA + Riluzole Treated Animals
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sanger, T.D.; Chen, D.; Fehlings, D.L.; Hallett, M.; Lang, A.E.; Mink, J.W.; Singer, H.S.; Alter, K.; Ben-Pazi, H.; Butler, E.E.; et al. Definition and classification of hyperkinetic movements in childhood. Mov. Disord. 2010, 25, 1538–1549. [Google Scholar] [CrossRef] [PubMed]
- Pandey, S.; Srivanitchapoom, P. Levodopa-induced Dyskinesia: Clinical Features, Pathophysiology, and Medical Management. Ann. Indian Acad. Neurol. 2017, 20, 190–198. [Google Scholar] [PubMed]
- Tran, T.N.; Vo, T.N.N.; Frei, K.; Truong, D.D. Levodopa-induced dyskinesia: Clinical features, incidence, and risk factors. J. Neural. Transm. 2018, 125, 1109–1117. [Google Scholar] [CrossRef] [PubMed]
- Bezard, E.; Hill, M.P.; Crossman, A.R.; Brotchie, J.M.; Michel, A.; Grimee, R.; Klitgaard, H. Levetiracetam improves choreic levodopa-induced dyskinesia in the MPTP-treated macaque. Eur. J. Pharmacol. 2004, 485, 159–164. [Google Scholar] [CrossRef]
- Du, H.; Nie, S.; Chen, G.; Ma, K.; Xu, Y.; Zhang, Z.; Papa, S.M.; Cao, X. Levetiracetam Ameliorates L-DOPA-Induced Dyskinesia in Hemiparkinsonian Rats Inducing Critical Molecular Changes in the Striatum. Parkinson’s Dis. 2015, 2015, 253878. [Google Scholar] [CrossRef]
- Kobylecki, C.; Burn, D.J.; Kass-Iliyya, L.; Kellett, M.W.; Crossman, A.R.; Silverdale, M.A. Randomized clinical trial of topiramate for levodopa-induced dyskinesia in Parkinson′s disease. Parkinsonism Relat. Disord. 2014, 20, 452–455. [Google Scholar] [CrossRef]
- Litim, N.; Morissette, M.; di Paolo, T. Metabotropic glutamate receptors as therapeutic targets in Parkinson’s disease: An update from the last 5 years of research. Neuropharmacology 2017, 115, 166–179. [Google Scholar] [CrossRef]
- Murata, M.; Hasegawa, K.; Kanazawa, I.; Fukasaka, J.; Kochi, K.; Shimazu, R.; Japan Zonisamide on, P.D.S.G. Zonisamide improves wearing-off in Parkinson′s disease: A randomized, double-blind study. Mov. Disord. 2015, 30, 1343–1350. [Google Scholar] [CrossRef]
- Durif, F.; Debilly, B.; Galitzky, M.; Morand, D.; Viallet, F.; Borg, M.; Thobois, S.; Broussolle, E.; Rascol, O. Clozapine improves dyskinesias in Parkinson disease: A double-blind, placebo-controlled study. Neurology 2004, 62, 381–388. [Google Scholar] [CrossRef]
- Borgohain, R.; Szasz, J.; Stanzione, P.; Meshram, C.; Bhatt, M.H.; Chirilineau, D.; Stocchi, F.; Lucini, V.; Giuliani, R.; Forrest, E.; et al. Two-year, randomized, controlled study of safinamide as add-on to levodopa in mid to late Parkinson’s disease. Mov. Disord. 2014, 29, 1273–1280. [Google Scholar] [CrossRef]
- Schapira, A.H.; Fox, S.H.; Hauser, R.A.; Jankovic, J.; Jost, W.H.; Kenney, C.; Kulisevsky, J.; Pahwa, R.; Poewe, W.; Anand, R. Assessment of Safety and Efficacy of Safinamide as a Levodopa Adjunct in Patients with Parkinson Disease and Motor Fluctuations: A Randomized Clinical Trial. JAMA Neurol. 2017, 74, 216–224. [Google Scholar] [CrossRef]
- Aum, D.J.; Tierney, T.S. Deep brain stimulation: Foundations and future trends. Front. Biosci. 2018, 23, 162–182. [Google Scholar]
- Zarzycki, M.Z.; Domitrz, I. Stimulation-induced side effects after deep brain stimulation—A systematic review. Acta Neuropsychiatr. 2020, 32, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Kalia, L.V.; Lang, A.E. Parkinson′s disease. Lancet 2015, 386, 896–912. [Google Scholar] [CrossRef]
