Interdependent Impact of Lipoprotein Receptors and Lipid-Lowering Drugs on HCV Infectivity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Lines
2.2. DNA Plasmids
2.3. Generation of Lipoprotein Receptor-Deficient Cell Lines
2.4. HCV Virus Production and Infection
2.5. HCV Kinetics in Primary Human Hepatocytes
2.6. RNA Extraction and RT-qPCR Quantification of Viral RNA
2.7. TCID50 Assay
2.8. Pseudoparticle Production and Transduction
2.9. HCV Replication Assay
2.10. Cell Viability Measurements
2.11. Flow Cytometry
2.12. Western Blot
2.13. Cholesterol Quantification Assay
2.14. Lipid-Lowering Drugs
2.15. Patients and Methods
2.16. Statistics
2.17. Ethics
3. Results
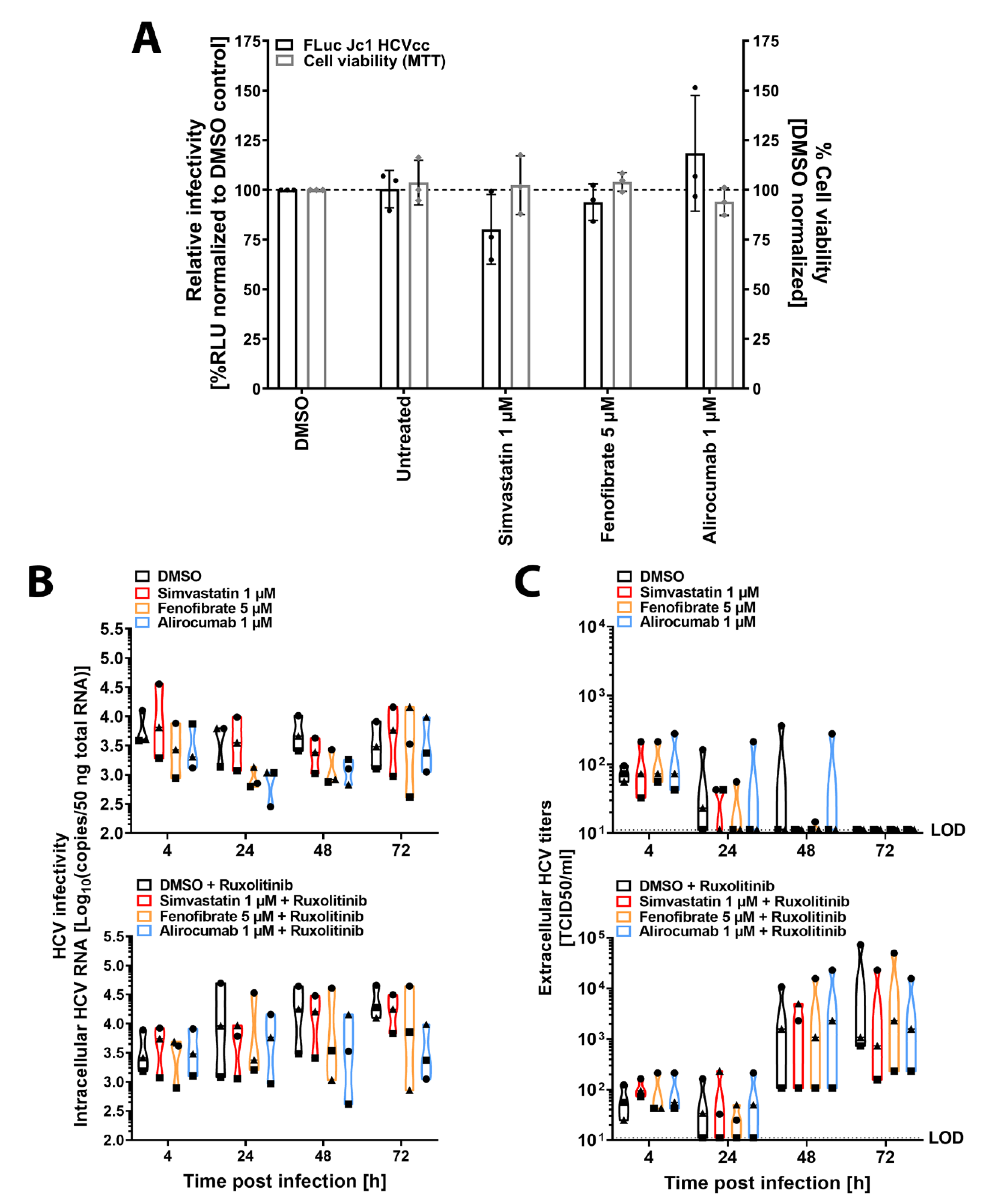
3.1. Glycoprotein-Driven HCV Entry Is Not Affected by Lipid-Lowering Drugs
3.2. Lipid-Lowering Drugs Do Not Impact HCV Infection of Human Immortalized or Primary Hepatocytes
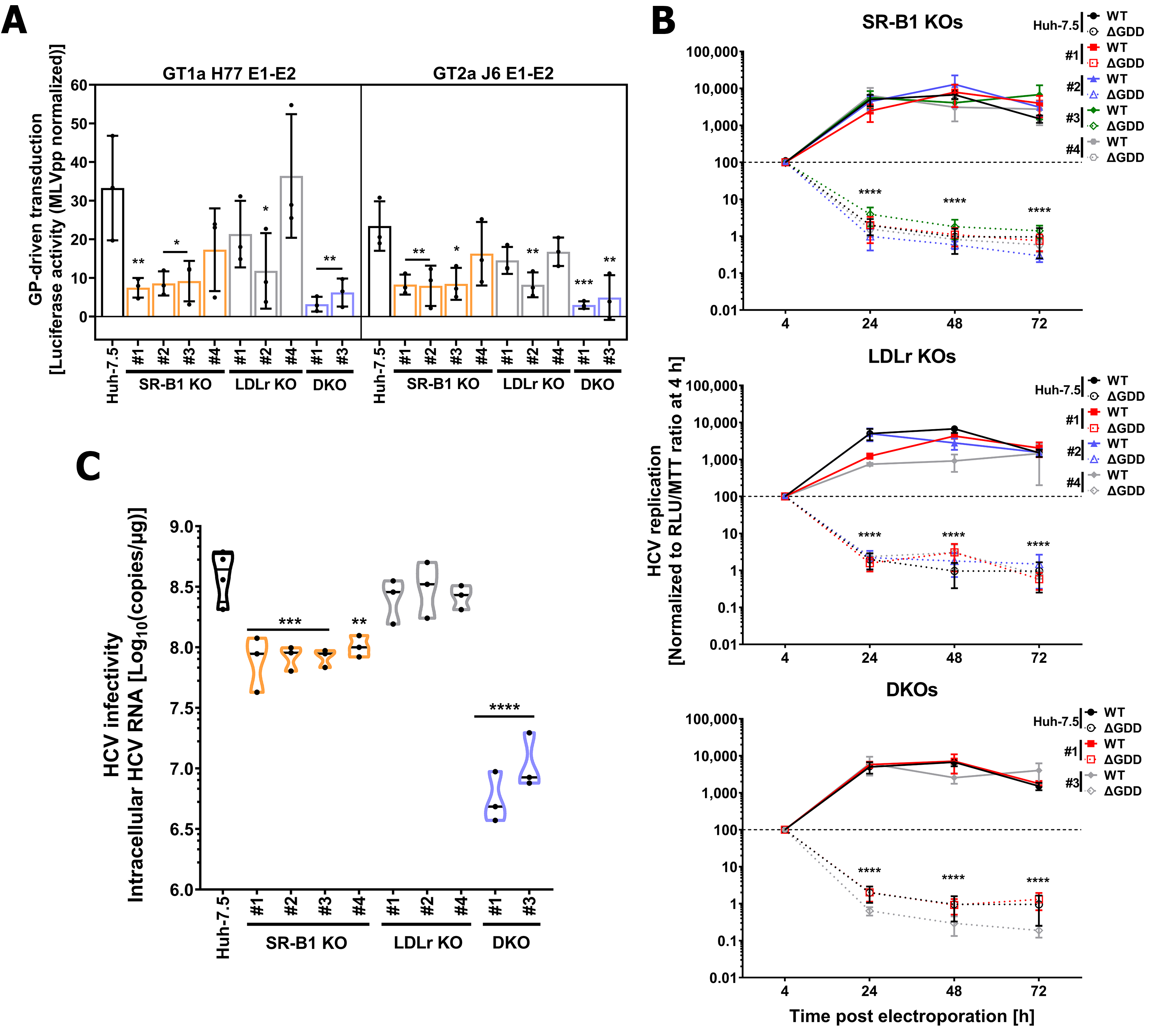
3.3. Genetic Ablation of Lipoprotein Receptors SR-B1 and LDLr Does Not Affect HCV Entry Factor Expression or Cholesterol Content
