DNA-RNA Hybrid (R-Loop): From a Unified Picture of the Mammalian Telomere to the Genome-Wide Profile
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. Sperm Preparation
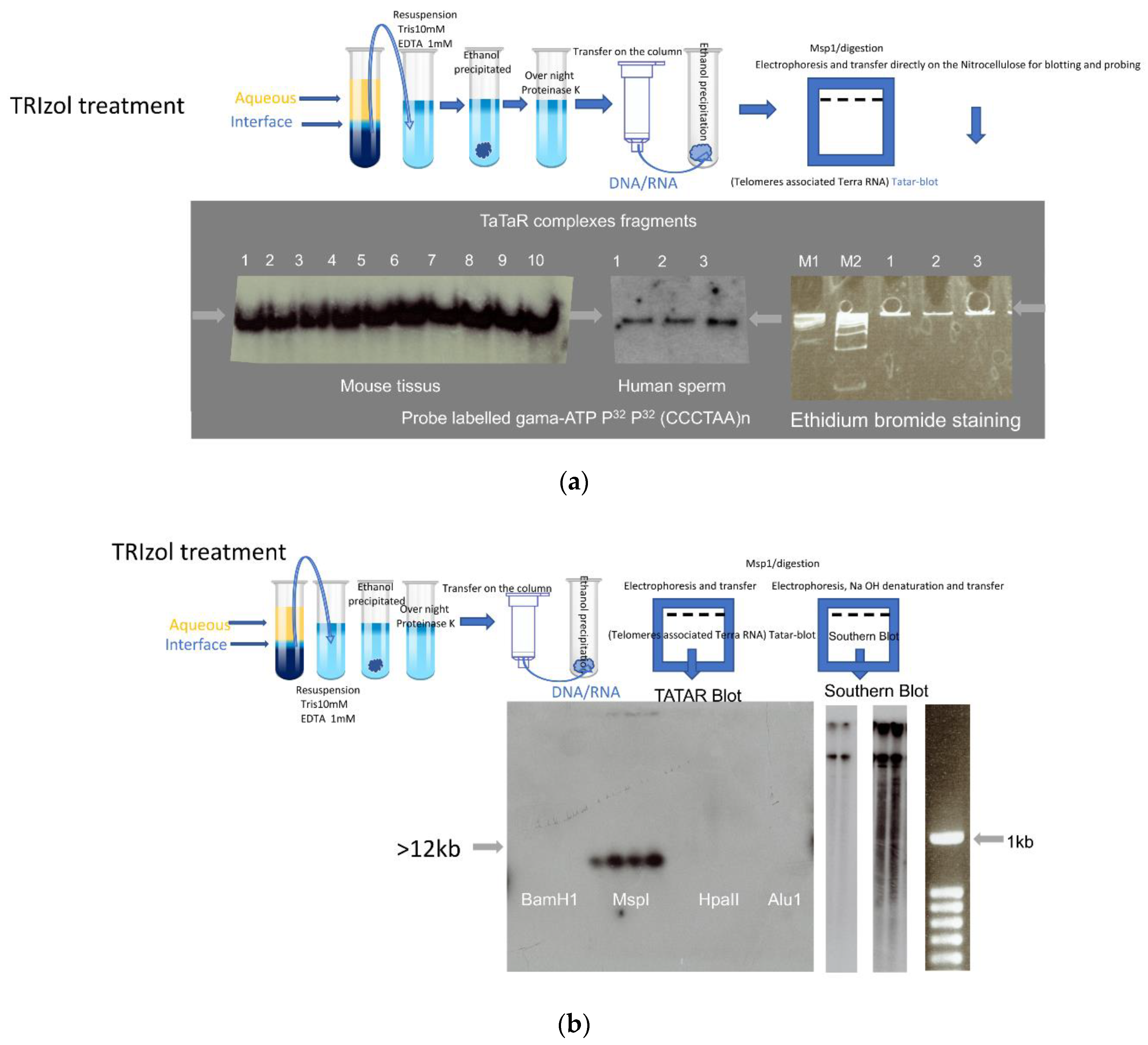
2.3. Gel Electrophoresis of the DNA/RNA Complex
2.4. Preparation of DNA-Bound RNA
2.5. Enzymatic Assays
2.6. Sequence Establishment
2.7. Sequence Analysis
3. Results
3.1. Antibody-Independent Profiling of R-Loop Complexes: A General Approach
3.2. The Telomeric TERRA Complex
3.3. Transcription of Individual Telomeres from Local Subtelomeric Promoters
3.4. Extension Analysis to Other DRNAs with UUAGGG Motif Repetition
3.5. DRNA Is Not Restricted to the UUAGGG Repeats
4. Discussion
4.1. Genomic Profiles
4.2. Genomic Profiles of the TERRA (Telomeric and Non-Telomeric)
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Costantino, L.; Koshland, D. The Yin and Yang of R-loop biology. Curr. Opin. Cell Biol. 2015, 34, 39–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boguslawski, S.J.; Smith, D.E.; Michalak, M.A.; Mickelson, K.E.; Yehle, C.O.; Patterson, W.L.; Carrico, R.J. Characterization of monoclonal antibody to DNA · RNA and its application to immunodetection of hybrids. J. Immunol. Methods 1986, 89, 123–130. [Google Scholar] [CrossRef]
- Diman, A.; Decottignies, A. Genomic origin and nuclear localization of TERRA telomeric repeat-containing RNA: From Darkness to Dawn. FEBS J. 2017, 285, 1389–1398. [Google Scholar] [CrossRef] [Green Version]
- Azzalin, C.M.; Reichenbach, P.; Khoriauli, L.; Giulotto, E.; Lingner, J. Telomeric Repeat Containing RNA and RNA Surveillance Factors at Mammalian Chromosome Ends. Science 2007, 318, 798–801. [Google Scholar] [CrossRef]
- Isken, O.; Maquat, L. Telomeric RNAs as a novel player in telomeric integrity. F1000 Biol. Rep. 2009, 1, 90. [Google Scholar] [CrossRef] [PubMed]
- Nergadze, S.G.; Farnung, B.O.; Wischnewski, H.; Khoriauli, L.; Vitelli, V.; Chawla, R.; Giulotto, E.; Azzalin, C.M. CpG-island promoters drive transcription of human telomeres. RNA 2009, 15, 2186–2194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luke, B.; Panza, A.; Redon, S.; Iglesias, N.; Li, Z.; Lingner, J. The Rat1p 5′ to 3′ Exonuclease Degrades Telomeric Repeat-Containing RNA and Promotes Telomere Elongation in Saccharomyces cerevisiae. Mol. Cell 2008, 32, 465–477. [Google Scholar] [CrossRef] [PubMed]
