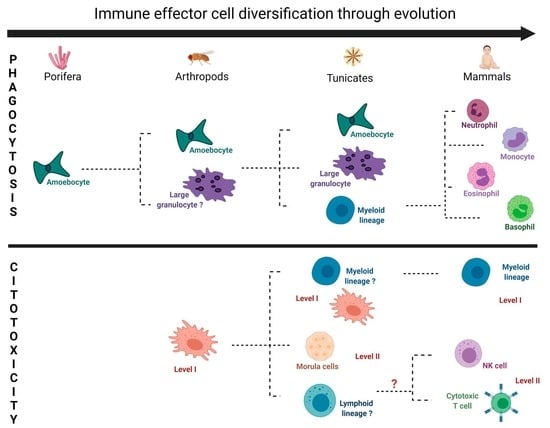
Evolution of Cellular Immunity Effector Cells; Perspective on Cytotoxic and Phagocytic Cellular Lineages
Abstract
1. Introduction
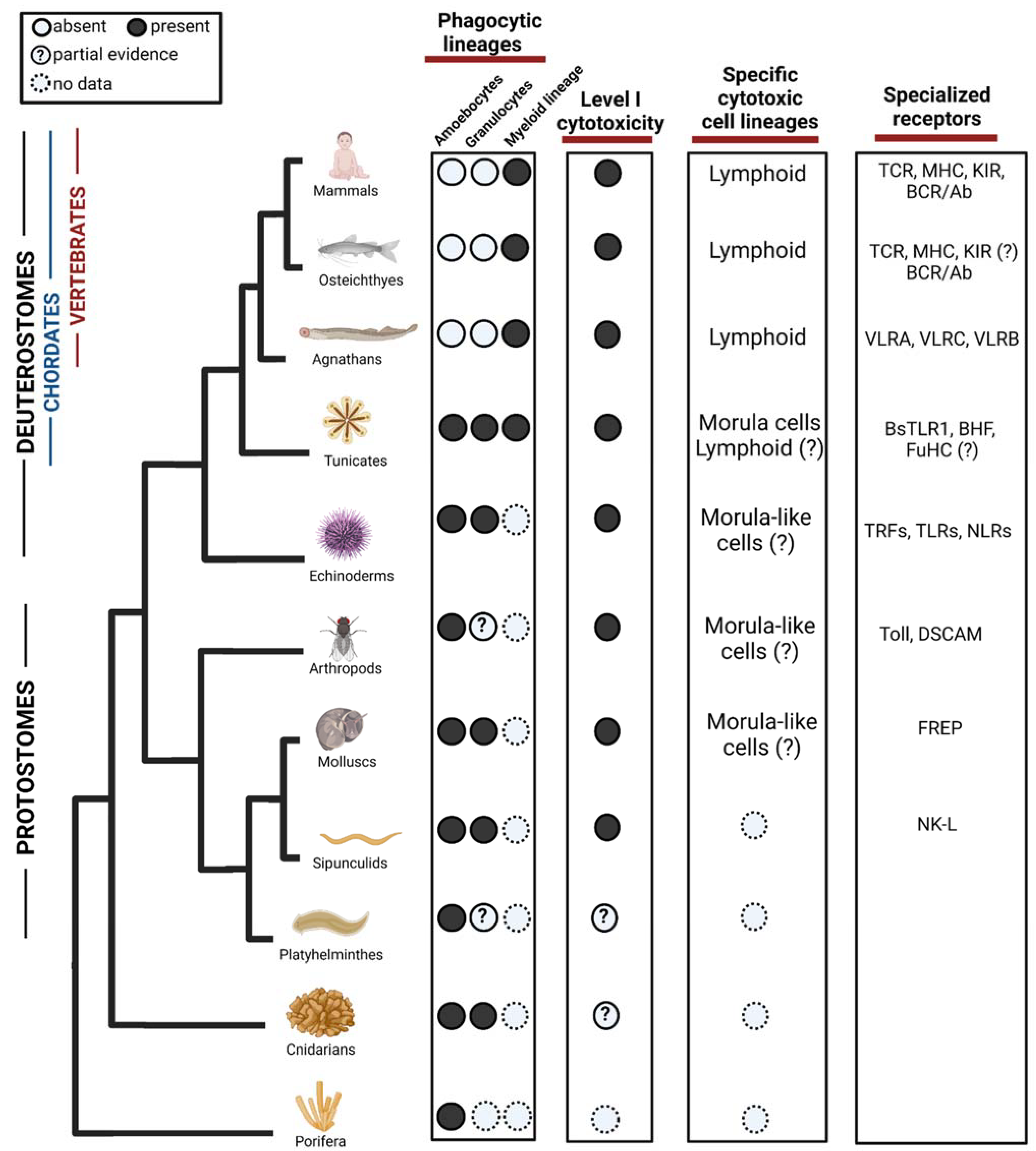
2. Phagocytosis in the Evolution of Immune Response
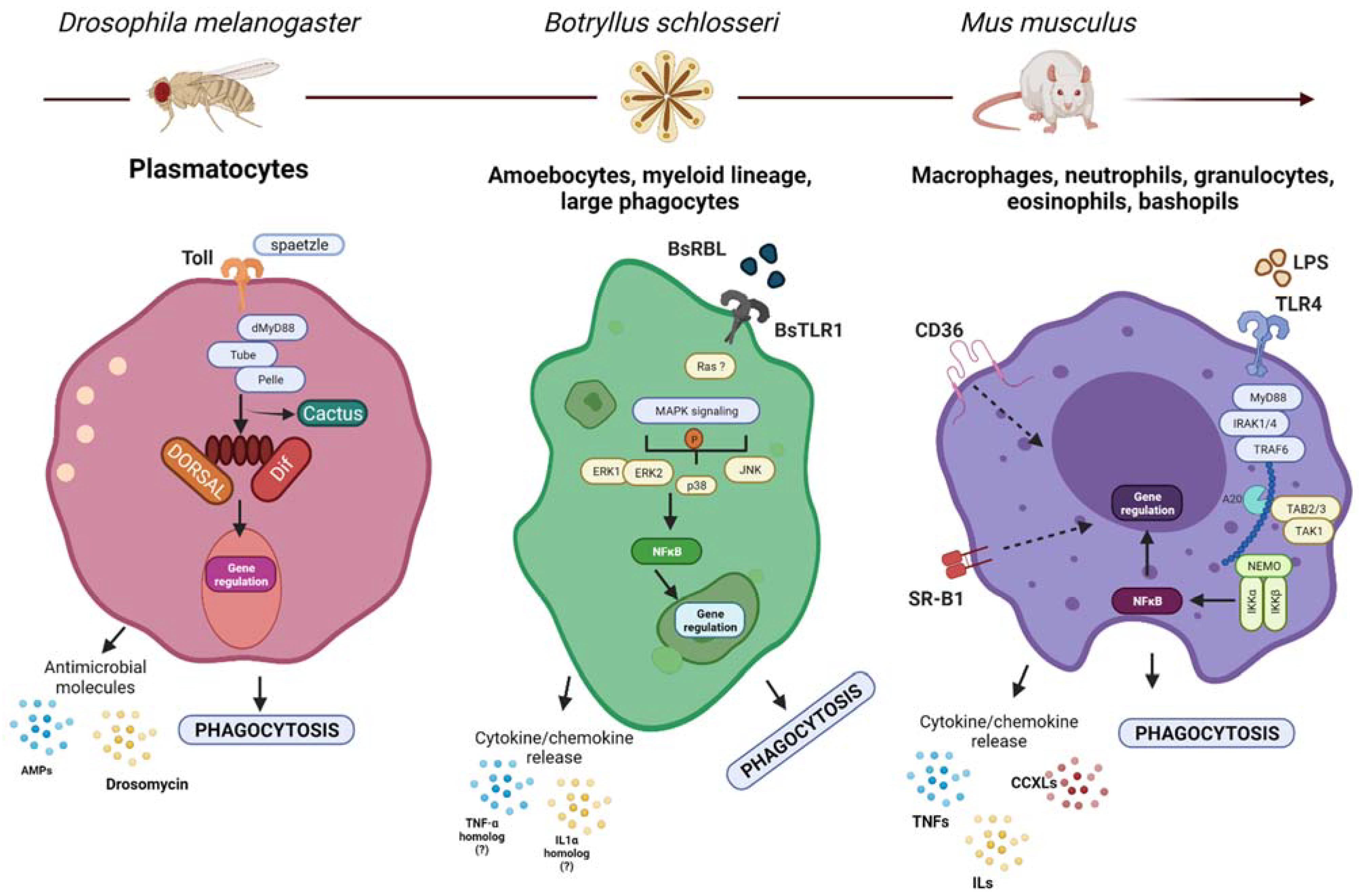
2.1. Phagocytic Effector Cells
2.2. Receptors
2.3. Effector Molecules
2.4. Signaling Pathways
3. Cellular Cytotoxicity in the Evolution of Immune Response against Abnormal Self and Pathogens
3.1. Effector Cells
3.2. Receptors
3.3. Effector Molecules
3.4. Signaling Pathways
4. Perspective on the Effect of the Immune System in the Evolution of Regenerative Capacities
5. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rolff, J. Why did the acquired immune system of vertebrates evolve? Dev. Comp. Immunol. 2007, 31, 476–482. [Google Scholar] [CrossRef] [PubMed]
- Buchmann, K. Evolution of Innate Immunity: Clues from Invertebrates via Fish to Mammals. Front. Immunol. 2014, 5, 459. [Google Scholar] [CrossRef] [PubMed]
- Kimbrell, D.A.; Beutler, B. The evolution and genetics of innate immunity. Nat. Rev. Genet. 2001, 2, 256–267. [Google Scholar] [CrossRef]
- Netea, M.G.; Schlitzer, A.; Placek, K.; Joosten, L.A.B.; Schultze, J.L. Innate and Adaptive Immune Memory: An Evolutionary Continuum in the Host’s Response to Pathogens. Cell Host Microbe 2019, 25, 13–26. [Google Scholar] [CrossRef]
- Gordon, S. Phagocytosis: An Immunobiologic Process. Immunity 2016, 44, 463–475. [Google Scholar] [CrossRef]
- Gordon, S. Elie Metchnikoff, the Man and the Myth. J. Innate Immun. 2016, 8, 223–227. [Google Scholar] [CrossRef]
- Metschnikoff, E. Memoirs: The Ancestral History of the Inflammatory Process. J. Cell Sci. 1884, s2–s24, 112–117. [Google Scholar] [CrossRef]
- Schulenburg, H.; Kurz, C.L.; Ewbank, J.J. Evolution of the innate immune system: The worm perspective. Immunol. Rev. 2004, 198, 36–58. [Google Scholar] [CrossRef]
- Mukherjee, S.; Ray, M.; Ray, S. Phagocytic efficiency and cytotoxic responses of Indian freshwater sponge (Eunapius carteri) cells isolated by density gradient centrifugation and flow cytometry: A morphofunctional analysis. Zoology 2015, 118, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Snyder, G.A.; Eliachar, S.; Connelly, M.T.; Talice, S.; Hadad, U.; Gershoni-Yahalom, O.; Browne, W.E.; Palmer, C.V.; Rosental, B.; Traylor-Knowles, N. Functional characterization of Hexacorallia phagocytic cells. Front. Immunol. 2021, 12, 2402. [Google Scholar]
- Morita, M. Phagocytic response of planarian reticular cells to heat-killed bacteria. Hydrobiologia 1991, 227, 193–199. [Google Scholar] [CrossRef]
- Maciel, E.I.; Oviedo, N.J. Platyhelminthes: Molecular Dissection of the Planarian Innate Immune System. In Advances in Comparative Immunology; Cooper, E.L., Ed.; Springer: Cham, Switzerland, 2018. [Google Scholar]
- Homa, J. Earthworm coelomocyte extracellular traps: Structural and functional similarities with neutrophil NETs. Cell Tissue Res. 2018, 371, 407–414. [Google Scholar] [CrossRef]
- Csordás, G.; Gábor, E.; Honti, V. There and back again: The mechanisms of differentiation and transdifferentiation in Drosophila blood cells. Dev. Biol. 2021, 469, 135–143. [Google Scholar] [CrossRef]
- Cattenoz, P.B.; Sakr, R.; Pavlidaki, A.; Delaporte, C.; Riba, A.; Molina, N.; Hariharan, N.; Mukherjee, T.; Giangrande, A. Temporal specificity and heterogeneity of Drosophila immune cells. EMBO J. 2020, 39, e104486. [Google Scholar] [CrossRef]
- Rosental, B.; Kowarsky, M.; Seita, J.; Corey, D.M.; Ishizuka, K.J.; Palmeri, K.J.; Chen, S.-Y.; Sinha, R.; Okamoto, J.; Mantalas, G.; et al. Complex mammalian-like haematopoietic system found in a colonial chordate. Nature 2018, 564, 425–429. [Google Scholar] [CrossRef]
- Rosental, B.; Raveh, T.; Voskoboynik, A.; Weissman, I.L. Evolutionary perspective on the hematopoietic system through a colonial chordate: Allogeneic immunity and hematopoiesis. Curr. Opin. Immunol. 2020, 62, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Grayfer, L.; Kerimoglu, B.; Yaparla, A.; Hodgkinson, J.W.; Xie, J.; Belosevic, M. Mechanisms of Fish Macrophage Antimicrobial Immunity. Front. Immunol. 2018, 9, 1105. [Google Scholar] [CrossRef]
