New Data on Organization and Spatial Localization of B-Chromosomes in Cell Nuclei of the Yellow-Necked Mouse Apodemus flavicollis
Abstract
1. Introduction
2. Materials and Methods
2.1. Specimens
2.2. Cell Cultures
2.3. Bone Marrow Suspension Preparations
2.4. Metaphase Chromosome Preparation and Karyotyping of Animals
2.5. DNA-Probe Generation from Pericentromeric Region of the Large Autosome of A. flavicollis Karyotype
2.6. Whole Chromosome Paints derived from the B-Chromosomes of A. flavicollis
2.7. Preparation of 3D-Preserved Interphase Nuclei for FISH Analysis
2.7.1. Suspension FISH (S-FISH)
2.7.2. 3D-FISH of Fibroblasts
2.7.3. 2D- and 3D-Microscopy
2.8. Evaluation
3. Results
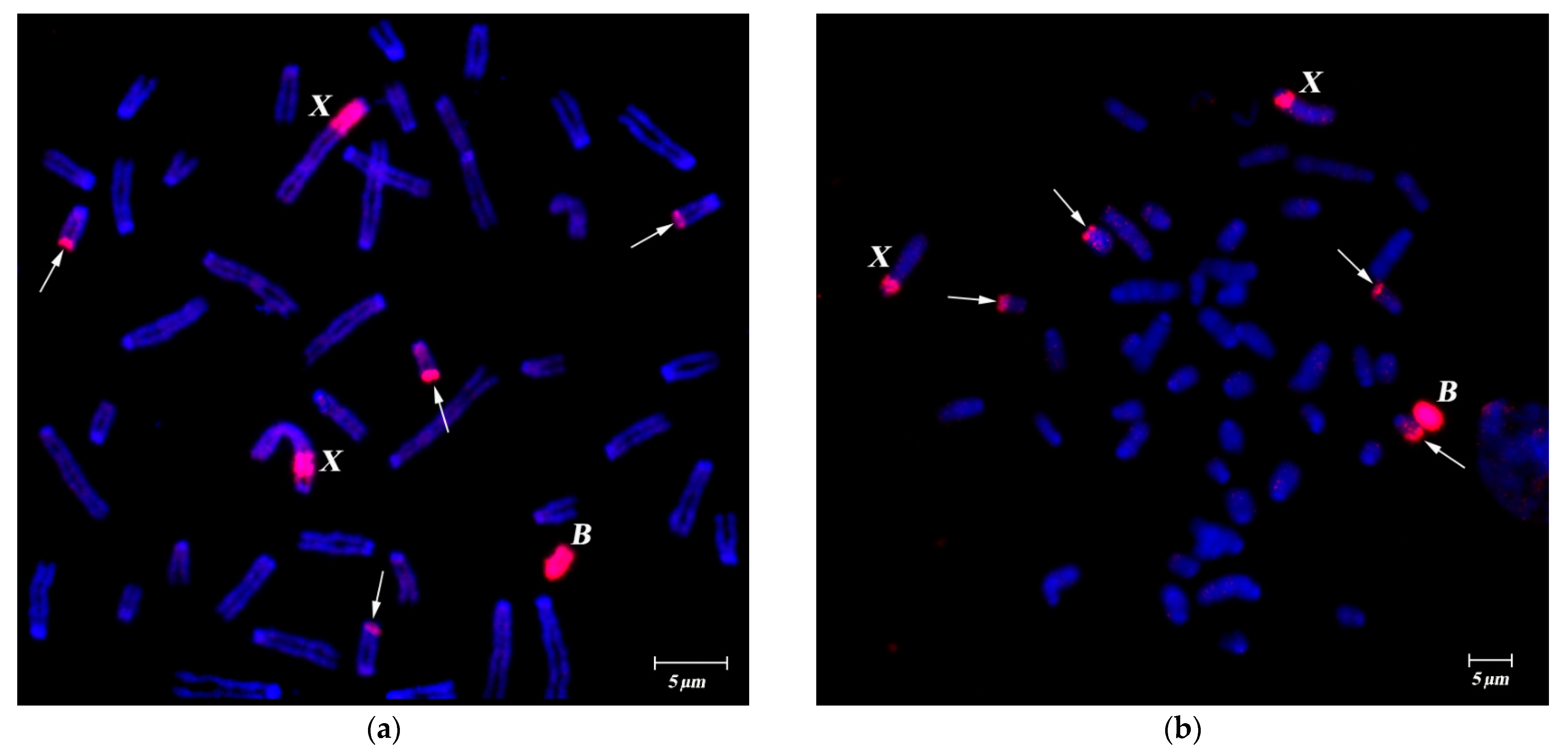
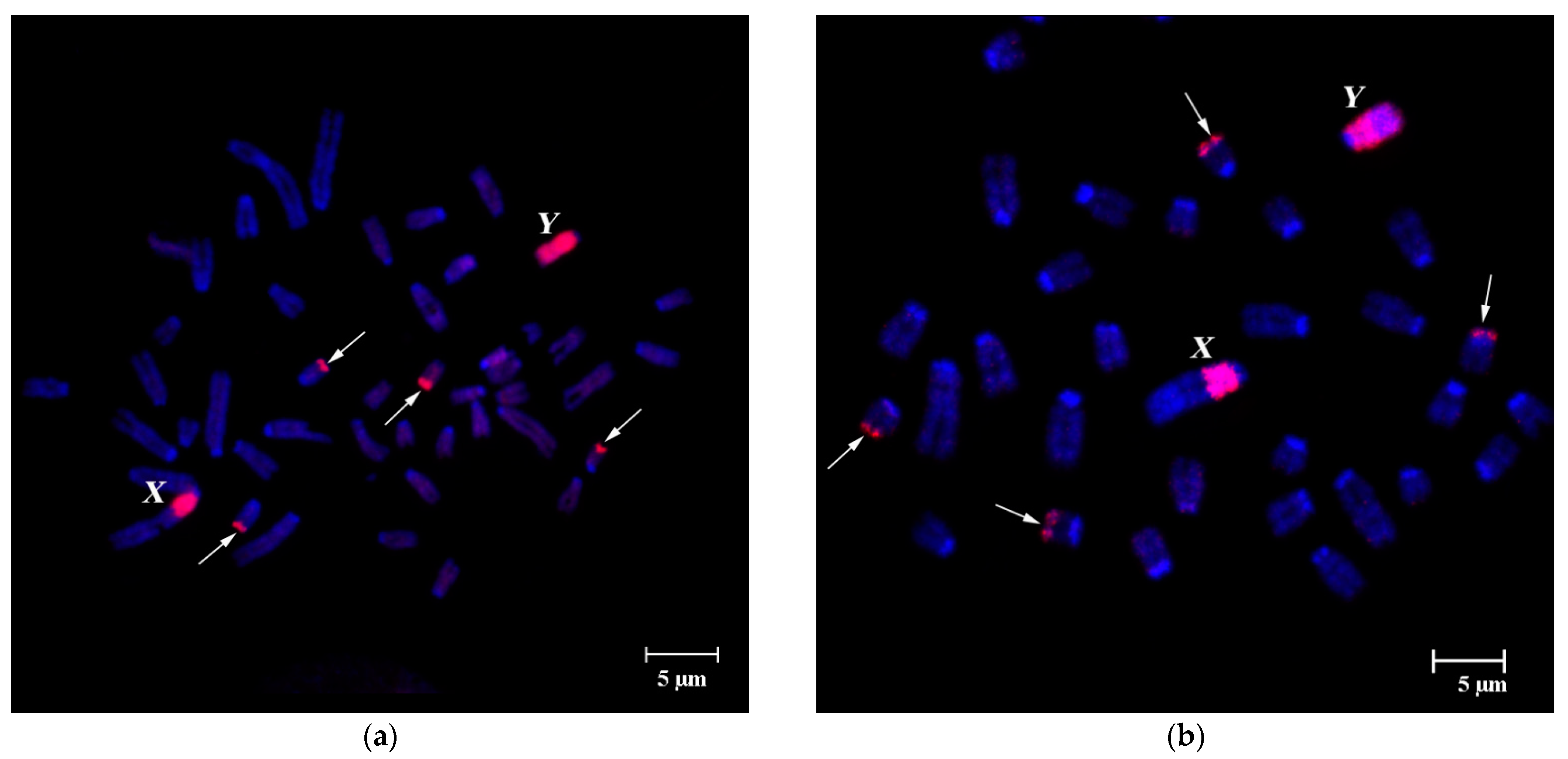
3.1. DNA Homology in Pericentromeric Regions of A- and B-Chromosomes in A. flavicollis
FISH of B-specific DNA Probes with A. flavicollis Chromosomes
3.2. FISH of DNA Probe Generated from the Pericentromeric Region of a Large Autosome of A. flavicollis
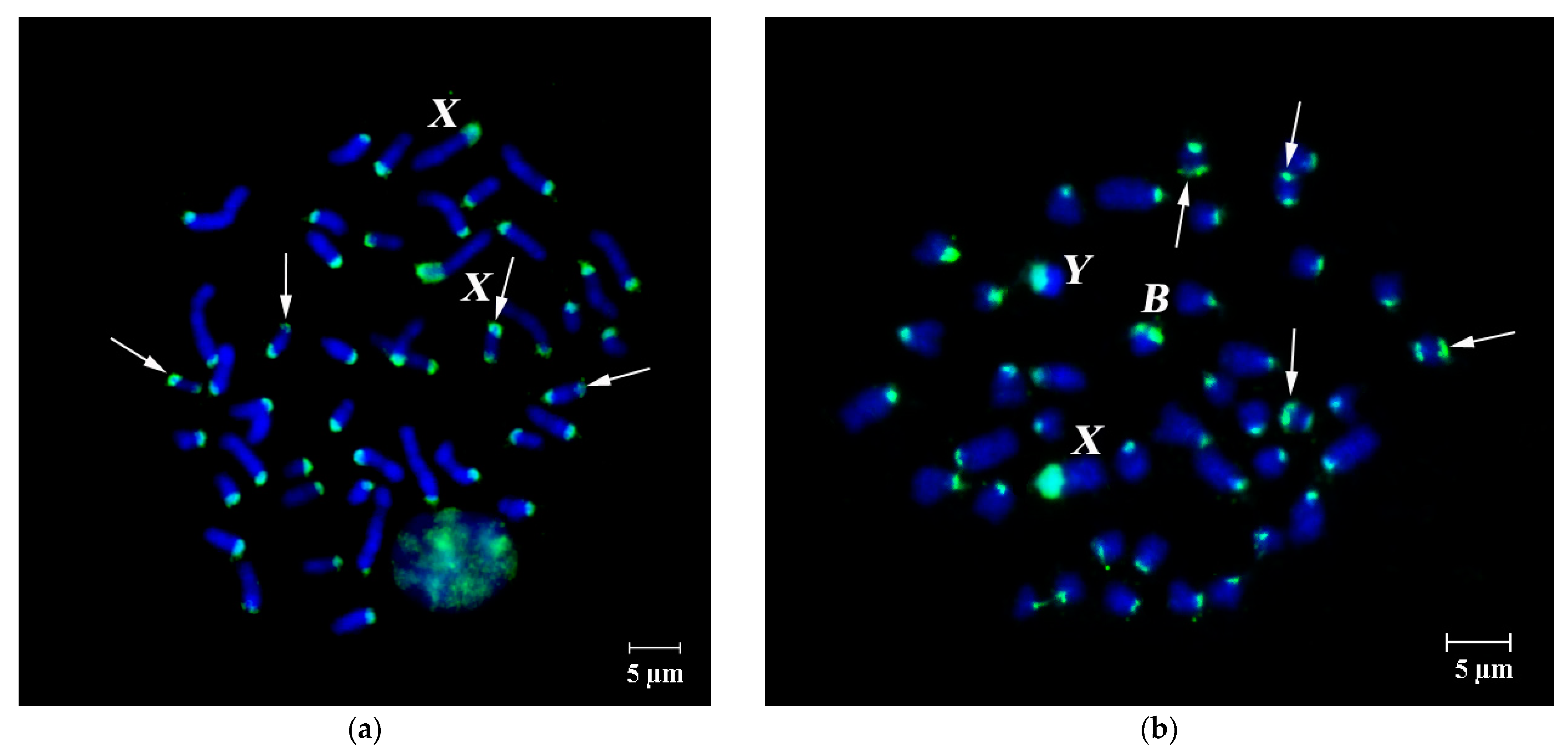
3.3. Dual-Color FISH of WCPBs and PCPAflaCEN Probes on Metaphase Plates of A. flavicollis
3.4. FISH Signal Distribution of WCPBs and PCPAflaCEN Probes in Interphase Nuclei of A. flavicollis
3.5. Spatial Organization of Interphase Nuclei in Bone Marrow Cells of A. flavicollis
3.6. Spatial Organization of Fibroblast Interphase Nuclei with Bs
4. Discussion
4.1. The Origin of Bs in A. flavicollis
4.2. Comparative Analysis of Bs Spatial Localization in Different Species of Mammals
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boveri, T. Uber Differenzierung der Zellkerne Wahrend der Furchung des Eies von Ascaris megalocephala. Anat. Anz. 1887, 2, 688–693. [Google Scholar]
- Satzinger, H. Theodor and Marcella Boveri: Chromosomes and Cytoplasm in Heredity and Development. Nat. Rev. Genet. 2008, 9, 231–238. [Google Scholar] [CrossRef]
- Pigozzi, M.I.; Solari, A.J. Germ Cell Restriction and Regular Transmission of an Accessory Chromosome that Mimics a Sex Body in the Zebra Finch, Taeniopygia Guttata. Chromosom. Res. Int. J. Mol. Supramol. Evol. Asp. Chromosom. Biol. 1998, 6, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Torgasheva, A.A.; Malinovskaya, L.P.; Zadesenets, K.S.; Karamysheva, T.V.; Kizilova, E.A.; Akberdina, E.A.; Pristyazhnyuk, I.E.; Shnaider, E.P.; Volodkina, V.A.; Saifitdinova, A.F.; et al. Germline-Restricted Chromosome ({GRC}) Is Widespread among Songbirds. Proc. Natl. Acad. Sci. USA 2019, 116, 11845–11850. [Google Scholar] [CrossRef] [PubMed]
- Valente, A.L.; Rummel, S.; Shriver, C.D.; Ellsworth, R.E. Sequence-Based Detection of Mutations in Cadherin 1 to Determine the Prevalence of Germline Mutations in Patients with Invasive Lobular Carcinoma of the Breast. Hered. Cancer Clin. Pract. 2014, 12, 17. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Singchat, W.; Jehangir, M.; Panthum, T.; Srikulnath, K. Consequence of Paradigm Shift with Repeat Landscapes in Reptiles: Powerful Facilitators of Chromosomal Rearrangements for Diversity and Evolution. Genes 2020, 11, 827. [Google Scholar] [CrossRef]