3.4. LDLr and SR-B1 Play a Redundant Role in HCV Infection but Not in Entry
3.5. Lipid-Lowering Drugs Affect HCV Infection in Lipoprotein Receptor-Deficient Cell Lines
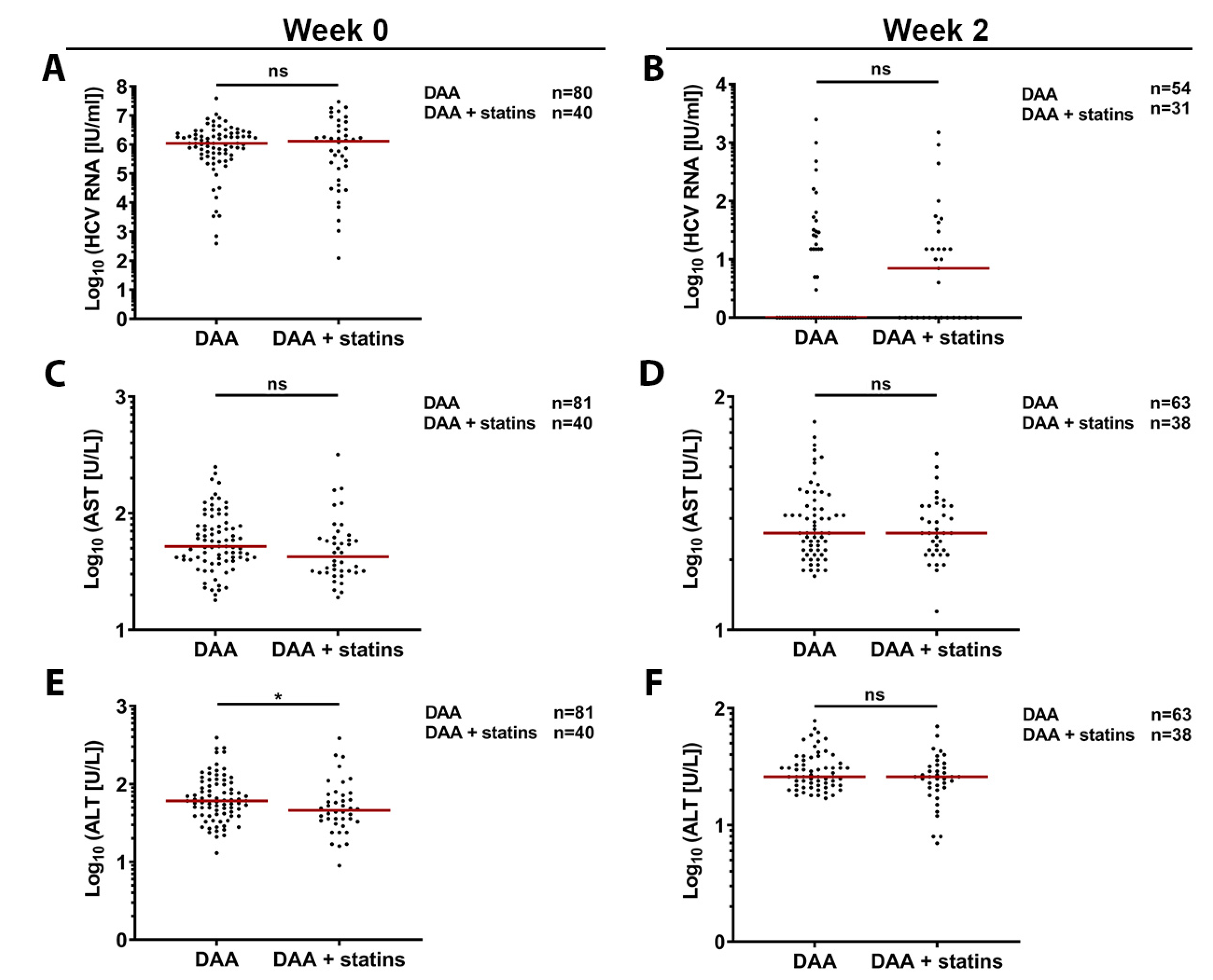
3.6. Retrospective In-Vivo Analysis of a HCV Patient Cohort Suggests a Slight Beneficial Effect of Statin Usage on Liver Health
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hepatitis C. Available online: https://www.who.int/news-room/fact-sheets/detail/hepatitis-c (accessed on 26 June 2020).