- Vandendriessche, S.; Cambier, S.; Proost, P.; Marques, P.E. Complement Receptors and Their Role in Leukocyte Recruitment and Phagocytosis. Front. Cell Dev. Biol. 2021, 9, 624025. [Google Scholar] [CrossRef] [PubMed]
- Hirayama, D.; Iida, T.; Nakase, H. The Phagocytic Function of Macrophage-Enforcing Innate Immunity and Tissue Homeostasis. Int. J. Mol. Sci. 2017, 1, 92. [Google Scholar] [CrossRef]
- Yuen, B.; Bayes, J.M.; Degnan, S.M. The characterization of sponge NLRs provides insight into the origin and evolution of this innate immune gene family in animals. Mol. Biol. Evol. 2014, 31, 106–120. [Google Scholar] [CrossRef] [PubMed]
- Wiens, M.; Korzhev, M.; Krasko, A.; Thakur, N.L.; Perović-Ottstadt, S.; Breter, H.J.; Ushijima, H.; Diehl-Seifert, B.; Müller, I.M.; Müller, W.E. Innate immune defense of the sponge Suberites domuncula against bacteria involves a MyD88-dependent signaling pathway. Induction of a perforin-like molecule. J. Biol. Chem. 2005, 280, 27949–27959. [Google Scholar] [CrossRef] [PubMed]
- Narbonne-Reveau, K.; Charroux, B.; Royet, J. Lack of an antibacterial response defect in Drosophila Toll-9 mutant. PLoS ONE 2011, 6, e17470. [Google Scholar] [CrossRef] [PubMed]
- Stuart, L.M.; Ezekowitz, R.A. Phagocytosis and comparative innate immunity: Learning on the fly. Nat. Rev. Immunol. 2008, 8, 131–141. [Google Scholar] [CrossRef]
- Peronato, A.; Franchi, N.; Loriano, B. BsTLR1: A new member of the TLR family of recognition proteins from the colonial ascidian Botryllus schlosseri. Fish Shellfish Immunol. 2020, 106, 967–974. [Google Scholar] [CrossRef]
- Buckley, K.M.; Rast, J.P. Dynamic evolution of toll-like receptor multigene families in echinoderms. Front. Immunol. 2012, 3, 136. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Zhang, H.; Zhao, C.; Zhang, H. Evolutionary History of the Toll-Like Receptor Gene Family across Vertebrates. Genome Biol. Evol. 2020, 12, 3615–3634. [Google Scholar] [CrossRef]
- Peiris, T.H.; Hoyer, K.K.; Oviedo, N.J. Innate immune system and tissue regeneration in planarians: An area ripe for exploration. Semin. Immunol. 2014, 26, 295–302. [Google Scholar] [CrossRef]
- Guillou, A.; Troha, K.; Wang, H.; Franc, N.C.; Buchon, N. The Drosophila CD36 Homologue croquemort is required to Maintain Immune and Gut Homeostasis during Development and Aging. PLoS Pathog. 2016, 12, e1005961. [Google Scholar] [CrossRef]
- Means, T.K.; Mylonakis, E.; Tampakakis, E.; Colvin, R.A.; Seung, E.; Puckett, L.; Tai, M.F.; Stewart, C.R.; Pukkila-Worley, R.; Hickman, S.E.; et al. Evolutionarily conserved recognition and innate immunity to fungal pathogens by the scavenger receptors SCARF1 and CD36. J. Exp. Med. 2009, 206, 637–653. [Google Scholar] [CrossRef]
- Febbraio, M.; Hajjar, D.P.; Silverstein, R.L. CD36: A class B scavenger receptor involved in angiogenesis, atherosclerosis, inflammation, and lipid metabolism. J. Clin. Investig. 2001, 108, 785–791. [Google Scholar] [CrossRef] [PubMed]
- Yakovenko, I.; Donnyo, A.; Ioscovich, O.; Rosental, B.; Oren, M. The Diverse Transformer (Trf) Protein Family in the Sea Urchin Paracentrotus lividus Acts through a Collaboration between Cellular and Humoral Immune Effector Arms. Int. J. Mol. Sci. 2021, 22, 6639. [Google Scholar] [CrossRef]
- Chou, H.-Y.; Lun, C.M.; Smith, L.C. SpTransformer proteins from the purple sea urchin opsonize bacteria, augment phagocytosis, and retard bacterial growth. PLoS ONE 2018, 13, e0196890. [Google Scholar] [CrossRef]
- Oren, M.; Barela Hudgell, M.A.; D’Allura, B.; Agronin, J.; Gross, A.; Podini, D.; Smith, L.C. Short tandem repeats, segmental duplications, gene deletion, and genomic instability in a rapidly diversified immune gene family. BMC Genom. 2016, 17, 900. [Google Scholar] [CrossRef]
- Smith, L.C.; Lun, C.M. The SpTransformer Gene Family (Formerly Sp185/333) in the Purple Sea Urchin and the Functional Diversity of the Anti-Pathogen rSpTransformer-E1 Protein. Front. Immunol. 2017, 8, 725. [Google Scholar] [CrossRef] [PubMed]