- Yang, F.; O’Brien, P.C.; Milne, B.S.; Graphodatsky, A.S.; Solanky, N.; Trifonov, V.; Rens, W.; Sargan, D.; Ferguson-Smith, M.A. A Complete Comparative Chromosome Map for the Dog, Red Fox, and Human and Its Integration with Canine Genetic Maps. Genomics 1999, 62, 189–202. [Google Scholar] [CrossRef]
- Camacho, J.P.; Sharbel, T.F.; Beukeboom, L.W. B-Chromosome Evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2000, 355, 163–178. [Google Scholar] [CrossRef]
- Rubtsov, N.B.; Karamysheva, T.V.; Andreenkova, O.V.; Bochkaerev, M.N.; Kartavtseva, I.V.; Roslik, G.V.; Borissov, Y.M. Comparative Analysis of Micro and Macro B Chromosomes in the Korean Field Mouse Apodemus Peninsulae (Rodentia, Murinae) Performed by Chromosome Microdissection and FISH. Cytogenet. Genome Res. 2004, 106, 289–294. [Google Scholar] [CrossRef]
- Itoh, Y.; Kampf, K.; Pigozzi, M.I.; Arnold, A.P. Molecular Cloning and Characterization of the Germline-Restricted Chromosome Sequence in the Zebra Finch. Chromosoma 2009, 118, 527–536. [Google Scholar] [CrossRef]
- Makunin, A.I.; Romanenko, S.A.; Beklemisheva, V.R.; Perelman, P.L.; Druzhkova, A.S.; Petrova, K.O.; Prokopov, D.Y.; Chernyaeva, E.N.; Johnson, J.L.; Kukekova, A.V.; et al. Sequencing of Supernumerary Chromosomes of Red Fox and Raccoon Dog Confirms a {Non-Random} Gene Acquisition by {B} Chromosomes. Genes 2018, 9, 405. [Google Scholar] [CrossRef]
- Matsubara, K.; Yamada, K.; Umemoto, S.; Tsuchiya, K.; Ikeda, N.; Nishida, C.; Chijiwa, T.; Moriwaki, K.; Matsuda, Y. Molecular Cloning and Characterization of the Repetitive {DNA} Sequences that Comprise the Constitutive Heterochromatin of the A and {B} Chromosomes of the Korean Field Mouse (Apodemus Peninsulae, Muridae, Rodentia). Chromosom. Res. Int. J. Mol. Supramol. Evol. Asp. Chromosom. Biol. 2008, 16, 1013–1026. [Google Scholar] [CrossRef]
- Trifonov, V.A.; Dementyeva, P.V.; Larkin, D.M.; O’Brien, P.C.M.; Perelman, P.L.; Yang, F.; Ferguson-Smith, M.A.; Graphodatsky, A.S. Transcription of a Protein-Coding Gene on {B} Chromosomes of the Siberian Roe Deer (\Textit{Capreolus Pygargus}). BMC Biol. 2013, 11, 90. [Google Scholar] [CrossRef] [PubMed]
- Biederman, M.K.; Nelson, M.M.; Asalone, K.C.; Pedersen, A.L.; Saldanha, C.J.; Bracht, J.R. Discovery of the First {Germline-Restricted} Gene by Subtractive Transcriptomic Analysis in the Zebra Finch, Taeniopygia Guttata. Curr. Biol. 2018, 28, 1620–1627.e5. [Google Scholar] [CrossRef] [PubMed]
- Dalla Benetta, E.; Akbari, O.S.; Ferree, P.M. Sequence Expression from Supernumerary {B} Chromosomes: Function or Fluff? Genes 2019, 10, 123. [Google Scholar] [CrossRef] [PubMed]
- Rubtsov, N.; Borisov, Y. Sequence Composition and Evolution of Mammalian B Chromosomes. Genes 2018, 9, 490. [Google Scholar] [CrossRef] [PubMed]
- Houben, A.; Jones, N.; Martins, C.; Trifonov, V. Evolution, Composition and Regulation of Supernumerary B Chromosomes. Genes 2019, 10, 161. [Google Scholar] [CrossRef] [PubMed]
- Vujošević, M.; Blagojević, J. B Chromosomes in Populations of Mammals. Cytogenet. Genome Res. 2004, 106, 247–256. [Google Scholar] [CrossRef] [PubMed]
- Vujošević, M.; Rajičić, M.; Blagojević, J. B Chromosomes in Populations of Mammals Revisited. Genes 2018, 9, 487. [Google Scholar] [CrossRef]