- Bhamidimarri, K.R.; Satapathy, S.K.; Martin, P. Hepatitis C virus and liver transplantation. Gastroenterol. Hepatol. 2017, 13, 214–220. [Google Scholar]
- Pecoraro, V.; Banzi, R.; Cariani, E.; Chester, J.; Villa, E.; D’Amico, R.; Bertele, V.; Trenti, T. New Direct-Acting Antivirals for the Treatment of Patients with Hepatitis C Virus Infection: A Systematic Review of Randomized Controlled Trials. J. Clin. Exp. Hepatol. 2019, 9, 522–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobson, I.M.; McHutchison, J.G.; Dusheiko, G.; Di Bisceglie, A.M.; Reddy, K.R.; Bzowej, N.H.; Marcellin, P.; Muir, A.J.; Ferenci, P.; Flisiak, R.; et al. ADVANCE Study Team Telaprevir for previously untreated chronic hepatitis C virus infection. N. Engl. J. Med. 2011, 364, 2405–2416. [Google Scholar] [CrossRef] [PubMed]
- Feld, J.J.; Jacobson, I.M.; Hézode, C.; Asselah, T.; Ruane, P.J.; Gruener, N.; Abergel, A.; Mangia, A.; Lai, C.-L.; Chan, H.L.Y.; et al. ASTRAL-1 Investigators Sofosbuvir and velpatasvir for HCV genotype 1, 2, 4, 5, and 6 infection. N. Engl. J. Med. 2015, 373, 2599–2607. [Google Scholar] [CrossRef] [Green Version]
- Thomssen, R.; Bonk, S.; Propfe, C.; Heermann, K.H.; Köchel, H.G.; Uy, A. Association of hepatitis C virus in human sera with beta-lipoprotein. Med. Microbiol. Immunol. 1992, 181, 293–300. [Google Scholar] [CrossRef]
- André, P.; Komurian-Pradel, F.; Deforges, S.; Perret, M.; Berland, J.L.; Sodoyer, M.; Pol, S.; Bréchot, C.; Paranhos-Baccalà, G.; Lotteau, V. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 2002, 76, 6919–6928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Egger, D.; Wölk, B.; Gosert, R.; Bianchi, L.; Blum, H.E.; Moradpour, D.; Bienz, K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J. Virol. 2002, 76, 5974–5984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miyanari, Y.; Atsuzawa, K.; Usuda, N.; Watashi, K.; Hishiki, T.; Zayas, M.; Bartenschlager, R.; Wakita, T.; Hijikata, M.; Shimotohno, K. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 2007, 9, 1089–1097. [Google Scholar] [CrossRef]
- Huang, H.; Sun, F.; Owen, D.M.; Li, W.; Chen, Y.; Gale, M.; Ye, J. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl. Acad. Sci. USA 2007, 104, 5848–5853. [Google Scholar] [CrossRef] [Green Version]
- Scarselli, E.; Ansuini, H.; Cerino, R.; Roccasecca, R.M.; Acali, S.; Filocamo, G.; Traboni, C.; Nicosia, A.; Cortese, R.; Vitelli, A. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002, 21, 5017–5025. [Google Scholar] [CrossRef] [Green Version]
- Agnello, V.; Abel, G.; Elfahal, M.; Knight, G.B.; Zhang, Q.X. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc. Natl. Acad. Sci. USA 1999, 96, 12766–12771. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamamoto, S.; Fukuhara, T.; Ono, C.; Uemura, K.; Kawachi, Y.; Shiokawa, M.; Mori, H.; Wada, M.; Shima, R.; Okamoto, T.; et al. Lipoprotein receptors redundantly participate in entry of hepatitis C virus. PLoS Pathog. 2016, 12, e1005610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dao Thi, V.L.; Granier, C.; Zeisel, M.B.; Guérin, M.; Mancip, J.; Granio, O.; Penin, F.; Lavillette, D.; Bartenschlager, R.; Baumert, T.F.; et al. Characterization of hepatitis C virus particle subpopulations reveals multiple usage of the scavenger receptor BI for entry steps. J. Biol. Chem. 2012, 287, 31242–31257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Albecka, A.; Belouzard, S.; Op de Beeck, A.; Descamps, V.; Goueslain, L.; Bertrand-Michel, J.; Tercé, F.; Duverlie, G.; Rouillé, Y.; Dubuisson, J. Role of low-density lipoprotein receptor in the hepatitis C virus life cycle. Hepatology 2012, 55, 998–1007. [Google Scholar] [CrossRef]
- Owen, D.M.; Huang, H.; Ye, J.; Gale, M. Apolipoprotein E on hepatitis C virion facilitates infection through interaction with low-density lipoprotein receptor. Virology 2009, 394, 99–108. [Google Scholar] [CrossRef] [Green Version]