- Majeske, A.J.; Oren, M.; Sacchi, S.; Smith, L.C. Single sea urchin phagocytes express messages of a single sequence from the diverse Sp185/333 gene family in response to bacterial challenge. J. Immunol. 2014, 193, 5678–5688. [Google Scholar] [CrossRef]
- Franchi, N.; Schiavon, F.; Carletto, M.; Gasparini, F.; Bertoloni, G.; Tosatto, S.C.; Ballarin, L. Immune roles of a rhamnose-binding lectin in the colonial ascidian Botryllus schlosseri. Immunobiology 2011, 216, 725–736. [Google Scholar] [CrossRef]
- Vanha-Aho, L.M.; Valanne, S.; Rämet, M. Cytokines in Drosophila immunity. Immunol. Lett. 2016, 170, 42–51. [Google Scholar] [CrossRef] [PubMed]
- Langenbacher, A.D.; De Tomaso, A.W. Temporally and spatially dynamic germ cell niches in Botryllus schlosseri revealed by expression of a TGF-beta family ligand and vasa. EvoDevo 2016, 7, 9. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Secombes, C.J. The Function of Fish Cytokines. Biology 2016, 5, 23. [Google Scholar] [CrossRef]
- Savan, R.; Ravichandran, S.; Collins, J.R.; Sakai, M.; Young, H.A. Structural conservation of interferon gamma among vertebrates. Cytokine Growth Factor Rev. 2009, 20, 115–124. [Google Scholar] [CrossRef]
- Williams, L.M.; Inge, M.M.; Mansfield, K.M.; Rasmussen, A.; Afghani, J.; Agrba, M.; Albert, C.; Andersson, C.; Babaei, M.; Babaei, M.; et al. Transcription factor NF-κB in a basal metazoan, the sponge, has conserved and unique sequences, activities, and regulation. Dev. Comp. Immunol. 2020, 104, 103559. [Google Scholar] [CrossRef] [PubMed]
- Cardoso, M.A.; Fontenele, M.; Lim, B.; Bisch, P.M.; Shvartsman, S.Y.; Araujo, H.M. A novel function for the IκB inhibitor Cactus in promoting Dorsal nuclear localization and activity in the Drosophila embryo. Development 2017, 144, 2907–2913. [Google Scholar]
- Franchi, N.; Schiavon, F.; Betti, M.; Canesi, L.; Ballarin, L. Insight on signal transduction pathways involved in phagocytosis in the colonial ascidian Botryllus schlosseri. J. Invertebr. Pathol. 2013, 112, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Dorrington, M.G.; Fraser, I.D.C. NF-κB Signaling in Macrophages: Dynamics, Crosstalk, and Signal Integration. Front. Immunol. 2019, 10, 705. [Google Scholar] [CrossRef] [PubMed]
- Valanne, S.; Wang, J.H.; Rämet, M. The Drosophila Toll signaling pathway. J. Immunol. 2011, 186, 649–656. [Google Scholar] [CrossRef] [PubMed]
- O’Neill, L.A.J.; Golenbock, D.; Bowie, A.G. The history of Toll-like receptors—Redefining innate immunity. Nat. Rev. Immunol. 2013, 13, 453–460. [Google Scholar] [CrossRef]
- Cooper, E.L. Phylogeny of cytotoxicity. Endeavour 1980, 4, 160–165. [Google Scholar] [CrossRef]
- Colantonio, A.D.; Bimber, B.N.; Neidermyer, W.J., Jr.; Reeves, R.K.; Alter, G.; Altfeld, M.; Johnson, R.P.; Carrington, M.; O’Connor, D.H.; Evans, D.T. KIR polymorphisms modulate peptide-dependent binding to an MHC class I ligand with a Bw6 motif. PLoS Pathog. 2011, 7, e1001316. [Google Scholar] [CrossRef]
- Rosental, B.; Appel, M.Y.; Yossef, R.; Hadad, U.; Brusilovsky, M.; Porgador, A. The effect of chemotherapy/radiotherapy on cancerous pattern recognition by NK cells. Curr. Med. Chem. 2012, 19, 1780–1791. [Google Scholar] [CrossRef]
- Franceschi, C.; Cossarizza, A.; Ortolani, C.; Monti, D.; Ottaviani, E. Natural cytotoxicity in a freshwater pulmonate mollusc: An unorthodox comparative approach. Adv. Neuroimmunol. 1991, 1, 99–113. [Google Scholar] [CrossRef]
- Boiledieu, D.; Valembois, P. Natural cytotoxic activity of sipunculid leukocytes on allogenic and xenogenic erythrocytes. Dev. Comp. Immunol. 1977, 1, 207–216. [Google Scholar] [CrossRef]
- Radomski, M.W.; Martin, J.F.; Moncada, S. Synthesis of Nitric Oxide by the Haemocytes of the American Horseshoe Crab (Limulus polyphemus). Philos. Trans. Biol. Sci. 1991, 334, 129–133. [Google Scholar]
- Söderhäll, K.; Wingren, A.; Johansson, M.W.; Bertheussen, K. The cytotoxic reaction of hemocytes from the freshwater crayfish, Astacus astacus. Cell. Immunol. 1985, 94, 326–332. [Google Scholar] [CrossRef]