- Makunin, A.I.; Rajičić, M.; Karamysheva, T.V.; Romanenko, S.A.; Druzhkova, A.S.; Blagojević, J.; Vujošević, M.; Rubtsov, N.B.; Graphodatsky, A.S.; Trifonov, V.A. Low-Pass Single-Chromosome Sequencing of Human Small Supernumerary Marker Chromosomes (Ssmcs) and Apodemus B Chromosomes. Chromosoma 2018, 127, 301–311. [Google Scholar] [CrossRef]
- Roslik, G.V.; Kartavtseva, I.V. Polymorphism and Mosaicism of B Chromosome Number in Korean Field Mouse Apodemus Peninsulae (Rodentia) in the Russian Far East. Cell Tissue Biol. 2010, 4, 77–89. [Google Scholar] [CrossRef]
- Perazzo, G.X.; Noleto, R.B.; Vicari, M.R.; Gava, A.; Cestari, M.M. B Chromosome Polymorphism in South American Cichlid. Neotrop. Biodivers. 2018, 4, 3–9. [Google Scholar] [CrossRef]
- Kolomiets, O.L.; Borbiev, T.E.; Safronova, L.D.; Borisov, Y.M.; Bogdanov, Y.F. Synaptonemal Complex Analysis of B-Chromosome Behavior in Meiotic Prophase I in the East-Asiatic Mouse Apodemus Peninsulae (Muridae, Rodentia). Cytogenet. Genome Res. 1988, 48, 183–187. [Google Scholar] [CrossRef]
- Sadoni, N.; Langer, S.; Fauth, C.; Bernardi, G.; Cremer, T.; Turner, B.M.; Zink, D. Nuclear Organization of Mammalian Genomes. Polar Chromosome Territories Build Up Functionally Distinct Higher Order Compartments. J. Cell Biol. 1999, 146, 1211–1226. [Google Scholar] [CrossRef] [PubMed]
- Dietzel, S.; Schiebel, K.; Little, G.; Edelmann, P.; Rappold, G.A.; Eils, R.; Cremer, C.; Cremer, T. The {3D} Positioning of {ANT2} and {ANT3} Genes within Female {X} Chromosome Territories Correlates with Gene Activity. Exp. Cell Res. 1999, 252, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.B.; Shen, J.; Yokota, H. Size-Dependent Positioning of Human Chromosomes in Interphase Nuclei. Biophys. J. 2000, 79, 184–190. [Google Scholar] [CrossRef]
- Boyle, S.; Gilchrist, S.; Bridger, J.M.; Mahy, N.L.; Ellis, J.A.; Bickmore, W.A. The Spatial Organization of Human Chromosomes within the Nuclei of Normal and Emerin-Mutant Cells. Hum. Mol. Genet. 2001, 10, 211–219. [Google Scholar] [CrossRef]
- Battulin, N.; Fishman, V.S.; Mazur, A.M.; Pomaznoy, M.; Khabarova, A.A.; Afonnikov, D.A.; Prokhortchouk, E.B.; Serov, O.L. Comparison of the Three-Dimensional Organization of Sperm and Fibroblast Genomes Using the Hi-C Approach. Genome Biol. 2015, 16, 77. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.O.J.; Oudelaar, A.M.; Higgs, D.R.; Hughes, J.R. How Best to Identify Chromosomal Interactions: A Comparison of Approaches. Nat. Methods 2017, 14, 125–134. [Google Scholar] [CrossRef]
- Dixon, J.R.; Selvaraj, S.; Yue, F.; Kim, A.; Li, Y.; Shen, Y.; Hu, M.; Liu, J.S.; Ren, B. Topological Domains in Mammalian Genomes Identified by Analysis of Chromatin Interactions. Nature 2012, 485, 376–380. [Google Scholar] [CrossRef] [PubMed]
- Makunin, A.I.; Kichigin, I.G.; Larkin, D.M.; O’Brien, P.C.M.; Ferguson-Smith, M.A.; Yang, F.; Proskuryakova, A.A.; Vorobieva, N.V.; Chernyaeva, E.N.; O’Brien, S.J.; et al. Contrasting Origin of B Chromosomes in Two Cervids (Siberian Roe Deer and Grey Brocket Deer) Unravelled by Chromosome-Specific DNA Sequencing. BMC Genom. 2016, 17, 618. [Google Scholar] [CrossRef] [PubMed]
- Bugarski-Stanojević, V.; Stamenković, G.; Blagojević, J.; Liehr, T.; Kosyakova, N.; Rajičić, M.; Vujošević, M. Exploring Supernumeraries—A New Marker for Screening of {B-Chromosomes} Presence in the Yellow Necked Mouse Apodemus flavicollis. PLoS ONE 2016, 11, e0160946. [Google Scholar] [CrossRef]