- Wünschmann, S.; Medh, J.D.; Klinzmann, D.; Schmidt, W.N.; Stapleton, J.T. Characterization of hepatitis C virus (HCV) and HCV E2 interactions with CD81 and the low-density lipoprotein receptor. J. Virol. 2000, 74, 10055–10062. [Google Scholar] [CrossRef] [Green Version]
- Chhetry, M.; Jialal, I. Lipid lowering drug therapy. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2020. [Google Scholar]
- Istvan, E.S.; Deisenhofer, J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science 2001, 292, 1160–1164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldstein, J.L.; Brown, M.S. Regulation of the mevalonate pathway. Nature 1990, 343, 425–430. [Google Scholar] [CrossRef]
- Ghonem, N.S.; Assis, D.N.; Boyer, J.L. Fibrates and cholestasis. Hepatology 2015, 62, 635–643. [Google Scholar] [CrossRef] [Green Version]
- Staels, B.; Dallongeville, J.; Auwerx, J.; Schoonjans, K.; Leitersdorf, E.; Fruchart, J.C. Mechanism of action of fibrates on lipid and lipoprotein metabolism. Circulation 1998, 98, 2088–2093. [Google Scholar] [CrossRef] [Green Version]
- Lagace, T.A. PCSK9 and LDLR degradation: Regulatory mechanisms in circulation and in cells. Curr. Opin. Lipidol. 2014, 25, 387–393. [Google Scholar] [CrossRef]
- Lagace, T.A.; Curtis, D.E.; Garuti, R.; McNutt, M.C.; Park, S.W.; Prather, H.B.; Anderson, N.N.; Ho, Y.K.; Hammer, R.E.; Horton, J.D. Secreted PCSK9 decreases the number of LDL receptors in hepatocytes and in livers of parabiotic mice. J. Clin. Investig. 2006, 116, 2995–3005. [Google Scholar] [CrossRef] [Green Version]
- Mullard, A. Cholesterol-lowering blockbuster candidates speed into Phase III trials. Nat. Rev. Drug. Discov. 2012, 11, 817–819. [Google Scholar] [CrossRef]
- Ye, J.; Wang, C.; Sumpter, R.; Brown, M.S.; Goldstein, J.L.; Gale, M. Disruption of hepatitis C virus RNA replication through inhibition of host protein geranylgeranylation. Proc. Natl. Acad. Sci. USA 2003, 100, 15865–15870. [Google Scholar] [CrossRef] [Green Version]
- Blanchet, M.; Le, Q.-T.; Seidah, N.G.; Labonté, P. Statins can exert dual, concentration dependent effects on HCV entry in vitro. Antivir. Res. 2016, 128, 43–48. [Google Scholar] [CrossRef]
- Wuestenberg, A.; Kah, J.; Singethan, K.; Sirma, H.; Keller, A.D.; Rosal, S.R.P.; Schrader, J.; Loscher, C.; Volz, T.; Bartenschlager, R.; et al. Matrix conditions and KLF2-dependent induction of heme oxygenase-1 modulate inhibition of HCV replication by fluvastatin. PLoS ONE 2014, 9, e96533. [Google Scholar] [CrossRef] [PubMed]
- Ikeda, M.; Abe, K.; Yamada, M.; Dansako, H.; Naka, K.; Kato, N. Different anti-HCV profiles of statins and their potential for combination therapy with interferon. Hepatology 2006, 44, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Delang, L.; Paeshuyse, J.; Vliegen, I.; Leyssen, P.; Obeid, S.; Durantel, D.; Zoulim, F.; Op de Beeck, A.; Neyts, J. Statins potentiate the in vitro anti-hepatitis C virus activity of selective hepatitis C virus inhibitors and delay or prevent resistance development. Hepatology 2009, 50, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Ramanathan, A.; Gusarova, V.; Stahl, N.; Gurnett-Bander, A.; Kyratsous, C.A. Alirocumab, a Therapeutic Human Antibody to PCSK9, Does Not Affect CD81 Levels or Hepatitis C Virus Entry and Replication into Hepatocytes. PLoS ONE 2016, 11, e0154498. [Google Scholar] [CrossRef] [Green Version]
- Sabile, A.; Perlemuter, G.; Bono, F.; Kohara, K.; Demaugre, F.; Kohara, M.; Matsuura, Y.; Miyamura, T.; Bréchot, C.; Barba, G. Hepatitis C virus core protein binds to apolipoprotein AII and its secretion is modulated by fibrates. Hepatology 1999, 30, 1064–1076. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Gao, Y.; Zhang, C.; Hu, H.; Guo, D.; Xu, Y.; Xu, Q.; Zhang, W.; Deng, S.; Lv, P.; et al. Quinolinate phosphoribosyltransferase is an antiviral host factor against hepatitis C virus infection. Sci. Rep. 2017, 7, 5876. [Google Scholar] [CrossRef]
- Bader, T.; Hughes, L.D.; Fazili, J.; Frost, B.; Dunnam, M.; Gonterman, A.; Madhoun, M.; Aston, C.E. A randomized controlled trial adding fluvastatin to peginterferon and ribavirin for naïve genotype 1 hepatitis C patients. J. Viral. Hepat. 2013, 20, 622–627. [Google Scholar] [CrossRef]