- Cárdenas, W.; Dankert, J.R. Cresolase, catecholase and laccase activities in haemocytes of the red swamp crayfish. Fish Shellfish Immunol. 2000, 10, 33–46. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Zhang, H.; Beck, G. Phylogeny of natural cytotoxicity: Cytotoxic activity of coelomocytes of the purple sea urchin, Arbacia punctulata. J. Exp. Zool. 2001, 290, 741–750. [Google Scholar] [CrossRef] [PubMed]
- Gross, P.S.; Al-Sharif, W.Z.; Clow, L.A.; Smith, L.C. Echinoderm immunity and the evolution of the complement system. Dev. Comp. Immunol. 1999, 23, 429–442. [Google Scholar] [CrossRef]
- Corey, D.M.; Rosental, B.; Kowarsky, M.; Sinha, R.; Ishizuka, K.J.; Palmeri, K.J.; Quake, S.R.; Voskoboynik, A.; Weissman, I.L. Developmental cell death programs license cytotoxic cells to eliminate histocompatible partners. Proc. Natl. Acad. Sci. USA 2016, 113, 6520–6525. [Google Scholar] [CrossRef]
- Voskoboynik, A.; Newman, A.M.; Corey, D.M.; Sahoo, D.; Pushkarev, D.; Neff, N.F.; Passarelli, B.; Koh, W.; Ishizuka, K.J.; Palmeri, K.J.; et al. Identification of a colonial chordate histocompatibility gene. Science 2013, 341, 384–387. [Google Scholar] [CrossRef]
- De Tomaso, A.W.; Nyholm, S.V.; Palmeri, K.J.; Ishizuka, K.J.; Ludington, W.B.; Mitchel, K.; Weissman, I.L. Isolation and characterization of a protochordate histocompatibility locus. Nature 2005, 438, 454–459. [Google Scholar] [CrossRef]
- Das, S.; Li, J.; Hirano, M.; Sutoh, Y.; Herrin, B.R.; Cooper, M.D. Evolution of two prototypic T cell lineages. Cell. Immunol. 2015, 296, 87–94. [Google Scholar] [CrossRef]
- Mayer, W.E.; Uinuk-ool, T.; Tichy, H.; Gartland, L.A.; Klein, J.; Cooper, M.D. Isolation and characterization of lymphocyte-like cells from a lamprey. Proc. Natl. Acad. Sci. USA 2002, 99, 14350–14355. [Google Scholar] [CrossRef]
- Uinuk-ool, T.; Mayer, W.E.; Sato, A.; Dongak, R.; Cooper, M.D.; Klein, J. Lamprey lymphocyte-like cells express homologs of genes involved in immunologically relevant activities of mammalian lymphocytes. Proc. Natl. Acad. Sci. USA 2002, 99, 14356–14361. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Das, S.; Herrin, B.R.; Hirano, M.; Cooper, M.D. Definition of a third VLR gene in hagfish. Proc. Natl. Acad. Sci. USA 2013, 110, 15013–15018. [Google Scholar] [CrossRef]
- Flajnik, M.F. A cold-blooded view of adaptive immunity. Nat. Rev. Immunol. 2018, 18, 438–453. [Google Scholar] [CrossRef]
- Schwartz, J.C.; Gibson, M.S.; Heimeier, D.; Koren, S.; Phillippy, A.M.; Bickhart, D.M.; Smith, T.P.; Medrano, J.F.; Hammond, J.A. The evolution of the natural killer complex; a comparison between mammals using new high-quality genome assemblies and targeted annotation. Immunogenetics 2017, 69, 255–269. [Google Scholar] [CrossRef]
- Shen, L.; Stuge, T.B.; Zhou, H.; Khayat, M.; Barker, K.S.; Quiniou, S.M.; Wilson, M.; Bengtén, E.; Chinchar, V.G.; Clem, L.W.; et al. Channel catfish cytotoxic cells: A mini-review. Dev. Comp. Immunol. 2002, 26, 141–149. [Google Scholar] [CrossRef]
- Morales, H.D.; Robert, J. Characterization of primary and memory CD8 T-cell responses against ranavirus (FV3) in Xenopus laevis. J. Virol. 2007, 81, 2240–2248. [Google Scholar] [CrossRef] [PubMed]
- Hirano, M.; Das, S.; Guo, P.; Cooper, M.D. The evolution of adaptive immunity in vertebrates. Adv. Immunol. 2011, 109, 125–157. [Google Scholar] [PubMed]
- Butenko, S.; Satyanarayanan, S.K.; Assi, S.; Schif-Zuck, S.; Barkan, D.; Sher, N.; Ariel, A. Transcriptomic Analysis of Monocyte-Derived Non-Phagocytic Macrophages Favors a Role in Limiting Tissue Repair and Fibrosis. Front. Immunol. 2020, 11, 405. [Google Scholar] [CrossRef] [PubMed]
- Geffner, J. Antibody-Dependent Cellular Cytotoxicity. In Encyclopedia of Immunotoxicology; Vohr, H.-W., Ed.; Springer: Berlin/Heidelberg, Germany, 2005; pp. 1–5. [Google Scholar]
- Hanton, G.; Pastoret, P.P. The reaction of antibody-dependent cell-mediated cytotoxicity (ADCC). Ann. Vet. Res. 1984, 15, 443–454. [Google Scholar]