- Crossley, M.P.; Bocek, M.; Cimprich, K.A. R-Loops as Cellular Regulators and Genomic Threats. Mol. Cell 2019, 73, 398–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montero, J.J.; De Silanes, I.L.; Graña, O.; Blasco, M.A. Telomeric RNAs are essential to maintain telomeres. Nat. Commun. 2016, 7, 12534. [Google Scholar] [CrossRef] [Green Version]
- Gibbons, H.R.; Aune, T.M. Immunoprecipitation of DNA:RNA Hybrids Using the S9.6 Antibody. In Methods in Molecular Biology; Clifton, N.J., Ed.; Humana Press: Totowa, NJ, USA, 2020; Volume 2116, pp. 195–207. [Google Scholar] [CrossRef]
- Sanz, L.A.; Chédin, F. High-resolution, strand-specific R-loop mapping via S9.6-based DNA–RNA immunoprecipitation and high-throughput sequencing. Nat. Protoc. 2019, 14, 1734–1755. [Google Scholar] [CrossRef] [PubMed]
- Kianmehr, L.; Khazali, H.; Rajabi-Maham, H.; Sharifi-Zarchi, A.; Cuzin, F.; Rassoulzadegan, M. Genome-Wide Distribution of Nascent Transcripts in Sperm DNA, Products of a Late Wave of General Transcription. Cells 2019, 8, 1196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 2011, 27, 2987–2993. [Google Scholar] [CrossRef] [Green Version]
- Heinz, S.; Benner, C.; Spann, N.; Bertolino, E.; Lin, Y.; Laslo, P.; Cheng, J.X.; Murre, C.; Singh, H.; Glass, C.K. Simple Combinations of Lineage-Determining Transcription Factors Prime cis-Regulatory Elements Required for Macrophage and B Cell Identities. Mol. Cell 2010, 38, 576–589. [Google Scholar] [CrossRef] [Green Version]
- Thorvaldsdóttir, H.; Robinson, J.T.; Mesirov, J.P. Integrative Genomics Viewer (IGV): High-performance genomics data visualization and exploration. Brief. Bioinform. 2013, 14, 178–192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [Green Version]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [Green Version]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2016. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Rio, D.C.; Ares, M.; Hannon, G.J.; Nilsen, T.W. Purification of RNA Using TRIzol (TRI Reagent). Cold Spring Harb. Protoc. 2010, 2010. [Google Scholar] [CrossRef]
- Schoeftner, S.; Blasco, M.A. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2007, 10, 228–236. [Google Scholar] [CrossRef]