- Forde, K.-A.; Law, C.; O’Flynn, R.; Kaplan, D.-E. Do statins reduce hepatitis C RNA titers during routine clinical use? World J. Gastroenterol. 2009, 15, 5020–5027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butt, A.A.; Yan, P.; Bonilla, H.; Abou-Samra, A.-B.; Shaikh, O.S.; Simon, T.G.; Chung, R.T.; Rogal, S.S. ERCHIVES (Electronically Retrieved Cohort of HCV Infected Veterans) Study Team Effect of addition of statins to antiviral therapy in hepatitis C virus-infected persons: Results from ERCHIVES. Hepatology 2015, 62, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Atsukawa, M.; Tsubota, A.; Kondo, C.; Itokawa, N.; Narahara, Y.; Nakatsuka, K.; Hashimoto, S.; Fukuda, T.; Matsushita, Y.; Kidokoro, H.; et al. Combination of fluvastatin with pegylated interferon/ribavirin therapy reduces viral relapse in chronic hepatitis C infected with HCV genotype 1b. J. Gastroenterol. Hepatol. 2013, 28, 51–56. [Google Scholar] [CrossRef] [PubMed]
- Fujita, N.; Kaito, M.; Kai, M.; Sugimoto, R.; Tanaka, H.; Horiike, S.; Konishi, M.; Iwasa, M.; Watanabe, S.; Adachi, Y. Effects of bezafibrate in patients with chronic hepatitis C virus infection: Combination with interferon and ribavirin. J. Viral. Hepat. 2006, 13, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Knop, V.; Bergk, A.; Schlosser, B.; Thieringer, J.; van Bömmel, F.; Frost, N.; Kintscher, U.; Berg, T. Bezafibrate maintenance therapy in patients with advanced chronic hepatitis C. Eur. J. Gastroenterol. Hepatol. 2013, 25, 594–600. [Google Scholar] [CrossRef]
- Labonté, P.; Begley, S.; Guévin, C.; Asselin, M.-C.; Nassoury, N.; Mayer, G.; Prat, A.; Seidah, N.G. PCSK9 impedes hepatitis C virus infection in vitro and modulates liver CD81 expression. Hepatology 2009, 50, 17–24. [Google Scholar] [CrossRef]
- Evans, M.J.; von Hahn, T.; Tscherne, D.M.; Syder, A.J.; Panis, M.; Wölk, B.; Hatziioannou, T.; McKeating, J.A.; Bieniasz, P.D.; Rice, C.M. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 2007, 446, 801–805. [Google Scholar] [CrossRef] [PubMed]
- Gunesch, A.P.; Zapatero-Belinchón, F.J.; Pinkert, L.; Steinmann, E.; Manns, M.P.; Schneider, G.; Pietschmann, T.; Brönstrup, M.; von Hahn, T. Filovirus Antiviral Activity of Cationic Amphiphilic Drugs Is Associated with Lipophilicity and Ability To Induce Phospholipidosis. Antimicrob. Agents Chemother. 2020, 64, e00143-20. [Google Scholar] [CrossRef]
- Bartosch, B.; Dubuisson, J.; Cosset, F.-L. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 2003, 197, 633–642. [Google Scholar] [CrossRef]
- Pietschmann, T.; Kaul, A.; Koutsoudakis, G.; Shavinskaya, A.; Kallis, S.; Steinmann, E.; Abid, K.; Negro, F.; Dreux, M.; Cosset, F.-L.; et al. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. USA 2006, 103, 7408–7413. [Google Scholar] [CrossRef] [Green Version]
- Koutsoudakis, G.; Kaul, A.; Steinmann, E.; Kallis, S.; Lohmann, V.; Pietschmann, T.; Bartenschlager, R. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 2006, 80, 5308–5320. [Google Scholar] [CrossRef] [Green Version]
- Sakuma, T.; Nishikawa, A.; Kume, S.; Chayama, K.; Yamamoto, T. Multiplex genome engineering in human cells using all-in-one CRISPR/Cas9 vector system. Sci. Rep. 2014, 4, 5400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheldon, J.; Beach, N.M.; Moreno, E.; Gallego, I.; Piñeiro, D.; Martínez-Salas, E.; Gregori, J.; Quer, J.; Esteban, J.I.; Rice, C.M.; et al. Increased replicative fitness can lead to decreased drug sensitivity of hepatitis C virus. J. Virol. 2014, 88, 12098–12111. [Google Scholar] [CrossRef] [Green Version]
- Ciesek, S.; Westhaus, S.; Wicht, M.; Wappler, I.; Henschen, S.; Sarrazin, C.; Hamdi, N.; Abdelaziz, A.I.; Strassburg, C.P.; Wedemeyer, H.; et al. Impact of intra- and interspecies variation of occludin on its function as coreceptor for authentic hepatitis C virus particles. J. Virol. 2011, 85, 7613–7621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleine, M.; Riemer, M.; Krech, T.; DeTemple, D.; Jäger, M.D.; Lehner, F.; Manns, M.P.; Klempnauer, J.; Borlak, J.; Bektas, H.; et al. Explanted diseased livers—A possible source of metabolic competent primary human hepatocytes. PLoS ONE 2014, 9, e101386. [Google Scholar] [CrossRef] [PubMed]