- Torben, W.; Ahmad, G.; Zhang, W.; Nash, S.; Le, L.; Karmakar, S.; Siddiqui, A.A. Role of antibody dependent cell mediated cytotoxicity (ADCC) in Sm-p80-mediated protection against Schistosoma mansoni. Vaccine 2012, 30, 6753–6758. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sun, Y.; Asmal, M.; Lane, S.; Permar, S.R.; Schmidt, S.D.; Mascola, J.R.; Letvin, N.L. Antibody-Dependent Cell-Mediated Cytotoxicity in Simian Immunodeficiency Virus-Infected Rhesus Monkeys. J. Virol. 2011, 85, 6906–6912. [Google Scholar] [CrossRef]
- Flajnik, M.F.; Du Pasquier, L. Evolution of innate and adaptive immunity: Can we draw a line? Trends Immunol. 2004, 25, 640–644. [Google Scholar] [CrossRef]
- Schultz, J.H.; Bu, L.; Adema, C.M. Comparative immunological study of the snail Physella acuta (Hygrophila, Pulmonata) reveals shared and unique aspects of gastropod immunobiology. Mol. Immunol. 2018, 101, 108–119. [Google Scholar] [CrossRef]
- Loker, E.S.; Adema, C.M.; Zhang, S.M.; Kepler, T.B. Invertebrate immune systems--not homogeneous, not simple, not well understood. Immunol. Rev. 2004, 198, 10–24. [Google Scholar] [CrossRef] [PubMed]
- Cárdenas, W.; Dankert, J.R.; Jenkins, J.A. Flow cytometric analysis of crayfish haemocytes activated by lipopolysaccharides. Fish Shellfish Immunol. 2004, 17, 223–233. [Google Scholar] [CrossRef]
- Ljunggren, H.-G.; Kärre, K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol. Today 1990, 11, 237–244. [Google Scholar] [CrossRef]
- Raulet, D.H. Missing self recognition and self tolerance of natural killer (NK) cells. Semin. Immunol. 2006, 18, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Borrego, F. The First Molecular Basis of the “Missing Self” Hypothesis. J. Immunol. 2006, 177, 5759–5760. [Google Scholar] [CrossRef]
- Khalturin, K.; Becker, M.; Rinkevich, B.; Bosch, T.C. Urochordates and the origin of natural killer cells: Identification of a CD94/NKR-P1-related receptor in blood cells of Botryllus. Proc. Natl. Acad. Sci. USA 2003, 100, 622–627. [Google Scholar] [CrossRef]
- Pancer, Z.; Amemiya, C.T.; Ehrhardt, G.R.A.; Ceitlin, J.; Larry Gartland, G.; Cooper, M.D. Somatic diversification of variable lymphocyte receptors in the agnathan sea lamprey. Nature 2004, 430, 174–180. [Google Scholar] [CrossRef]
- Kishishita, N.; Nagawa, F. Evolution of adaptive immunity: Implications of a third lymphocyte lineage in lampreys. BioEssays News Rev. Mol. Cell. Dev. Biol. 2014, 36, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Hirano, M.; Aghaallaei, N.; Bajoghli, B.; Boehm, T.; Cooper, M.D. Organization of lamprey variable lymphocyte receptor C locus and repertoire development. Proc. Natl. Acad. Sci. USA 2013, 110, 6043–6048. [Google Scholar] [CrossRef]
- Kasahara, M. Variable Lymphocyte Receptors: A Current Overview. Results Probl. Cell Differ. 2015, 57, 175–192. [Google Scholar]
- Croft, N.P.; Smith, S.A.; Pickering, J.; Sidney, J.; Peters, B.; Faridi, P.; Witney, M.J.; Sebastian, P.; Flesch, I.E.A.; Heading, S.L.; et al. Most viral peptides displayed by class I MHC on infected cells are immunogenic. Proc. Natl. Acad. Sci. USA 2019, 116, 3112–3117. [Google Scholar] [CrossRef]
- Marino, J.; Paster, J.; Benichou, G. Allorecognition by T Lymphocytes and Allograft Rejection. Front. Immunol. 2016, 7, 582. [Google Scholar] [CrossRef] [PubMed]
- Bosselut, R. T cell antigen recognition: Evolution-driven affinities. Proc. Natl. Acad. Sci. USA 2019, 116, 21969–21971. [Google Scholar] [CrossRef] [PubMed]
- Kasahara, M.; Flajnik, M.F. Origin and evolution of the specialized forms of proteasomes involved in antigen presentation. Immunogenetics 2019, 71, 251–261. [Google Scholar] [CrossRef]
- Castro, R.; Bernard, D.; Lefranc, M.P.; Six, A.; Benmansour, A.; Boudinot, P. T cell diversity and TcR repertoires in teleost fish. Fish Shellfish Immunol. 2011, 31, 644–654. [Google Scholar] [CrossRef]
- Foulkrod, A.M.; Appasamy, P.M. Expression of TCR genes in adult and larval Xenopus laevis. Dev. Comp. Immunol. 2019, 96, 78–82. [Google Scholar] [CrossRef]
- Jiravanichpaisal, P.; Lee, S.Y.; Kim, Y.A.; Andrén, T.; Söderhäll, I. Antibacterial peptides in hemocytes and hematopoietic tissue from freshwater crayfish Pacifastacus leniusculus: Characterization and expression pattern. Dev. Comp. Immunol. 2007, 31, 441–455. [Google Scholar] [CrossRef] [PubMed]