- Cenci, M.A.; Lundblad, M. Ratings of L-DOPA-induced dyskinesia in the unilateral 6-OHDA lesion model of Parkinson’s disease in rats and mice. Curr. Protoc. Neurosci. 2007. [Google Scholar] [CrossRef]
- Pagliaroli, L.; Widomska, J.; Nespoli, E.; Hildebrandt, T.; Barta, C.; Glennon, J.; Hengerer, B.; Poelmans, G. Riluzole Attenuates L-DOPA-Induced Abnormal Involuntary Movements Through Decreasing CREB1 Activity: Insights from a Rat Model. Mol. Neurobiol. 2019, 56, 5111–5121. [Google Scholar] [CrossRef] [PubMed]
- Cenci, M.A. Dopamine dysregulation of movement control in L-DOPA-induced dyskinesia. Trends Neurosci 2007, 30, 236–243. [Google Scholar] [CrossRef] [PubMed]
- Brotchie, J.M. Nondopaminergic mechanisms in levodopa-induced dyskinesia. Mov. Disord. 2005, 20, 919–931. [Google Scholar] [CrossRef] [PubMed]
- Nadjar, A.; Gerfen, C.R.; Bezard, E. Priming for L-dopa-induced dyskinesia in Parkinson’s disease: A feature inherent to the treatment or the disease? Prog. Neurobiol. 2009, 87, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Eskow, K.L.; Gupta, V.; Alam, S.; Park, J.Y.; Bishop, C. The partial 5-HT(1A) agonist buspirone reduces the expression and development of l-DOPA-induced dyskinesia in rats and improves l-DOPA efficacy. Pharmacol. Biochem. Behav. 2007, 87, 306–314. [Google Scholar] [CrossRef]
- Jenner, P. Molecular mechanisms of L-DOPA-induced dyskinesia. Nat. Rev. Neurosci. 2008, 9, 665–677. [Google Scholar] [CrossRef]
- Picconi, B.; Centonze, D.; Hakansson, K.; Bernardi, G.; Greengard, P.; Fisone, G.; Cenci, M.A.; Calabresi, P. Loss of bidirectional striatal synaptic plasticity in L-DOPA-induced dyskinesia. Nat. Neurosci. 2003, 6, 501–506. [Google Scholar] [CrossRef]
- Santini, E.; Alcacer, C.; Cacciatore, S.; Heiman, M.; Herve, D.; Greengard, P.; Girault, J.A.; Valjent, E.; Fisone, G. L-DOPA activates ERK signaling and phosphorylates histone H3 in the striatonigral medium spiny neurons of hemiparkinsonian mice. J. Neurochem. 2009, 108, 621–633. [Google Scholar] [CrossRef] [PubMed]
- Dekundy, A.; Lundblad, M.; Danysz, W.; Cenci, M.A. Modulation of L-DOPA-induced abnormal involuntary movements by clinically tested compounds: Further validation of the rat dyskinesia model. Behav. Brain Res. 2007, 179, 76–89. [Google Scholar] [CrossRef] [PubMed]
- Lundblad, M.; Usiello, A.; Carta, M.; Hakansson, K.; Fisone, G.; Cenci, M.A. Pharmacological validation of a mouse model of L-DOPA-induced dyskinesia. Ex. Neurol. 2005, 194, 66–75. [Google Scholar] [CrossRef]
- Marin, C.; Jimenez, A.; Bonastre, M.; Chase, T.N.; Tolosa, E. Non-NMDA receptor-mediated mechanisms are involved in levodopa-induced motor response alterations in Parkinsonian rats. Synapse 2000, 36, 267–274. [Google Scholar] [CrossRef]
- Liu, J.; Wang, L.N. The efficacy and safety of riluzole for neurodegenerative movement disorders: A systematic review with meta-analysis. Drug. Deliv. 2018, 25, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Schiefer, J.; Landwehrmeyer, G.B.; Luesse, H.G.; Sprunken, A.; Puls, C.; Milkereit, A.; Milkereit, E.; Kosinski, C.M. Riluzole prolongs survival time and alters nuclear inclusion formation in a transgenic mouse model of Huntington’s disease. Mov. Disord. 2002, 17, 748–757. [Google Scholar] [CrossRef]
- Mignani, S.; Majoral, J.P.; Desaphy, J.F.; Lentini, G. From Riluzole to Dexpramipexole via Substituted-Benzothiazole Derivatives for Amyotrophic Lateral Sclerosis Disease Treatment: Case Studies. Molecules 2020, 25, 3320. [Google Scholar] [CrossRef] [PubMed]