- Rubtsov, N.B.; Kartavtseva, I.V.; Roslik, G.V.; Karamysheva, T.V.; Pavlenko, M.V.; Iwasa, M.A.; Koh, H.S. Features of the (B) Chromosome in Korean Field Mouse Apodemus Peninsulae (Thomas, 1906) from Transbaikalia and the Far East Identified by the {FISH} Method. Russ. J. Genet. 2015, 51, 278–288. [Google Scholar] [CrossRef]
- Silva, D.M.Z.d.A.; Pansonato-Alves, J.C.; Utsunomia, R.; Araya-Jaime, C.; Ruiz-Ruano, F.J.; Daniel, S.N.; Hashimoto, D.T.; Oliveira, C.; Porto-Foresti, F.; Foresti, F.; et al. Delimiting the Origin of a (B) Chromosome by {FISH} Mapping, Chromosome Painting and {DNA} Sequence Analysis in Astyanax Paranae (Teleostei, Characiformes). PLoS ONE 2014, 9, e94896. [Google Scholar] [CrossRef]
- Martis, M.M.; Klemme, S.; Banaei-Moghaddam, A.M.; Blattner, F.R.; Macas, J.; Schmutzer, T.; Scholz, U.; Gundlach, H.; Wicker, T.; Šimková, H.; et al. Selfish Supernumerary Chromosome Reveals Its Origin as a Mosaic of Host Genome and Organellar Sequences. Proc. Natl. Acad. Sci. USA 2012, 109, 13343–13346. [Google Scholar] [CrossRef] [PubMed]
- Hirning, U.; Schulz, W.A.; Just, W.; Adolph, S.; Vogel, W. A Comparative Study of the Heterochromatin of Apodemus Sylvaticus and Apodemus Flavicollis. Chromosoma 1989, 98, 450–455. [Google Scholar] [CrossRef]
- Karamysheva, T.V.; Andreenkova, O.V.; Bochkaerev, M.N.; Borissov, Y.M.; Bogdanchikova, N.; Borodin, P.M.; Rubtsov, N.B. (B) Chromosomes of Korean Field Mouse {Apodemus Peninsulae} (Rodentia, Murinae) Analysed by Microdissection and (FISH). Cytogenet. Genome Res. 2002, 96, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Karamysheva, T.V.; Torgasheva, A.A.; Yefremov, Y.R.; Bogomolov, A.G.; Liehr, T.; Borodin, P.M.; Rubtsov, N.B. Spatial Organization of Fibroblast and Spermatocyte Nuclei with Different B-Chromosome Content in Korean Field Mouse, Apodemus Peninsulae (Rodentia, Muridae). Genome 2017, 60, 815–824. [Google Scholar] [CrossRef] [PubMed]
- Wójcik, J.M.; Wójcik, A.M.; Macholán, M.; Piálek, J.; Zima, J. The Mammalian Model for Population Studies of (B) Chromosomes: The Wood Mouse (Apodemus). Cytogenet. Genome Res. 2004, 106, 264–270. [Google Scholar] [CrossRef]
- Rajičić, M.; Romanenko, S.A.; Karamysheva, T.V.; Blagojević, J.; Adna\djević, T.; Budinski, I.; Bogdanov, A.S.; Trifonov, V.A.; Rubtsov, N.B.; Vujošević, M. The Origin of (B) Chromosomes in Yellow-Necked Mice (Apodemus Flavicollis)-Break Rules but Keep Playing the Game. PLoS ONE 2017, 12, e0172704. [Google Scholar] [CrossRef]
- Rubtsov, N.B.; Karamysheva, T.V.; Bogdanov, A.S.; Likhoshvay, T.V.; Kartavtseva, I. V Comparative (FISH) Analysis of C-Positive Regions of Chromosomes of Wood Mice (Rodentia, Muridae, Sylvaemus). Russ. J. Genet. 2011, 47, 1096. [Google Scholar] [CrossRef]
- Stanyon, R.; Galleni, L. A Rapid Fibroblast Culture Technique for High Resolution Karyotypes. Boll. Zool. 1991, 58, 81–83. [Google Scholar] [CrossRef]
- Romanenko, S.A.; Biltueva, L.S.; Serdyukova, N.A.; Kulemzina, A.I.; Beklemisheva, V.R.; Gladkikh, O.L.; Lemskaya, N.A.; Interesova, E.A.; Korentovich, M.A.; Vorobieva, N.V.; et al. Segmental Paleotetraploidy Revealed in Sterlet (Acipenser Ruthenus) Genome by Chromosome Painting. Mol. Cytogenet. 2015, 8, 1. [Google Scholar] [CrossRef]