- Perales, C.; Beach, N.M.; Gallego, I.; Soria, M.E.; Quer, J.; Esteban, J.I.; Rice, C.; Domingo, E.; Sheldon, J. Response of hepatitis C virus to long-term passage in the presence of alpha interferon: Multiple mutations and a common phenotype. J. Virol. 2013, 87, 7593–7607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carpentier, A.; Sheldon, J.; Vondran, F.W.R.; Brown, R.J.; Pietschmann, T. Efficient acute and chronic infection of stem cell-derived hepatocytes by hepatitis C virus. Gut 2020, 69, 1659–1666. [Google Scholar] [CrossRef] [Green Version]
- Costa, R.; Todt, D.; Zapatero-Belinchón, F.; Schenk, C.; Anastasiou, O.E.; Walker, A.; Hertel, B.; Timmer, L.; Bojkova, D.; Ruckert, M.; et al. SEC14L2, a lipid-binding protein, regulates HCV replication in culture with inter- and intra-genotype variations. J. Hepatol. 2019, 70, 603–614. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M. UGENE team Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef] [Green Version]
- Vieyres, G.; Pietschmann, T. Entry and replication of recombinant hepatitis C viruses in cell culture. Methods 2013, 59, 233–248. [Google Scholar] [CrossRef]
- Zapatero-Belinchón, F.J.; Dietzel, E.; Dolnik, O.; Döhner, K.; Costa, R.; Hertel, B.; Veselkova, B.; Kirui, J.; Klintworth, A.; Manns, M.P.; et al. Characterization of the Filovirus-Resistant Cell Line SH-SY5Y Reveals Redundant Role of Cell Surface Entry Factors. Viruses 2019, 11, 275. [Google Scholar] [CrossRef] [Green Version]
- Bruening, J.; Lasswitz, L.; Banse, P.; Kahl, S.; Marinach, C.; Vondran, F.W.; Kaderali, L.; Silvie, O.; Pietschmann, T.; Meissner, F.; et al. Hepatitis C virus enters liver cells using the CD81 receptor complex proteins calpain-5 and CBLB. PLoS Pathog. 2018, 14, e1007111. [Google Scholar] [CrossRef]
- Riss, T.L.; Moravec, R.A.; Niles, A.L.; Benink, H.A.; Worzella, T.J.; Minor, L. Cell Viability Assays. In Assay Guidance Manual; Sittampalam, G.S., Coussens, N.P., Nelson, H., Arkin, M., Auld, D., Austin, C., Bejcek, B., Glicksman, M., Inglese, J., Iversen, P.W., et al., Eds.; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2004. [Google Scholar]
- Maasoumy, B.; Vermehren, J.; Welker, M.-W.; Bremer, B.; Perner, D.; Höner Zu Siederdissen, C.; Deterding, K.; Lehmann, P.; Cloherty, G.; Reinhardt, B.; et al. Clinical value of on-treatment HCV RNA levels during different sofosbuvir-based antiviral regimens. J. Hepatol. 2016, 65, 473–482. [Google Scholar] [CrossRef] [PubMed]
- Pileri, P.; Uematsu, Y.; Campagnoli, S.; Galli, G.; Falugi, F.; Petracca, R.; Weiner, A.J.; Houghton, M.; Rosa, D.; Grandi, G.; et al. Binding of hepatitis C virus to CD81. Science 1998, 282, 938–941. [Google Scholar] [CrossRef]
- Knopf, H.C.; Busch, M.A.; Du, Y.; Truthmann, J.; Schienkiewitz, A.; Scheidt-Nave, C. Changes in the prevalence of statin use in Germany—Findings from national health interview and examination surveys 1997–1999 and 2008–2011. Z. Evid. Fortbild. Qual. Gesundhwes. 2017, 122, 22–31. [Google Scholar] [CrossRef] [PubMed]
- Westhaus, S.; Deest, M.; Nguyen, A.T.X.; Stanke, F.; Heckl, D.; Costa, R.; Schambach, A.; Manns, M.P.; Berg, T.; Vondran, F.W.R.; et al. Scavenger receptor class B member 1 (SCARB1) variants modulate hepatitis C virus replication cycle and viral load. J. Hepatol. 2017, 67, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Steba, G.S.; Koekkoek, S.M.; Tanck, M.W.T.; Vanhommerig, J.W.; van der Meer, J.T.M.; Kwa, D.; Brinkman, K.; Prins, M.; Berkhout, B.; Pollakis, G.; et al. MOSAIC (MSM observational Study of Acute infection with Hepatitis C) Study Group and the ACS (Amsterdam Cohort Studies) SNP rs688 within the low-density lipoprotein receptor (LDL-R) gene associates with HCV susceptibility. Liver Int. 2019, 39, 463–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Witteveldt, J.; Evans, M.J.; Bitzegeio, J.; Koutsoudakis, G.; Owsianka, A.M.; Angus, A.G.N.; Keck, Z.-Y.; Foung, S.K.H.; Pietschmann, T.; Rice, C.M.; et al. CD81 is dispensable for hepatitis C virus cell-to-cell transmission in hepatoma cells. J. Gen. Virol. 2009, 90, 48–58. [Google Scholar] [CrossRef]