- Ammal Kaidery, N.; Tarannum, S.; Thomas, B. Epigenetic landscape of Parkinson′s disease: Emerging role in disease mechanisms and therapeutic modalities. Neurotherapeutics 2013, 10, 698–708. [Google Scholar] [CrossRef]
- Figge, D.A.; Standaert, D.G. Dysregulation of BET proteins in levodopa-induced dyskinesia. Neurobiol. Dis. 2017, 102, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Figge, D.A.; Jaunarajs, K.L.E.; Standaert, D.G. Dynamic DNA Methylation Regulates Levodopa-Induced Dyskinesia. J. Neurosci. 2016, 36, 6514–6524. [Google Scholar] [CrossRef] [PubMed]
- Miranda-Morales, E.; Meier, K.; Sandoval-Carrillo, A.; Salas-Pacheco, J.; Vazquez-Cardenas, P.; Arias-Carrion, O. Implications of DNA Methylation in Parkinson′s Disease. Front. Mol. Neurosci. 2017, 10, 225. [Google Scholar] [CrossRef] [PubMed]
- Pagliaroli, L.; Veto, B.; Aranyi, T.; Barta, C. From Genetics to Epigenetics: New Perspectives in Tourette Syndrome Research. Front. Neurosci. 2016, 10, 277. [Google Scholar] [CrossRef]
- Ziller, M.J.; Muller, F.; Liao, J.; Zhang, Y.; Gu, H.; Bock, C.; Boyle, P.; Epstein, C.B.; Bernstein, B.E.; Lengauer, T.; et al. Genomic distribution and inter-sample variation of non-CpG methylation across human cell types. PLoS Genet 2011, 7, e1002389. [Google Scholar] [CrossRef]
- Bird, A.P. CpG-rich islands and the function of DNA methylation. Nature 1986, 321, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Deaton, A.M.; Bird, A. CpG islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef]
- Hackett, J.A.; Surani, M.A. DNA methylation dynamics during the mammalian life cycle. Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20110328. [Google Scholar] [CrossRef] [PubMed]
- Aranyi, T.; Faucheux, B.A.; Khalfallah, O.; Vodjdani, G.; Biguet, N.F.; Mallet, J.; Meloni, R. The tissue-specific methylation of the human tyrosine hydroxylase gene reveals new regulatory elements in the first exon. J. Neurochem. 2005, 94, 129–139. [Google Scholar] [CrossRef]
- Aranyi, T.; Paldi, A. The constant variation: DNA methylation changes during preimplantation development. FEBS Lett. 2006, 580, 6521–6526. [Google Scholar] [CrossRef]
- Yamagata, Y.; Szabo, P.; Szuts, D.; Bacquet, C.; Aranyi, T.; Paldi, A. Rapid turnover of DNA methylation in human cells. Epigenetics 2012, 7, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Ziller, M.J.; Gu, H.; Muller, F.; Donaghey, J.; Tsai, L.T.; Kohlbacher, O.; De Jager, P.L.; Rosen, E.D.; Bennett, D.A.; Bernstein, B.E.; et al. Charting a dynamic DNA methylation landscape of the human genome. Nature 2013, 500, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Winkler, C.; Kirik, D.; Bjorklund, A.; Cenci, M.A. L-DOPA-induced dyskinesia in the intrastriatal 6-hydroxydopamine model of parkinson′s disease: Relation to motor and cellular parameters of nigrostriatal function. Neurobiol. Dis. 2002, 10, 165–186. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Liu, H.; Ma, Z.; Li, S.Y.; Park, J.; Sheng, X.; Susztak, K. Dnmt3a and Dnmt3b-Decommissioned Fetal Enhancers are Linked to Kidney Disease. J. Am. Soc. Nephrol. 2020, 31, 765–782. [Google Scholar] [CrossRef]
- Li, B.; Feng, Z.H.; Sun, H.; Zhao, Z.H.; Yang, S.B.; Yang, P. The blood genome-wide DNA methylation analysis reveals novel epigenetic changes in human heart failure. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 1828–1836. [Google Scholar] [CrossRef]