- Baker, R.J.; Haiduk, M.W.; Robbins, L.W.; Cadena, A.; Koop, B.F. Chromosomal Studies of South American Bats and Their Systematic Implications. Mamm. Biol. S. Am. 1982, 4, 303–327. [Google Scholar]
- Yang, F.; Trifonov, V.; Ng, B.L.; Kosyakova, N.; Carter, N.P. Generation of Paint Probes from {Flow-Sorted} and Microdissected Chromosomes. In Fluorescence In Situ Hybridization (FISH) Application Guide; Springer Protocols Handbooks; Liehr, T., Ed.; Springer: Berlin/Heidelberg, Germany, 2017; pp. 63–79. [Google Scholar]
- Karamysheva, T.B.; Matveeva, V.G.; Shorina, A.P.; Rubtsov, N.B. Clinical and Molecular Cytogenetic Analysis of a Rare Case of Mosaicism for Partial Trisomy 3p and Partial Trisomy 10q in Humans. Russ. J. Genet. 2001, 37, 666–670. [Google Scholar] [CrossRef]
- Rubtsov, N.; Senger, G.; Neumann, A.; Kelbova, C.; Junker, K.; Beensen, V.; Claussen, U.; Rubtsov, N.; Kuzcera, H. Interstitial Deletion of Chromosome 6q: Precise Definition of the Breakpoints by Microdissection, DNA Amplification, and Reverse Painting. Hum. Genet. 1996, 97, 705–709. [Google Scholar] [CrossRef]
- Rubtsov, N.B.; Karamisheva, T.V.; Astakhova, N.M.; Liehr, T.; Claussen, U.; Zhdanova, N.S. Zoo-FISH with Region-Specific Paints for Mink Chromosome 5q: Delineation of Inter- and Intrachromosomal Rearrangements in Human, Pig, and Fox. Cytogenet. Genome Res. 2000, 90, 268–270. [Google Scholar] [CrossRef] [PubMed]
- Klein, E.; Manvelyan, M.; Simonyan, I.; Hamid, A.B.; Guilherme, R.S.; Liehr, T.; Karamysheva, T. Centromeric Association of Small Supernumerary Marker Chromosomes with Their Sister-Chromosomes Detected by Three Dimensional Molecular Cytogenetics. Mol. Cytogenet. 2012, 5, 15. [Google Scholar] [CrossRef] [PubMed]
- Manvelyan, M.; Hunstig, F.; Mrasek, K.; Bhatt, S.; Pellestor, F.; Weise, A.; Liehr, T. Position of Chromosomes 18, 19, 21 and 22 in 3D-Preserved Interphase Nuclei of Human and Gorilla and White Hand Gibbon. Mol. Cytogenet. 2008, 1, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Iourov, I.Y.; Liehr, T.; Vorsanova, S.G.; Yurov, Y.B. Interphase Chromosome-Specific Multicolor Banding (ICS-MCB): A New Tool for Analysis of Interphase Chromosomes in Their Integrity. Biomol. Eng. 2007, 24, 415–417. [Google Scholar] [CrossRef]
- Steinhaeuser, U.; Starke, H.; Nietzel, A.; Lindenau, J.; Ullmann, P.; Claussen, U.; Liehr, T. Suspension (S)-FISH, a New Technique for Interphase Nuclei. J. Histochem. Cytochem. Off. J. Histochem. Soc. 2002, 50, 1697–1698. [Google Scholar] [CrossRef] [PubMed]
- Solovei, I.; Cavallo, A.; Schermelleh, L.; Jaunin, F.; Scasselati, C.; Cmarko, D.; Cremer, C.; Fakan, S.; Cremer, T. Spatial Preservation of Nuclear Chromatin Architecture during Three-Dimensional Fluorescence In Situ Hybridization (3D-FISH). Exp. Cell Res. 2002, 276, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Cremer, T.; Cremer, C. Chromosome Territories, Nuclear Architecture and Gene Regulation in Mammalian Cells. Nat. Rev. Genet. 2001, 2, 292–301. [Google Scholar] [CrossRef]
