Activation of Nrf2 in Astrocytes Suppressed PD-Like Phenotypes via Antioxidant and Autophagy Pathways in Rat and Drosophila Models
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. 6-OHDA Rat PD Model and CDDO-Me Treatment
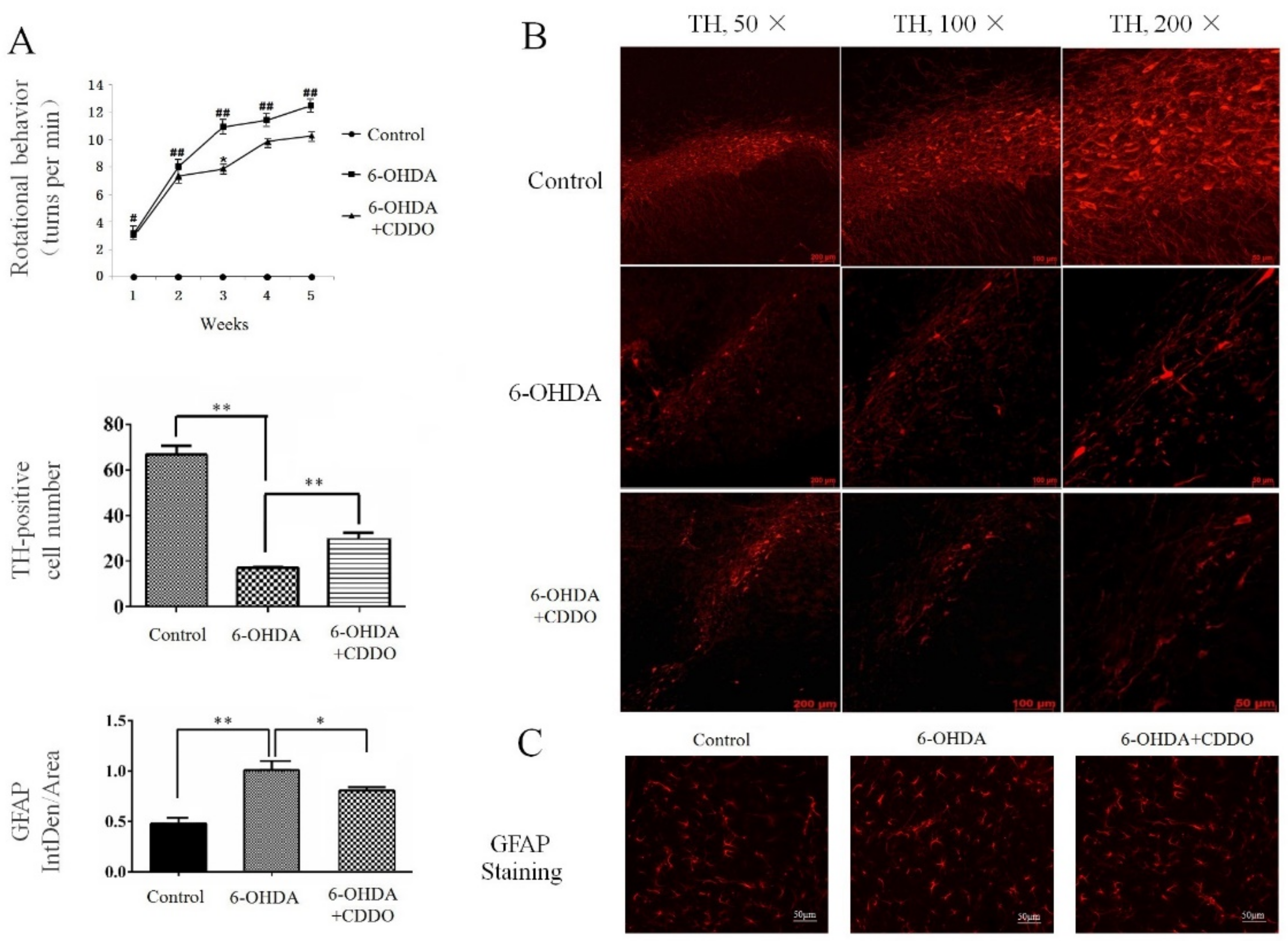
2.3. Apomorphine-Induced Rotational Experiment
2.4. Tissue Sampling and Immunohistochemistry
2.5. Drosophila Model and Drug Treatment
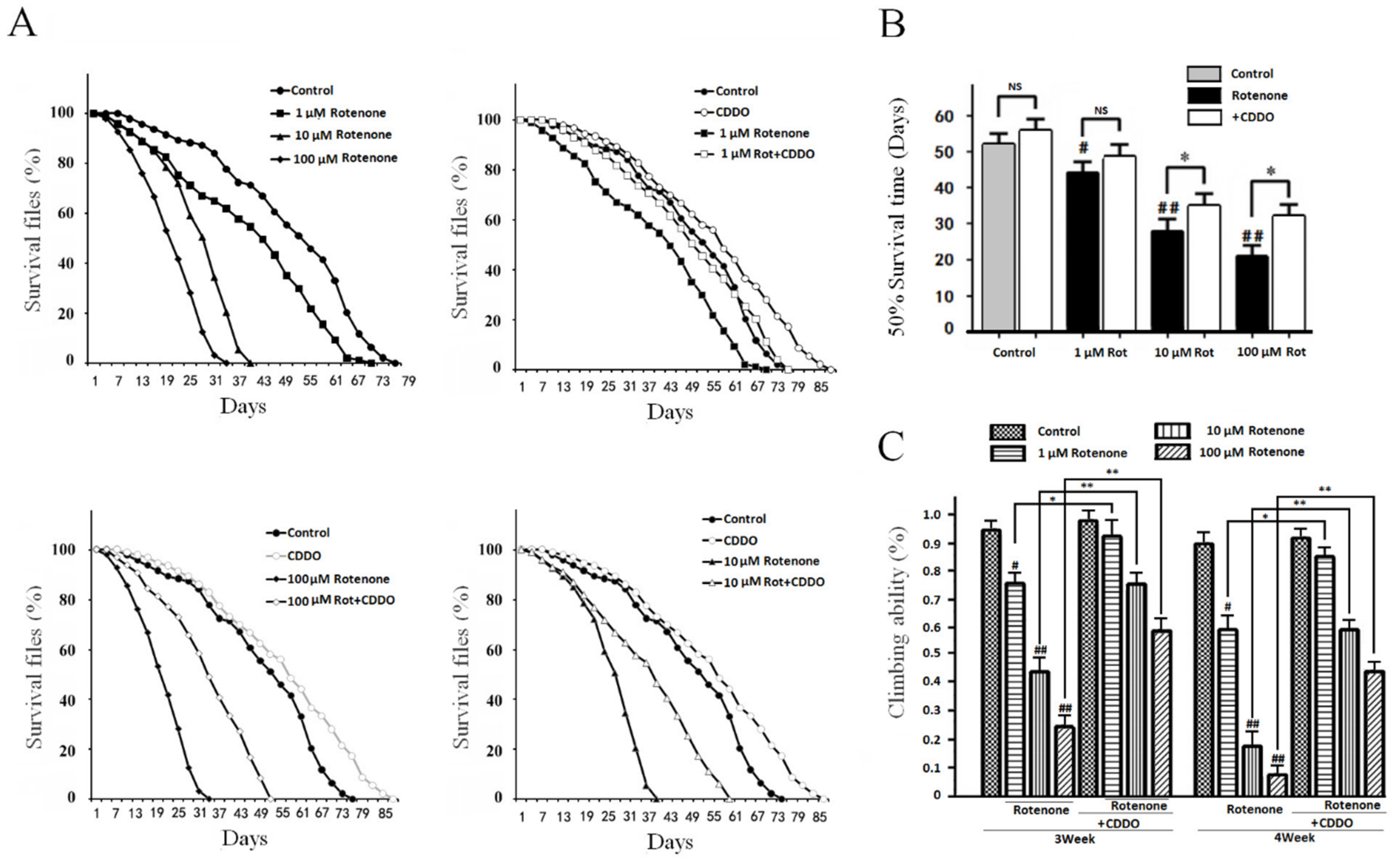
2.6. The Drosophila Lifespan Test
2.7. Climbing Ability Test
2.8. Redox State Analysis of the Drosophila Brains
2.9. Tissue Preparation and Western Blotting
2.10. Quantitative Real Time Polymerase Chain Reaction (qRT-PCR)
2.11. Statistical Analysis
3. Results
3.1. CDDO-Me Significantly Attenuated 6-OHDA-Induced PD-Like Phenotypes in Rats
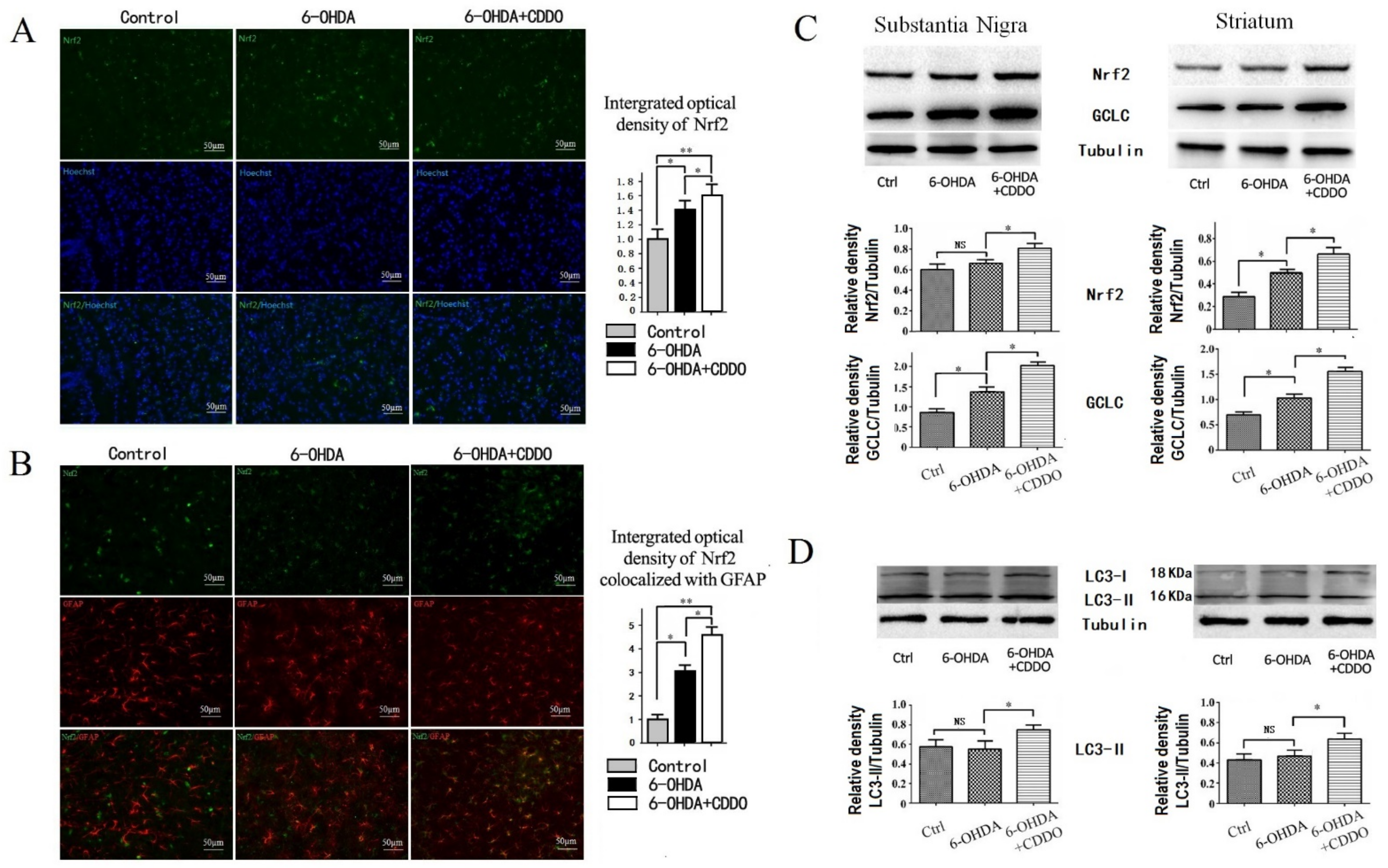
3.2. CDDO-Me Induced Activation of Nrf2 Pathway in Astrocytes and Increased Autophagy in 6-OHDA Rat Model
3.3. CDDO-Me Significantly Improved the Flies’ Lifespans and Motility in a PD Drosophila Model
3.4. Overexpression of Nrf2 in Glial Cells Displayed Antioxidant Effects and Rescued Rotenone-Induced PD-Like Phenotypes in Flies
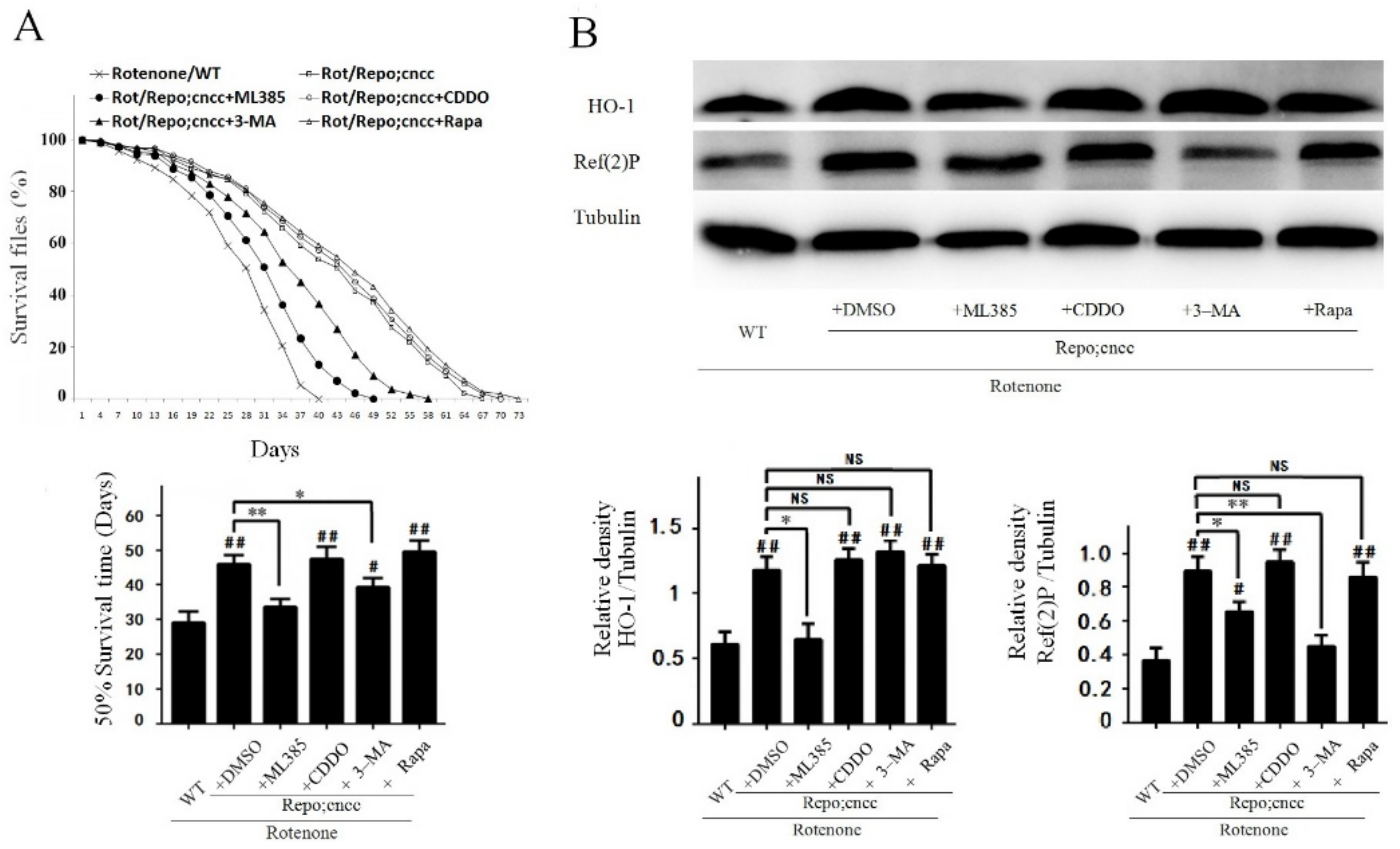
3.5. Activation of the Nrf2 Signaling Pathway Enhanced Autophagy
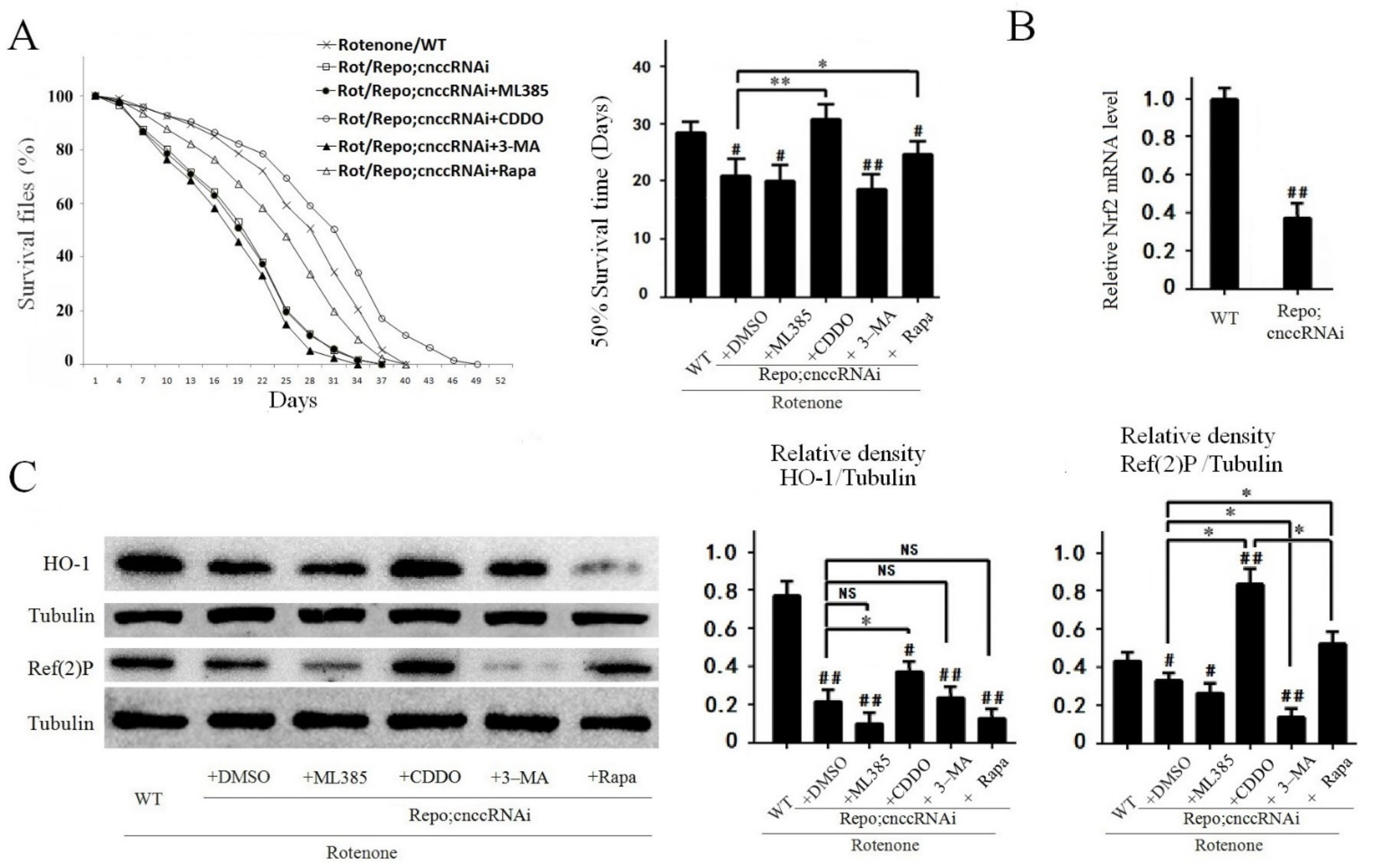
3.6. Knockdown of Nrf2 in Fly Glial Cells Worsened Rotenone-Induced PD-Like Phenotypes in Flies