- Arora, R.; Lee, Y.; Wischnewski, H.; Brun, C.M.; Schwarz, T.; Azzalin, C.M. RNaseH1 regulates TERRA-telomeric DNA hybrids and telomere maintenance in ALT tumour cells. Nat. Commun. 2014, 5, 5220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sagie, S.; Toubiana, S.; Hartono, S.R.; Katzir, H.; Tzur-Gilat, A.; Havazelet, S.; Francastel, C.; Velasco, G.; Chedin, F.; Selig, S. Telomeres in ICF syndrome cells are vulnerable to DNA damage due to elevated DNA:RNA hybrids. Nat. Commun. 2017, 8, 14015. [Google Scholar] [CrossRef] [Green Version]
- De Silanes, I.L.; Graña, O.; De Bonis, M.L.; Dominguez, O.; Pisano, D.; Blasco, M.A. Identification of TERRA locus unveils a telomere protection role through association to nearly all chromosomes. Nat. Commun. 2014, 5, 4723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buxton, J.L.; Suderman, M.; Pappas, J.J.; Borghol, N.; McArdle, W.; Blakemore, A.I.F.; Hertzman, C.; Power, C.; Szyf, M.; Pembrey, M. Human leukocyte telomere length is associated with DNA methylation levels in multiple subtelomeric and imprinted loci. Sci. Rep. 2014, 4, 4954. [Google Scholar] [CrossRef] [Green Version]
- Blasco, M.A.; Lee, H.-W.; Hande, M.P.; Samper, E.; Lansdorp, P.M.; DePinho, R.A.; Greider, C.W. Telomere Shortening and Tumor Formation by Mouse Cells Lacking Telomerase RNA. Cell 1997, 91, 25–34. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-W.; Blasco, M.A.; Gottlieb, G.J.; Ii, J.W.H.; Greider, C.W.; DePinho, R.A. Essential role of mouse telomerase in highly proliferative organs. Nat. Cell Biol. 1998, 392, 569–574. [Google Scholar] [CrossRef] [PubMed]
- Müller, S.; Rodriguez, R. G-quadruplex interacting small molecules and drugs: From bench toward bedside. Expert Rev. Clin. Pharmacol. 2014, 7, 663–679. [Google Scholar] [CrossRef]
- Kent, W.J. BLAT—The BLAST-Like Alignment Tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, H.-P.; Cifuentes-Rojas, C.; Kesner, B.; Aeby, E.; Lee, H.-G.; Wei, C.; Oh, H.J.; Boukhali, M.; Haas, W.; Lee, J.T. TERRA RNA Antagonizes ATRX and Protects Telomeres. Cell 2017, 170, 86–101.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, H.-P.; Froberg, J.E.; Kesner, B.; Oh, H.J.; Ji, F.; Sadreyev, R.; Pinter, S.F.; Lee, J.T. PAR-TERRA directs homologous sex chromosome pairing. Nat. Struct. Mol. Biol. 2017, 24, 620–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanz, L.A.; Hartono, S.R.; Lim, Y.W.; Steyaert, S.; Rajpurkar, A.; Ginno, P.A.; Xu, X.; Chédin, F. Prevalent, Dynamic, and Conserved R-Loop Structures Associate with Specific Epigenomic Signatures in Mammals. Mol. Cell 2016, 63, 167–178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rassoulzadegan, M.; Grandjean, V.; Gounon, P.; Vincent, S.; Gillot, I.; Cuzin, F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nat. Cell Biol. 2006, 441, 469–474. [Google Scholar] [CrossRef] [PubMed]