- Bolzer, A.; Kreth, G.; Solovei, I.; Koehler, D.; Saracoglu, K.; Fauth, C.; Müller, S.; Eils, R.; Cremer, C.; Speicher, M.R.; et al. Three-dimensional Maps of All Chromosomes in Human Male Fibroblast Nuclei and Prometaphase Rosettes. PLoS Biol. 2005, 3, e157. [Google Scholar] [CrossRef]
- Quinodoz, S.A.; Ollikainen, N.; Tabak, B.; Palla, A.; Schmidt, J.M.; Detmar, E.; Lai, M.M.; Shishkin, A.A.; Bhat, P.; Takei, Y.; et al. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell 2018, 174, 744–757.e24. [Google Scholar] [CrossRef] [PubMed]
- Rubtsov, N.B.; Karamysheva, T.V.; Bogdanov, A.S.; Kartavtseva, I.V.; Bochkarev, M.N.; Iwasa, M.A. Comparative Analysis of (DNA) Homology in Pericentric Regions of Chromosomes of Wood Mice from Genera Apodemus and Sylvaemus. Russ. J. Genet. 2015, 51, 1233–1242. [Google Scholar] [CrossRef]
- Rubtzov, N.B.; Borissov, Y.M.; Karamysheva, T.V.; Bochkarev, M.N. The Mechanisms of Formation and Evolution of (B) Chromosomes in Korean Field Mice Apodemus Peninsulae (Mammalia, Rodentia). Russ. J. Genet. 2009, 45, 389–396. [Google Scholar] [CrossRef]
- Chen, C.-P.; Lin, S.-P.; Lin, C.-C.; Li, Y.-C.; Chern, S.-R.; Chen, W.-M.; Lee, C.-C.; Hsieh, L.-J.; Wang, W. Perinatal Findings and Molecular Cytogenetic Analysis of De Novo Partial Trisomy 16q (16q22.1→qter) and Partial Monosomy 20q (20q13.3→qter). Prenat. Diagn. Publ. Affil. Int. Soc. Prenat. Diagn. 2005, 25, 112–118. [Google Scholar]
- Weise, A.; Liehr, T.; Efferth, T.; Kuechler, A.; Gebhart, E. Comparative (M-FISH) and (CGH) Analyses in Sensitive and Drug-Resistant Human T-Cell Acute Leukemia Cell Lines. Cytogenet. Genome Res. 2002, 98, 118–125. [Google Scholar] [CrossRef] [PubMed]
- Schröck, E.; Veldman, T.; Padilla-Nash, H.; Ning, Y.; Spurbeck, J.; Jalal, S.; Shaffer, L.G.; Papenhausen, P.; Kozma, C.; Phelan, M.C.; et al. Spectral Karyotyping Refines Cytogenetic Diagnostics of Constitutional Chromosomal Abnormalities. Hum. Genet. 1997, 101, 255–262. [Google Scholar] [CrossRef]
- Liehr, T.; Mrasek, K.; Kosyakova, N.; Ogilvie, C.; Vermeesch, J.; Trifonov, V.; Rubtsov, N. Small Supernumerary Marker Chromosomes (Ssmc) in Humans; Are There B Chromosomes Hidden among Them. Mol. Cytogenet. 2008, 1, 12. [Google Scholar] [CrossRef]
- Horvath, J.E. Lessons from the Human Genome: Transitions between Euchromatin and Heterochromatin. Hum. Mol. Genet. 2001, 10, 2215–2223. [Google Scholar] [CrossRef] [PubMed]
- Gerstel, D.U.; Burns, J.A. The Effect of the (NICOTIANA) (OTOPHORA) Genome on Chromosome Breakage and Megachromosomes in N. (TABACUMxN). (OTOPHORA) Derivatives. Genetics 1970, 66, 331–338. [Google Scholar] [CrossRef]
- Cowell, J.K. Double Minutes and Homogeneously Staining Regions: Gene Amplification in Mammalian Cells. Annu. Rev. Genet. 1982, 16, 21–59. [Google Scholar] [CrossRef] [PubMed]
- Stark, G.R.; Debatisse, M.; Giulotto, E.; Wahl, G.M. Recent Progress in Understanding Mechanisms of Mammalian (DNA) Amplification. Cell 1989, 57, 901–908. [Google Scholar] [CrossRef]
- L’Abbate, A.; Macchia, G.; D’Addabbo, P.; Lonoce, A.; Tolomeo, D.; Trombetta, D.; Kok, K.; Bartenhagen, C.; Whelan, C.W.; Palumbo, O.; et al. Genomic Organization and Evolution of Double Minutes/Homogeneously Staining Regions with (MYC) Amplification in Human Cancer. Nucleic Acids Res. 2014, 42, 9131–9145. [Google Scholar] [CrossRef]