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bologna, M.; Suppa, A.; Di Stasio, F.; Conte, A.; Fabbrini, G.; Berardelli, A. Neurophysiological studies on atypical parkinsonian syndromes. Parkinsonism Relat. Disord. 2017, 42, 12–21. [Google Scholar] [CrossRef] [PubMed]
- Akbar, M.; Essa, M.M.; Daradkeh, G.; Abdelmegeed, M.A.; Choi, Y.; Mahmood, L.; Song, B.J. Mitochondrial dysfunction and cell death in neurodegenerative diseases through nitroxidative stress. Brain Res. 2016, 1637, 34–55. [Google Scholar] [CrossRef] [PubMed]
- Chang, K.H.; Chen, C.M. The role of oxidative stress in Parkinson’s disease. Antioxidants 2020, 9, 597. [Google Scholar] [CrossRef]
- Liddell, J.R. Are astrocytes the predominant cell type for activation of Nrf2 in aging and neurodegeneration? Antioxidants 2017, 6, 65. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.L.; Santoro, M.; Mustafa, S.; Riedel, G.; Forrester, J.V.; Teismann, P. Evidence for a role of adaptive immune response in the disease pathogenesis of the MPTP mouse model of Parkinson’s disease. Glia 2016, 64, 386–395. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.; Sherratt, P.J.; Nioi, P.; Yang, C.S.; Pickett, C.B. Nrf2 controls constitutive and inducible expression of ARE-driven genes through a dynamic pathway involving nucleocytoplasmic shuttling by Keap1. J. Biol. Chem. 2005, 280, 32485–32492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masaki, Y.; Izumi, Y.; Matsumura, A.; Akaike, A.; Kume, T. Protective effect of Nrf2-ARE activator isolated from green perilla leaves on dopaminergic neuronal loss in a Parkinson’s disease model. Eur. J. Pharmacol. 