- Nappi, A.J.; Ottaviani, E. Cytotoxicity and cytotoxic molecules in invertebrates. BioEssays News Rev. Mol. Cell. Dev. Biol. 2000, 22, 469–480. [Google Scholar] [CrossRef]
- Wink, D.A.; Hines, H.B.; Cheng, R.Y.; Switzer, C.H.; Flores-Santana, W.; Vitek, M.P.; Ridnour, L.A.; Colton, C.A. Nitric oxide and redox mechanisms in the immune response. J. Leukoc. Biol. 2011, 89, 873–891. [Google Scholar] [CrossRef] [PubMed]
- Lisi, F.; Zelikin, A.N.; Chandrawati, R. Nitric Oxide to Fight Viral Infections. Adv. Sci. 2021, 8, 2003895. [Google Scholar] [CrossRef] [PubMed]
- Weber, K.; Schilling, J.D. Distinct lysosome phenotypes influence inflammatory function in peritoneal and bone marrow-derived macrophages. Int. J. Inflamm. 2014, 2014, 154936. [Google Scholar] [CrossRef]
- Fauriat, C.; Long, E.O.; Ljunggren, H.G.; Bryceson, Y.T. Regulation of human NK-cell cytokine and chemokine production by target cell recognition. Blood 2010, 115, 2167–2176. [Google Scholar] [CrossRef]
- Schultz, J.H.; Adema, C.M. Comparative immunogenomics of molluscs. Dev. Comp. Immunol. 2017, 75, 3–15. [Google Scholar] [CrossRef]
- Peters, P.J.; Borst, J.; Oorschot, V.; Fukuda, M.; Krähenbühl, O.; Tschopp, J.; Slot, J.W.; Geuze, H.J. Cytotoxic T lymphocyte granules are secretory lysosomes, containing both perforin and granzymes. J. Exp. Med. 1991, 173, 1099–1109. [Google Scholar] [CrossRef]
- Perussia, B. Signaling for cytotoxicity. Nat. Immunol. 2000, 1, 372–374. [Google Scholar] [CrossRef]
- Kabanova, A.; Zurli, V.; Baldari, C.T. Signals Controlling Lytic Granule Polarization at the Cytotoxic Immune Synapse. Front. Immunol. 2018, 9, 307. [Google Scholar] [CrossRef]
- Humphries, J.E.; Yoshino, T.P. Cellular receptors and signal transduction in molluscan hemocytes: Connections with the innate immune system of vertebrates. Integr. Comp. Biol. 2003, 43, 305–312. [Google Scholar] [CrossRef] [PubMed]
- Franchi, N.; Ballarin, L. Immunity in Protochordates: The Tunicate Perspective. Front. Immunol. 2017, 8, 674. [Google Scholar] [CrossRef] [PubMed]
- Wolff, L.; Humeniuk, R. Concise review: Erythroid versus myeloid lineage commitment: Regulating the master regulators. Stem cells 2013, 31, 1237–1244. [Google Scholar] [CrossRef]
- Potts, K.S.; Bowman, T.V. Modeling Myeloid Malignancies Using Zebrafish. Front. Oncol. 2017, 7, 297. [Google Scholar] [CrossRef] [PubMed]
- Wei, S.; Zhou, J.M.; Chen, X.; Shah, R.N.; Liu, J.; Orcutt, T.M.; Traver, D.; Djeu, J.Y.; Litman, G.W.; Yoder, J.A. The zebrafish activating immune receptor Nitr9 signals via Dap12. Immunogenetics 2007, 59, 813–821. [Google Scholar] [CrossRef] [PubMed]
- Moss, L.D.; Monette, M.M.; Jaso-Friedmann, L.; Leary, J.H., 3rd; Dougan, S.T.; Krunkosky, T.; Evans, D.L. Identification of phagocytic cells, NK-like cytotoxic cell activity and the production of cellular exudates in the coelomic cavity of adult zebrafish. Dev. Comp. Immunol. 2009, 33, 1077–1087. [Google Scholar] [CrossRef] [PubMed]
- Tang, Q.; Iyer, S.; Lobbardi, R.; Moore, J.C.; Chen, H.; Lareau, C.; Hebert, C.; Shaw, M.L.; Neftel, C.; Suva, M.L.; et al. Dissecting hematopoietic and renal cell heterogeneity in adult zebrafish at single-cell resolution using RNA sequencing. J. Exp. Med. 2017, 214, 2875–2887. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Das, S.; Hirano, M.; Holland, S.J.; McCurley, N.; Guo, P.; Rosenberg, C.S.; Boehm, T.; Cooper, M.D. Characterization of Lamprey IL-17 Family Members and Their Receptors. J. Immunol. 2015, 195, 5440–5451. [Google Scholar] [CrossRef] [PubMed]
- Hirano, M.; Guo, P.; McCurley, N.; Schorpp, M.; Das, S.; Boehm, T.; Cooper, M.D. Evolutionary implications of a third lymphocyte lineage in lampreys. Nature 2013, 501, 435–438. [Google Scholar] [CrossRef]
- Nyholm, S.V.; Passegue, E.; Ludington, W.B.; Voskoboynik, A.; Mitchel, K.; Weissman, I.L.; De Tomaso, A.W. Fester, a candidate allorecognition receptor from a primitive chordate. Immunity 2006, 25, 163–173. [Google Scholar] [CrossRef]