- Li, S.Y.; Park, J.; Guan, Y.; Chung, K.; Shrestha, R.; Palmer, M.B.; Susztak, K. DNMT1 in Six2 Progenitor Cells Is Essential for Transposable Element Silencing and Kidney Development. J. Am. Soc. Nephrol. 2019, 30, 594–609. [Google Scholar] [CrossRef]
- Lim, Y.C.; Chia, S.Y.; Jin, S.; Han, W.; Ding, C.; Sun, L. Dynamic DNA methylation landscape defines brown and white cell specificity during adipogenesis. Mol. Metab. 2016, 5, 1033–1041. [Google Scholar] [CrossRef]
- Reizel, Y.; Sabag, O.; Skversky, Y.; Spiro, A.; Steinberg, B.; Bernstein, D.; Wang, A.; Kieckhaefer, J.; Li, C.; Pikarsky, E.; et al. Postnatal DNA demethylation and its role in tissue maturation. Nat. Commun. 2018, 9, 2040. [Google Scholar] [CrossRef]
- Schmidt, M.; Van Bel, M.; Woloszynska, M.; Slabbinck, B.; Martens, C.; De Block, M.; Coppens, F.; Van Lijsebettens, M. Plant-RRBS, a bisulfite and next-generation sequencing-based methylome profiling method enriching for coverage of cytosine positions. BMC Plant Biol. 2017, 17, 115. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, Y.; Chen, M.; Sun, L.; Han, J.; Elena, V.K.; Qiao, H. CXCL12 methylation-mediated epigenetic regulation of gene expression in papillary thyroid carcinoma. Sci. Re 2017, 7, 44033. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, S.; Ma, L.; Jiang, E.; Xu, H.; Chen, R.; Yang, Q.; Chen, H.; Li, Z.; Lan, X. Reduced representation bisulfite sequencing (RRBS) of dairy goat mammary glands reveals DNA methylation profiles of integrated genome-wide and critical milk-related genes. Oncotarget 2017, 8, 115326–115344. [Google Scholar] [CrossRef]
- Meissner, A.; Gnirke, A.; Bell, G.W.; Ramsahoye, B.; Lander, E.S.; Jaenisch, R. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nucleic. Acids Res. 2005, 33, 5868–5877. [Google Scholar] [CrossRef]
- Veillard, A.-C.; Datlinger, P.; Laczik, M.; Squazzo, S.; Bock, C. Diagenode® Premium RRBS technology: Cost-effective DNA methylation mapping with superior coverage. Nature Methods 2016, 13, 184. [Google Scholar] [CrossRef]
- Wreczycka, K.; Gosdschan, A.; Yusuf, D.; Gruning, B.; Assenov, Y.; Akalin, A. Strategies for analyzing bisulfite sequencing data. J. Biotechnol. 2017, 261, 105–115. [Google Scholar] [CrossRef]
- Eden, E.; Navon, R.; Steinfeld, I.; Lipson, D.; Yakhini, Z. GOrilla: A tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinform. 2009, 10, 48. [Google Scholar] [CrossRef]
- Chertkow-Deutsher, Y.; Cohen, H.; Klein, E.; Ben-Shachar, D. DNA methylation in vulnerability to post-traumatic stress in rats: Evidence for the role of the post-synaptic density protein Dlgap2. Int. J. Neuropsychopharmacol. 2010, 13, 347–359. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Payen, C.; Guillot, A.; Paillat, L.; Fothi, A.; Dib, A.; Bourreau, J.; Schmitt, F.; Loufrani, L.; Aranyi, T.; Henrion, D.; et al. Pathophysiological adaptations of resistance arteries in rat offspring exposed in utero to maternal obesity is associated with sex-specific epigenetic alterations. Int. J. Obes. 2021. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.M.; Cho, S.Y.; Kim, M.W.; Roh, S.R.; Shin, H.S.; Suh, Y.H.; Geum, D.; Lee, M.A. Genome-Wide Analysis Identifies NURR1-Controlled Network of New Synapse Formation and Cell Cycle Arrest in Human Neural Stem Cells. Mol. Cells 2020, 43, 551–571. [Google Scholar]