2017, 798, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Indu Viswanath, A.N.; Park, J.H.; Lee, H.E.; Park, A.Y.; Choi, J.W.; Kim, H.J.; Londhe, A.M.; Jang, B.K.; Lee, J.; et al. Nrf2 activator via interference of Nrf2-Keap1 interaction has antioxidant and anti-inflammatory properties in Parkinson’s disease animal model. Neuropharmacology 2020, 167, 107989. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Liu, Q.; Shan, H.; Xia, C.; Liu, Z. Nrf2 inducer and cncC overexpression attenuates neurodegeneration due to α-synuclein in Drosophila. Biochem. Cell Biol. 2015, 93, 351–358. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Teschemacher, A.G.; Kasparov, S. Astroglia as a cellular target for neuroprotection and treatment of neuro-psychiatric disorders. Glia 2017, 65, 1205–1226. [Google Scholar] [CrossRef] [Green Version]
- Pistollato, F.; Canovas-Jorda, D.; Zagoura, D.; Bal-Price, A. Nrf2 pathway activation upon rotenone treatment in human iPSC-derived neural stem cells undergoing differentiation towards neurons and astrocytes. Neurochem. Int. 2017, 108, 457–471. [Google Scholar] [CrossRef]
- Wang, B.; Su, C.J.; Liu, T.T.; Zhou, Y.; Feng, Y.; Huang, Y.; Liu, X.; Wang, Z.H.; Chen, L.H.; Luo, W.F.; et al. The neuroprotection of low-dose morphine in cellular and animal models of Parkinson’s disease through ameliorating endoplasmic reticulum (ER) stress and activating autophagy. Front. Mol. Neurosci. 2018, 11, 120. [Google Scholar] [CrossRef] [PubMed]
- Montava-Garriga, L.; Ganley, I.G. Outstanding questions in mitophagy: What we do and do not know. J. Mol. Biol. 2020, 432, 206–230. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Pitoniak, A.; Wang, J.; Bohmann, D. Preserving transcriptional stress responses as an anti-aging strategy. Aging Cell 2021, 20, e13297. [Google Scholar] [CrossRef] [PubMed]
- Aryal, B.; Lee, Y. Disease model organism for Parkinson disease: Drosophila melanogaster. BMB Rep. 2019, 52, 250–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fao, L.; Mota, S.I.; Rego, A.C. Shaping the Nrf2-ARE-related pathways in Alzheimer’s and Parkinson’s diseases. Ageing Res. Rev. 2019, 54, 100942. [Google Scholar] [CrossRef] [PubMed]
- Mottaghi, S.; Buchholz, O.; Hofmann, U.G. Systematic evaluation of DBS parameters in the hemi-parkinsonian rat model. Front. Neurosci. 2020, 14, 561008. [Google Scholar] [CrossRef]
- Doktor, B.; Damulewicz, M.; Pyza, E. Overexpression of mitochondrial ligases reverses rotenone-induced effects in a Drosophila model of Parkinson’s disease. Front. Neurosci. 2019, 13, 94. [Google Scholar] [CrossRef]
- Markiewicz, I.; Lukomska, B. The role of astrocytes in the physiology and pathology of the central nervous system. Acta Neurobiol. Exp. 2006, 66, 343–358. [Google Scholar]