- Rassoulzadegan, M.; Cuzin, F. From paramutation to human disease: RNA-mediated heredity. Semin. Cell Dev. Biol. 2015, 44, 47–50. [Google Scholar] [CrossRef]
- Yan, Q.; Shields, E.J.; Bonasio, R.; Sarma, K. Mapping Native R-Loops Genome-wide Using a Targeted Nuclease Approach. Cell Rep. 2019, 29, 1369–1380.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Chen, J.-Y.; Zhang, X.; Gu, Y.; Xiao, R.; Shao, C.; Tang, P.; Qian, H.; Luo, D.; Li, H.; et al. R-ChIP Using Inactive RNase H Reveals Dynamic Coupling of R-loops with Transcriptional Pausing at Gene Promoters. Mol. Cell 2017, 68, 745–757. [Google Scholar] [CrossRef] [Green Version]
- Estill, M.S.; Hauser, R.; Krawetz, S.A. RNA element discovery from germ cell to blastocyst. Nucleic Acids Res. 2018, 47, 2263–2275. [Google Scholar] [CrossRef]
- Yan, P.; Liu, Z.; Song, M.; Wu, Z.; Xu, W.; Li, K.; Ji, Q.; Wang, S.; Liu, X.; Yan, K.; et al. Genome-wide R-loop Landscapes during Cell Differentiation and Reprogramming. Cell Rep. 2020, 32, 107870. [Google Scholar] [CrossRef] [PubMed]






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rassoulzadegan, M.; Sharifi-Zarchi, A.; Kianmehr, L. DNA-RNA Hybrid (R-Loop): From a Unified Picture of the Mammalian Telomere to the Genome-Wide Profile. Cells 2021, 10, 1556. https://doi.org/10.3390/cells10061556
Rassoulzadegan M, Sharifi-Zarchi A, Kianmehr L. DNA-RNA Hybrid (R-Loop): From a Unified Picture of the Mammalian Telomere to the Genome-Wide Profile. Cells. 2021; 10(6):1556. https://doi.org/10.3390/cells10061556
Chicago/Turabian StyleRassoulzadegan, Minoo, Ali Sharifi-Zarchi, and Leila Kianmehr. 2021. "DNA-RNA Hybrid (R-Loop): From a Unified Picture of the Mammalian Telomere to the Genome-Wide Profile" Cells 10, no. 6: 1556. https://doi.org/10.3390/cells10061556
APA StyleRassoulzadegan, M., Sharifi-Zarchi, A., & Kianmehr, L. (2021). DNA-RNA Hybrid (R-Loop): From a Unified Picture of the Mammalian Telomere to the Genome-Wide Profile. Cells, 10(6), 1556. https://doi.org/10.3390/cells10061556