- Arizza, V.; Giaramita, F.T.; Parrinello, D.; Cammarata, M.; Parrinello, N. Cell cooperation in coelomocyte cytotoxic activity of Paracentrotus lividus coelomocytes. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2007, 147, 389–394. [Google Scholar] [CrossRef]
- Muñoz-Chápuli, R.; Carmona, R.; Guadix, J.A.; Macías, D.; Pérez-Pomares, J.M. The origin of the endothelial cells: An evo-devo approach for the invertebrate/vertebrate transition of the circulatory system. Evol. Dev. 2005, 7, 351–358. [Google Scholar] [CrossRef]
- Melcarne, C.; Lemaitre, B.; Kurant, E. Phagocytosis in Drosophila: From molecules and cellular machinery to physiology. Insect Biochem. Mol. Biol. 2019, 109, 1–12. [Google Scholar] [CrossRef]
- Lanot, R.; Zachary, D.; Holder, F.; Meister, M. Postembryonic Hematopoiesis in Drosophila. Dev. Biol. 2001, 230, 243–257. [Google Scholar] [CrossRef] [PubMed]
- Meister, M.; Lagueux, M. Drosophila blood cells. Cell. Microbiol. 2003, 5, 573–580. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, K.; Nomoto, A.M.; Nishijima, M.; Maruyama, T. Morphological and Functional Characterization of Hemocytes in the Giant ClamTridacna crocea. J. Invertebr. Pathol. 1997, 69, 105–111. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, M.S.; Lopes, K.A.R.; Leite, P.M.S.C.M.; Morais, F.V.; de Campos Velho, N.M.R. Physiological evaluation of the behavior and epidermis of freshwater planarians (Girardia tigrina and Girardia sp.) exposed to stressors. Biol. Open 2018, 7, 6. [Google Scholar] [CrossRef] [PubMed]
- Pita, L.; Hoeppner, M.P.; Ribes, M.; Hentschel, U. Differential expression of immune receptors in two marine sponges upon exposure to microbial-associated molecular patterns. Sci. Rep. 2018, 8, 16081. [Google Scholar] [CrossRef] [PubMed]
- Iismaa, S.E.; Kaidonis, X.; Nicks, A.M.; Bogush, N.; Kikuchi, K.; Naqvi, N.; Harvey, R.P.; Husain, A.; Graham, R.M. Comparative regenerative mechanisms across different mammalian tissues. NPJ Regen. Med. 2018, 3, 6. [Google Scholar] [CrossRef]
- Abarca-Buis, R.F.; Mandujano-Tinoco, E.A.; Cabrera-Wrooman, A.; Krötzsch, E. The complexity of TGFβ/activin signaling in regeneration. J. Cell Commun. Signal. 2021, 15, 7–23. [Google Scholar] [CrossRef]
- Abnave, P.; Ghigo, E. Role of the immune system in regeneration and its dynamic interplay with adult stem cells. Semin. Cell Dev. Biol. 2019, 87, 160–168. [Google Scholar] [CrossRef]
- Harty, M.; Neff, A.W.; King, M.W.; Mescher, A.L. Regeneration or scarring: An immunologic perspective. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2003, 226, 268–279. [Google Scholar] [CrossRef] [PubMed]
- Mescher, A.L.; Neff, A.W. Regenerative capacity and the developing immune system. Adv. Biochem. Eng. Biotechnol. 2005, 93, 39–66. [Google Scholar]
- Adolph, V.R.; DiSanto, S.K.; Bleacher, J.C.; Dillon, P.W.; Krummel, T.M. The potential role of the lymphocyte in fetal wound healing. J. Pediatric Surg. 1993, 28, 1316–1320. [Google Scholar] [CrossRef]
- Jameson, J.; Ugarte, K.; Chen, N.; Yachi, P.; Fuchs, E.; Boismenu, R.; Havran, W.L. A role for skin gammadelta T cells in wound repair. Science 2002, 296, 747–749. [Google Scholar] [CrossRef]
- Kimura, Y.; Madhavan, M.; Call, M.K.; Santiago, W.; Tsonis, P.A.; Lambris, J.D.; Del Rio-Tsonis, K. Expression of Complement 3 and Complement 5 in Newt Limb and Lens Regeneration. J. Immunol. 2003, 170, 2331–2339. [Google Scholar] [CrossRef] [PubMed]
- Godwin, J.W.; Rosenthal, N. Scar-free wound healing and regeneration in amphibians: Immunological influences on regenerative success. Differ. Res. Biol. Divers. 2014, 87, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Abarca-Buis, R.F.; Martínez-Jiménez, A.; Vera-Gómez, E.; Contreras-Figueroa, M.E.; Garciadiego-Cázares, D.; Paus, R.; Robles-Tenorio, A.; Krötzsch, E. Mechanisms of epithelial thickening due to IL-1 signalling blockade and TNF-α administration differ during wound repair and regeneration. Differ. Res. Biol. Divers. 2018, 99, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Mescher, A.L.; Neff, A.W.; King, M.W. Changes in the inflammatory response to injury and its resolution during the loss of regenerative capacity in developing Xenopus limbs. PLoS ONE 2013, 8, e80477. [Google Scholar] [CrossRef] [PubMed]