- Xia, R.; Ji, C.; Zhang, L. Neuroprotective effects of pycnogenol against oxygen-glucose deprivation/reoxygenation-induced injury in primary rat astrocytes via NF-κB and ERK1/2 MAPK pathways. Cell. Physiol. Biochem. 2017, 42, 987–998. [Google Scholar] [CrossRef]
- Zgorzynska, E.; Dziedzic, B.; Gorzkiewicz, A.; Stulczewski, D.; Bielawska, K.; Su, K.P.; Walczewska, A. Omega-3 polyunsaturated fatty acids improve the antioxidative defense in rat astrocytes via an Nrf2-dependent mechanism. Pharmacol. Rep. 2017, 69, 935–942. [Google Scholar] [CrossRef] [PubMed]
- Serapide, M.F.; L’Episcopo, F.; Tirolo, C.; Testa, N.; Caniglia, S.; Giachino, C.; Marchetti, B. Boosting antioxidant self-defenses by grafting astrocytes rejuvenates the aged microenvironment and mitigates nigrostriatal toxicity in Parkinsonian brain via an Nrf2-driven Wnt/β-catenin prosurvival axis. Front. Aging Neurosci. 2020, 12, 24. [Google Scholar] [CrossRef] [PubMed]
- Rappold, P.M.; Tieu, K. Astrocytes and therapeutics for Parkinson’s disease. Neurotherapeutics 2010, 7, 413–423. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leyns, C.E.G.; Holtzman, D.M. Glial contributions to neurodegeneration in tauopathies. Mol. Neurodegener. 2017, 12, 50. [Google Scholar] [CrossRef] [PubMed]