- L’Abbate, A.; Tolomeo, D.; Cifola, I.; Severgnini, M.; Turchiano, A.; Augello, B.; Squeo, G.; D’Addabbo, P.; Traversa, D.; Daniele, G.; et al. (MYC-containing) Amplicons in Acute Myeloid Leukemia: Genomic Structures, Evolution, and Transcriptional Consequences. Leukemia 2018, 32, 2152–2166. [Google Scholar] [CrossRef] [PubMed]
- Raudsepp, T.; Chowdhary, B.P. The Eutherian Pseudoautosomal Region. Cytogenet. Genome Res. 2015, 147, 81–94. [Google Scholar] [CrossRef]
- Kociucka, B.; Sosnowski, J.; Kubiak, A.; Nowak, A.; Pawlak, P.; Szczerbal, I. Three-Dimensional Positioning of (B) Chromosomes in Fibroblast Nuclei of the Red Fox and the Chinese Raccoon Dog. Cytogenet. Genome Res. 2013, 139, 243–249. [Google Scholar] [CrossRef]
- Nie, W.; Wang, J.; Perelman, P.; Graphodatsky, A.S.; Yang, F. Comparative Chromosome Painting Defines the Karyotypic Relationships among the Domestic Dog, Chinese Raccoon Dog and Japanese Raccoon Dog. Chromosom. Res. Int. J. Mol. Supramol. Evol. Asp. Chromosom. Biol. 2003, 11, 735–740. [Google Scholar] [CrossRef]
- Croft, J.A.; Bridger, J.M.; Boyle, S.; Perry, P.; Teague, P.; Bickmore, W.A. Differences in the Localization and Morphology of Chromosomes in the Human Nucleus. J. Cell Biol. 1999, 145, 1119–1131. [Google Scholar] [CrossRef]
- Tanabe, H.; Küpper, K.; Ishida, T.; Neusser, M.; Mizusawa, H. Inter- and Intra-Specific Gene-Density-Correlated Radial Chromosome Territory Arrangements Are Conserved in Old World Monkeys. Cytogenet. Genome Res. 2005, 108, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Lanctôt, C.; Cheutin, T.; Cremer, M.; Cavalli, G.; Cremer, T. Dynamic Genome Architecture in the Nuclear Space: Regulation of Gene Expression in Three Dimensions. Nat. Rev. Genet. 2007, 8, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Solovei, I.; Thanisch, K.; Feodorova, Y. How to Rule the Nucleus: Divide et Impera. Curr. Opin. Cell Biol. 2016, 40, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Cremer, T.; Cremer, M. Chromosome Territories. Cold Spring Harb. Perspect. Biol. 2010, 2, a003889. [Google Scholar] [CrossRef] [PubMed]
- Fritz, A.J.; Barutcu, A.R.; Martin-Buley, L.; van Wijnen, A.J.; Zaidi, S.K.; Imbalzano, A.N.; Lian, J.B.; Stein, J.L.; Stein, G.S. Chromosomes at Work: Organization of Chromosome Territories in the Interphase Nucleus. J. Cell. Biochem. 2016, 117, 9–19. [Google Scholar] [CrossRef]
- Schooley, A.; Vollmer, B.; Antonin, W. Building a Nuclear Envelope at the End of Mitosis: Coordinating Membrane Reorganization, Nuclear Pore Complex Assembly, and Chromatin De-Condensation. Chromosoma 2012, 121, 539–554. [Google Scholar] [CrossRef]






| Species | Collection Number of Specimen, Sex | Karyotype | Cells for Metaphase Chromosome Preparation | Locality of Specimen Collection | Libraries | Remark |
|---|---|---|---|---|---|---|
| A. flavicollis | 3656, ♀ | 50,XX,+2B | Cells of bone marrow | Milošev Do, Serbia | used for FISH | |
| 3727, ♂ | 49,XY,+1B | Gonad tissue | Milošev Do, Serbia | WCP3727B | used for microdissection of B-like chromosomes | |
| 3795, ♂ | 51,XY,+3B | Cells of bone marrow | Bosilegrad, Serbia | used for FISH | ||
| 3854, ♀ | 51,XX,+3B | Cells of bone marrow | Misača, Serbia | used for FISH 3D-FISH | ||