- Bitzegeio, J.; Bankwitz, D.; Hueging, K.; Haid, S.; Brohm, C.; Zeisel, M.B.; Herrmann, E.; Iken, M.; Ott, M.; Baumert, T.F.; et al. Adaptation of hepatitis C virus to mouse CD81 permits infection of mouse cells in the absence of human entry factors. PLoS Pathog. 2010, 6, e1000978. [Google Scholar] [CrossRef] [Green Version]
- Janicko, M.; Drazilova, S.; Pella, D.; Fedacko, J.; Jarcuska, P. Pleiotropic effects of statins in the diseases of the liver. World J. Gastroenterol. 2016, 22, 6201–6213. [Google Scholar] [CrossRef]
- Liao, J.K.; Laufs, U. Pleiotropic effects of statins. Annu. Rev. Pharmacol. Toxicol. 2005, 45, 89–118. [Google Scholar] [CrossRef] [Green Version]
- Björkhem-Bergman, L.; Lindh, J.D.; Bergman, P. What is a relevant statin concentration in cell experiments claiming pleiotropic effects? Br. J. Clin. Pharmacol. 2011, 72, 164–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adorni, M.P.; Cipollari, E.; Favari, E.; Zanotti, I.; Zimetti, F.; Corsini, A.; Ricci, C.; Bernini, F.; Ferri, N. Inhibitory effect of PCSK9 on Abca1 protein expression and cholesterol efflux in macrophages. Atherosclerosis 2017, 256, 1–6. [Google Scholar] [CrossRef]
- Martin, G.; Schoonjans, K.; Lefebvre, A.M.; Staels, B.; Auwerx, J. Coordinate regulation of the expression of the fatty acid transport protein and acyl-CoA synthetase genes by PPARalpha and PPARgamma activators. J. Biol. Chem. 1997, 272, 28210–28217. [Google Scholar] [CrossRef] [Green Version]
- Lamb, R.G.; Koch, J.C.; Bush, S.R. An enzymatic explanation of the differential effects of oleate and gemfibrozil on cultured hepatocyte triacylglycerol and phosphatidylcholine biosynthesis and secretion. Biochim. Biophys. Acta BBA Lipids Lipid Metab. 1993, 1165, 299–305. [Google Scholar] [CrossRef]
- Maillard, P.; Walic, M.; Meuleman, P.; Roohvand, F.; Huby, T.; Le Goff, W.; Leroux-Roels, G.; Pécheur, E.-I.; Budkowska, A. Lipoprotein lipase inhibits hepatitis C virus (HCV) infection by blocking virus cell entry. PLoS ONE 2011, 6, e26637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pose, E.; Trebicka, J.; Mookerjee, R.P.; Angeli, P.; Ginès, P. Statins: Old drugs as new therapy for liver diseases? J. Hepatol. 2019, 70, 194–202. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zapatero-Belinchón, F.J.; Ötjengerdes, R.; Sheldon, J.; Schulte, B.; Carriquí-Madroñal, B.; Brogden, G.; Arroyo-Fernández, L.M.; Vondran, F.W.R.; Maasoumy, B.; von Hahn, T.; et al. Interdependent Impact of Lipoprotein Receptors and Lipid-Lowering Drugs on HCV Infectivity. Cells 2021, 10, 1626. https://doi.org/10.3390/cells10071626
Zapatero-Belinchón FJ, Ötjengerdes R, Sheldon J, Schulte B, Carriquí-Madroñal B, Brogden G, Arroyo-Fernández LM, Vondran FWR, Maasoumy B, von Hahn T, et al. Interdependent Impact of Lipoprotein Receptors and Lipid-Lowering Drugs on HCV Infectivity. Cells. 2021; 10(7):1626. https://doi.org/10.3390/cells10071626
Chicago/Turabian StyleZapatero-Belinchón, Francisco J., Rina Ötjengerdes, Julie Sheldon, Benjamin Schulte, Belén Carriquí-Madroñal, Graham Brogden, Laura M. Arroyo-Fernández, Florian W. R. Vondran, Benjamin Maasoumy, Thomas von Hahn, and et al. 2021. "Interdependent Impact of Lipoprotein Receptors and Lipid-Lowering Drugs on HCV Infectivity" Cells 10, no. 7: 1626. https://doi.org/10.3390/cells10071626
APA StyleZapatero-Belinchón, F. J., Ötjengerdes, R., Sheldon, J., Schulte, B., Carriquí-Madroñal, B., Brogden, G., Arroyo-Fernández, L. M., Vondran, F. W. R., Maasoumy, B., von Hahn, T., & Gerold, G. (2021). Interdependent Impact of Lipoprotein Receptors and Lipid-Lowering Drugs on HCV Infectivity. Cells, 10(7), 1626. https://doi.org/10.3390/cells10071626