- Mena, M.A.; Garcia de Yebenes, J. Glial cells as players in parkinsonism: The “good,” the “bad,” and the “mysterious” glia. Neuroscientist 2008, 14, 544–560. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, J.K.; Gardaneh, M.; Iwasiow, R.; Lanthier, P.; Gangaraju, S.; Ribecco-Lutkiewicz, M.; Tremblay, R.; Kiuchi, K.; Sikorska, M. Astrocyte-secreted GDNF and glutathione antioxidant system protect neurons against 6OHDA cytotoxicity. Neurobiol. Dis. 2009, 33, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Fenoy, A.J.; Goetz, L.; Chabardes, S.; Xia, Y. Deep brain stimulation: Are astrocytes a key driver behind the scene? CNS Neurosci. Ther. 2014, 20, 191–201. [Google Scholar] [CrossRef] [Green Version]
- Kalvala, A.K.; Kumar, R.; Sherkhane, B.; Gundu, C.; Arruri, V.K.; Kumar, A. Bardoxolone methyl ameliorates hyperglycemia induced mitochondrial dysfunction by activating the keap1-Nrf2-ARE pathway in experimental diabetic neuropathy. Mol. Neurobiol. 2020, 57, 3616–3631. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, A.; Rojo, A.I.; Wells, G.; Hayes, J.D.; Cousin, S.P.; Rumsey, W.L.; Attucks, O.C.; Franklin, S.; Levonen, A.L.; Kensler, T.W.; et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019, 18, 295–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Petrillo, S.; Schirinzi, T.; Di Lazzaro, G.; D’Amico, J.; Colona, V.L.; Bertini, E.; Pierantozzi, M.; Mari, L.; Mercuri, N.B.; Piemonte, F.; et al. Systemic activation of Nrf2 pathway in Parkinson’s disease. Mov. Disord. 