- Cruces-Sande, A.; Rodriguez-Perez, A.I.; Herbello-Hermelo, P.; Bermejo-Barrera, P.; Mendez-Alvarez, E.; Labandeira-Garcia, J.L.; Soto-Otero, R. Copper Increases Brain Oxidative Stress and Enhances the Ability of 6-Hydroxydopamine to Cause Dopaminergic Degeneration in a Rat Model of Parkinson′s Disease. Mol. Neurobiol. 2019, 56, 2845–2854. [Google Scholar] [CrossRef]
- Vonk, W.I.; Kakkar, V.; Bartuzi, P.; Jaarsma, D.; Berger, R.; Hofker, M.H.; Klomp, L.W.; Wijmenga, C.; Kampinga, H.H.; van de Sluis, B. The Copper Metabolism MURR1 domain protein 1 (COMMD1) modulates the aggregation of misfolded protein species in a client-specific manner. PLoS ONE 2014, 9, e92408. [Google Scholar] [CrossRef]
- Cirnaru, M.D.; Melis, C.; Fanutza, T.; Naphade, S.; Tshilenge, K.T.; Muntean, B.S.; Martemyanov, K.A.; Plotkin, J.L.; Ellerby, L.M.; Ehrlich, M.E. Nuclear Receptor Nr4a1 Regulates Striatal Striosome Development and Dopamine D1 Receptor Signaling. eNeuro 2019, 6. [Google Scholar] [CrossRef]
- Gilbert, F.; Morissette, M.; St-Hilaire, M.; Paquet, B.; Rouillard, C.; Di Paolo, T.; Levesque, D. Nur77 gene knockout alters dopamine neuron biochemical activity and dopamine turnover. Biol. Psychiatry 2006, 60, 538–547. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, S.; Blanchet, P.J.; Levesque, D. Haloperidol-induced striatal Nur77 expression in a non-human primate model of tardive dyskinesia. Eur. J. Neurosci. 2013, 38, 2192–2198. [Google Scholar] [CrossRef][Green Version]
- Chang, F.C.; Westenberger, A.; Dale, R.C.; Smith, M.; Pall, H.S.; Perez-Duenas, B.; Grattan-Smith, P.; Ouvrier, R.A.; Mahant, N.; Hanna, B.C.; et al. Phenotypic insights into ADCY5-associated disease. Mov. Disord. 2016, 31, 1033–1040. [Google Scholar] [CrossRef]
- Doyle, T.B.; Hayes, M.P.; Chen, D.H.; Raskind, W.H.; Watts, V.J. Functional characterization of AC5 gain-of-function variants: Impact on the molecular basis of ADCY5-related dyskinesia. Biochem. Pharmacol. 2019, 163, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Niccolini, F.; Mencacci, N.E.; Yousaf, T.; Rabiner, E.A.; Salpietro, V.; Pagano, G.; Balint, B.; Efthymiou, S.; Houlden, H.; Gunn, R.N.; et al. PDE10A and ADCY5 mutations linked to molecular and microstructural basal ganglia pathology. Mov. Disord. 2018, 33, 1961–1965. [Google Scholar] [CrossRef]
- Beck, G.; Maehara, S.; Chang, P.L.; Papa, S.M. A Selective Phosphodiesterase 10A Inhibitor Reduces L-Dopa-Induced Dyskinesias in Parkinsonian Monkeys. Mov. Disord. 2018, 33, 805–814. [Google Scholar] [CrossRef]
- Niccolini, F.; Foltynie, T.; Reis Marques, T.; Muhlert, N.; Tziortzi, A.C.; Searle, G.E.; Natesan, S.; Kapur, S.; Rabiner, E.A.; Gunn, R.N.; et al. Loss of phosphodiesterase 10A expression is associated with progression and severity in Parkinson’s disease. Brain 2015, 138, 3003–3015. [Google Scholar] [CrossRef]
- Wilson, J.M.; Ogden, A.M.; Loomis, S.; Gilmour, G.; Baucum, A.J., 2nd; Belecky-Adams, T.L.; Merchant, K.M. Phosphodiesterase 10A inhibitor, MP-10 (PF-2545920), produces greater induction of c-Fos in dopamine D2 neurons than in D1 neurons in the neostriatum. Neuropharmacology 2015, 99, 379–386. [Google Scholar] [CrossRef]
- Franco, R.; Aguinaga, D.; Reyes, I.; Canela, E.I.; Lillo, J.; Tarutani, A.; Hasegawa, M.; Del Ser-Badia, A.; Del Rio, J.A.; Kreutz, M.R.; et al. N-Methyl-D-Aspartate Receptor Link to the MAP Kinase Pathway in Cortical and Hippocampal Neurons and Microglia Is Dependent on Calcium Sensors and Is Blocked by alpha-Synuclein, Tau, and Phospho-Tau in Non-transgenic and Transgenic APPSw, Ind Mice. Front. Mol. Neurosci. 2018, 11, 273. [Google Scholar] [CrossRef] [PubMed]