| 3977, ♀ | 49,XX,+1B | Primary cultured fibroblasts | Petnica, Serbia | WCP3977B | used for FISH | |
| 3978, ♂ | 48,XY | Primary cultured fibroblasts | Orašac, Serbia | used for FISH | ||
| 3979, ♂ | 48,XY | Primary cultured fibroblasts | Orašac, Serbia | used for FISH | ||
| 3980 | 49,XX,+1B | Primary cultured fibroblasts | Orašac, Serbia | WCP3980B | used for microdissection of B-like chromosomes and for 3D-FISH | |
| 24448, ♂ | 49,XY,+1B | Cells of bone marrow | Novgorod region, Soletsky district, 60 km S from Veliky Novgorod, bank of Shelon river, Russia | used for S-FISH | ||
| 24123, ♂ | 48,XY | Penza region, Belinsky district, vicinities of Shiryaevo village, Russia | used for FISH | |||
| 24943, ♂ | 49,XY,+1B | Cells of bone marrow | Minsk region, vicinities of Sadovy train station, Republic of Belarus | used for FISH | ||
| 24944, ♂ | 49,XY,+1B | Cells of bone marrow | Minsk region, vicinities of Sadovy train station, Republic of Belarus | used for 3D-FISH | ||
| 24985, ♀ | 51,XX,+3B | Cells of bone marrow | Rostov-on-Don region, Donskoy national park, Russia | WCP24985Ba WCP24985Bb | used for microdissection of B-like chromosomes and for FISH | |
| 26368, ♀ | 49,XX,+1B | Cells of bone marrow | Ulyanovsk region, Kuzovatovsky district, vicinities of the Naleyka railway station, Russia | WCP26368B | used for FISH | |
| 1851 | 48,XX | Cells of bone marrow | Novgorod region, vicinities of the town of Malaya Vishera, Russia | PCPAflaCEN | used for FISH | |
| A. peninsulae | A. peninsulae # 5, ♂ | 54,XY,+6B | Cells of bone marrow | outskirts of Novosibirsk city, Russia | Ape_mB | used for 3D-FISH |
| A. peninsulae # 9, ♂ | 54,XY,+6B micro | Cells of bone marrow | Krasnoyarsk region, near Novokargino settlement, Russia | Ape_dB | used for 3D-FISH |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karamysheva, T.; Romanenko, S.; Makunin, A.; Rajičić, M.; Bogdanov, A.; Trifonov, V.; Blagojević, J.; Vujošević, M.; Orishchenko, K.; Rubtsov, N. New Data on Organization and Spatial Localization of B-Chromosomes in Cell Nuclei of the Yellow-Necked Mouse Apodemus flavicollis. Cells 2021, 10, 1819. https://doi.org/10.3390/cells10071819
Karamysheva T, Romanenko S, Makunin A, Rajičić M, Bogdanov A, Trifonov V, Blagojević J, Vujošević M, Orishchenko K, Rubtsov N. New Data on Organization and Spatial Localization of B-Chromosomes in Cell Nuclei of the Yellow-Necked Mouse Apodemus flavicollis. Cells. 2021; 10(7):1819. https://doi.org/10.3390/cells10071819
Chicago/Turabian StyleKaramysheva, Tatyana, Svetlana Romanenko, Alexey Makunin, Marija Rajičić, Alexey Bogdanov, Vladimir Trifonov, Jelena Blagojević, Mladen Vujošević, Konstantin Orishchenko, and Nikolay Rubtsov. 2021. "New Data on Organization and Spatial Localization of B-Chromosomes in Cell Nuclei of the Yellow-Necked Mouse Apodemus flavicollis" Cells 10, no. 7: 1819. https://doi.org/10.3390/cells10071819
APA StyleKaramysheva, T., Romanenko, S., Makunin, A., Rajičić, M., Bogdanov, A., Trifonov, V., Blagojević, J., Vujošević, M., Orishchenko, K., & Rubtsov, N. (2021). New Data on Organization and Spatial Localization of B-Chromosomes in Cell Nuclei of the Yellow-Necked Mouse Apodemus flavicollis. Cells, 10(7), 1819. https://doi.org/10.3390/cells10071819