2020, 35, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.H.; Lee, S.R.; Park, Y.J.; Ra, M.; Lee, Y.; Pang, C.; Kim, K.H. Suppression of 6-hydroxydopamine-induced oxidative stress by hyperoside via activation of Nrf2/HO-1 signaling in dopaminergic neurons. Int. J. Mol. Sci. 2019, 20, 5832. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baluchnejadmojarad, T.; Rabiee, N.; Zabihnejad, S.; Roghani, M. Ellagic acid exerts protective effect in intrastriatal 6-hydroxydopamine rat model of Parkinson’s disease: Possible involvement of ERβ/Nrf2/HO-1 signaling. Brain Res. 2017, 1662, 23–30. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Ocak, U.; Gao, L.; Tu, S.; Lenahan, C.J.; Zhang, J.; Shao, A. Selective autophagy as a therapeutic target for neurological diseases. Cell. Mol. Life Sci. 2021, 78, 1369–1392. [Google Scholar] [CrossRef] [PubMed]
- Moors, T.E.; Hoozemans, J.J.; Ingrassia, A.; Beccari, T.; Parnetti, L.; Chartier-Harlin, M.C.; van de Berg, W.D. Therapeutic potential of autophagy-enhancing agents in Parkinson’s disease. Mol. Neurodegener. 2017, 12, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, K.E.; Gysbers, A.M.; Abbott, S.K.; Spiro, A.S.; Furuta, A.; Cooper, A.; Garner, B.; Kabuta, T.; Halliday, G.M. Lysosomal-associated membrane protein 2 isoforms are differentially affected in early Parkinson’s disease. Mov. Disord. 2015, 30, 1639–1647. [Google Scholar] [CrossRef] [PubMed]