- Caracci, M.O.; Fuentealba, L.M.; Marzolo, M.P. Golgi Complex Dynamics and Its Implication in Prevalent Neurological Disorders. Front. Cell Dev. Biol. 2019, 7, 75. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, A.; Rashidi, E.; Amooeian, V.G. Brain, blood, cerebrospinal fluid, and serum biomarkers in schizophrenia. Psychiatry Res. 2018, 265, 25–38. [Google Scholar] [CrossRef]
- Sanchez-Sanchez, S.M.; Magdalon, J.; Griesi-Oliveira, K.; Yamamoto, G.L.; Santacruz-Perez, C.; Fogo, M.; Passos-Bueno, M.R.; Sertie, A.L. Rare RELN variants affect Reelin-DAB1 signal transduction in autism spectrum disorder. Hum Mutat. 2018, 39, 1372–1383. [Google Scholar] [CrossRef]
- Zilhao, N.R.; Padmanabhuni, S.S.; Pagliaroli, L.; Barta, C.; Consortium, B.; Smit, D.J.; Cath, D.; Nivard, M.G.; Baselmans, B.M.; van Dongen, J.; et al. Epigenome-Wide Association Study of Tic Disorders. Twin Res. Hum. Genet 2015, 18, 699–709. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.Y.; Yu, D.; Davis, L.K.; Sul, J.H.; Tsetsos, F.; Ramensky, V.; Zelaya, I.; Ramos, E.M.; Osiecki, L.; Chen, J.A.; et al. Rare Copy Number Variants in NRXN1 and CNTN6 Increase Risk for Tourette Syndrome. Neuron 2017, 94, 1101–1111.e1107. [Google Scholar] [CrossRef]
- Paschou, P.; Yu, D.; Gerber, G.; Evans, P.; Tsetsos, F.; Davis, L.K.; Karagiannidis, I.; Chaponis, J.; Gamazon, E.; Mueller-Vahl, K.; et al. Genetic association signal near NTN4 in Tourette syndrome. Ann. Neurol. 2014, 76, 310–315. [Google Scholar] [CrossRef]
- Liu, X.S.; Wu, H.; Krzisch, M.; Wu, X.; Graef, J.; Muffat, J.; Hnisz, D.; Li, C.H.; Yuan, B.; Xu, C.; et al. Rescue of Fragile X Syndrome Neurons by DNA Methylation Editing of the FMR1 Gene. Cell 2018, 172, 979–992.e976. [Google Scholar] [CrossRef]
- Park, J.; Guan, Y.; Sheng, X.; Gluck, C.; Seasock, M.J.; Hakimi, A.A.; Qiu, C.; Pullman, J.; Verma, A.; Li, H.; et al. Functional methylome analysis of human diabetic kidney disease. JCI Insight 2019, 4, e128886. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pagliaroli, L.; Fothi, A.; Nespoli, E.; Liko, I.; Veto, B.; Devay, P.; Szeri, F.; Hengerer, B.; Barta, C.; Aranyi, T. Riluzole Administration to Rats with Levodopa-Induced Dyskinesia Leads to Loss of DNA Methylation in Neuronal Genes. Cells 2021, 10, 1442. https://doi.org/10.3390/cells10061442
Pagliaroli L, Fothi A, Nespoli E, Liko I, Veto B, Devay P, Szeri F, Hengerer B, Barta C, Aranyi T. Riluzole Administration to Rats with Levodopa-Induced Dyskinesia Leads to Loss of DNA Methylation in Neuronal Genes. Cells. 2021; 10(6):1442. https://doi.org/10.3390/cells10061442
Chicago/Turabian StylePagliaroli, Luca, Abel Fothi, Ester Nespoli, Istvan Liko, Borbala Veto, Piroska Devay, Flora Szeri, Bastian Hengerer, Csaba Barta, and Tamas Aranyi. 2021. "Riluzole Administration to Rats with Levodopa-Induced Dyskinesia Leads to Loss of DNA Methylation in Neuronal Genes" Cells 10, no. 6: 1442. https://doi.org/10.3390/cells10061442
APA StylePagliaroli, L., Fothi, A., Nespoli, E., Liko, I., Veto, B., Devay, P., Szeri, F., Hengerer, B., Barta, C., & Aranyi, T. (2021). Riluzole Administration to Rats with Levodopa-Induced Dyskinesia Leads to Loss of DNA Methylation in Neuronal Genes. Cells, 10(6), 1442. https://doi.org/10.3390/cells10061442