- Chia, S.J.; Tan, E.K.; Chao, Y.X. Historical perspective: Models of Parkinson’s disease. Int. J. Mol. Sci. 2020, 21, 2464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Degtyarev, M.; De Maziere, A.; Orr, C.; Lin, J.; Lee, B.B.; Tien, J.Y.; Prior, W.W.; van Dijk, S.; Wu, H.; Gray, D.C.; et al. Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J. Cell Biol. 2008, 183, 101–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malpartida, A.B.; Williamson, M.; Narendra, D.P.; Wade-Martins, R.; Ryan, B.J. Mitochondrial dysfunction and mitophagy in Parkinson’s disease: From mechanism to therapy. Trends Biochem. Sci. 2021, 46, 329–343. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, Q.; Wang, B.; Wang, X.; Smith, W.W.; Zhu, Y.; Liu, Z. Activation of Nrf2 in Astrocytes Suppressed PD-Like Phenotypes via Antioxidant and Autophagy Pathways in Rat and Drosophila Models. Cells 2021, 10, 1850. https://doi.org/10.3390/cells10081850
Guo Q, Wang B, Wang X, Smith WW, Zhu Y, Liu Z. Activation of Nrf2 in Astrocytes Suppressed PD-Like Phenotypes via Antioxidant and Autophagy Pathways in Rat and Drosophila Models. Cells. 2021; 10(8):1850. https://doi.org/10.3390/cells10081850
Chicago/Turabian StyleGuo, Qing, Bing Wang, Xiaobo Wang, Wanli W. Smith, Yi Zhu, and Zhaohui Liu. 2021. "Activation of Nrf2 in Astrocytes Suppressed PD-Like Phenotypes via Antioxidant and Autophagy Pathways in Rat and Drosophila Models" Cells 10, no. 8: 1850. https://doi.org/10.3390/cells10081850