- Martin, P.; D’Souza, D.; Martin, J.; Grose, R.; Cooper, L.; Maki, R.; McKercher, S.R. Wound Healing in the PU.1 Null Mouse—Tissue Repair is Not Dependent on Inflammatory Cells. Curr. Biol. 2003, 13, 1122–1128. [Google Scholar] [CrossRef]
- Redd, M.J.; Cooper, L.; Wood, W.; Stramer, B.; Martin, P. Wound healing and inflammation: Embryos reveal the way to perfect repair. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2004, 359, 777–784. [Google Scholar] [CrossRef]
- Liechty, K.W.; Kim, H.B.; Adzick, N.S.; Crombleholme, T.M. Fetal wound repair results in scar formation in interleukin-10-deficient mice in a syngeneic murine model of scarless fetal wound repair. J. Pediatr. Surg. 2000, 35, 866–872, discussion 872–873. [Google Scholar] [CrossRef] [PubMed]
- Hopkinson-Woolley, J.; Hughes, D.; Gordon, S.; Martin, P. Macrophage recruitment during limb development and wound healing in the embryonic and foetal mouse. J. Cell Sci. 1994, 107, 1159–1167. [Google Scholar] [CrossRef] [PubMed]
- Yates, C.C.; Hebda, P.; Wells, A. Skin Wound Healing and Scarring: Fetal Wounds and Regenerative Restitution. Birth Defects Res. Part C Embryo Today Rev. 2012, 96, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Wilgus, T.A. Inflammation as an orchestrator of cutaneous scar formation: A review of the literature. Plast. Aesthetic Res. 2020, 7, 54. [Google Scholar] [CrossRef]
- Mescher, A.L.; Neff, A.W.; King, M.W. Inflammation and immunity in organ regeneration. Dev. Comp. Immunol. 2017, 66, 98–110. [Google Scholar] [CrossRef]
- Gourevitch, D.; Kossenkov, A.V.; Zhang, Y.; Clark, L.; Chang, C.; Showe, L.C.; Heber-Katz, E. Inflammation and Its Correlates in Regenerative Wound Healing: An Alternate Perspective. Adv. Wound Care 2014, 3, 592–603. [Google Scholar] [CrossRef] [PubMed]
- Canhamero, T.; Garcia, L.V.; De Franco, M. Acute Inflammation Loci are Involved in Wound Healing in the Mouse Ear Punch Model. Adv. Wound Care 2014, 3, 582–591. [Google Scholar] [CrossRef] [PubMed]
- Seifert, A.W.; Monaghan, J.R.; Voss, S.R.; Maden, M. Skin regeneration in adult axolotls: A blueprint for scar-free healing in vertebrates. PLoS ONE 2012, 7, e32875. [Google Scholar] [CrossRef]
- Mathew, L.K.; Sengupta, S.; Kawakami, A.; Andreasen, E.A.; Löhr, C.V.; Loynes, C.A.; Renshaw, S.A.; Peterson, R.T.; Tanguay, R.L. Unraveling tissue regeneration pathways using chemical genetics. J. Biol. Chem. 2007, 282, 35202–35210. [Google Scholar] [CrossRef]
- Li, L.; Yan, B.; Shi, Y.Q.; Zhang, W.Q.; Wen, Z.L. Live imaging reveals differing roles of macrophages and neutrophils during zebrafish tail fin regeneration. J. Biol. Chem. 2012, 287, 25353–25360. [Google Scholar] [CrossRef]
- Petrie, T.A.; Strand, N.S.; Yang, C.T.; Rabinowitz, J.S.; Moon, R.T. Macrophages modulate adult zebrafish tail fin regeneration. Development 2014, 141, 2581–2591. [Google Scholar] [CrossRef]
- King, M.W.; Neff, A.W.; Mescher, A.L. The Developing Xenopus Limb as a Model for Studies on the Balance between Inflammation and Regeneration. Anat. Rec. 2012, 295, 1552–1561. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Janeway, C.A., Jr. Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 1989, 54, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Lemaitre, B.; Nicolas, E.; Michaut, L.; Reichhart, J.M.; Hoffmann, J.A. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell 1996, 86, 973–983. [Google Scholar] [CrossRef]
- Laird, D.J.; De Tomaso, A.W.; Weissman, I.L. Stem Cells are Units of Natural Selection in a Colonial Ascidian. Cell 2005, 123, 1351–1360. [Google Scholar] [CrossRef] [PubMed]
- Weissman, I.L. Stem cells are units of natural selection for tissue formation, for germline development, and in cancer development. Proc. Natl. Acad. Sci. USA 2015, 112, 8922–8928. [Google Scholar] [CrossRef]
- Meselson, M.; Yuan, R. DNA Restriction Enzyme from E. coli. Nature 1968, 217, 1110–1114. [Google Scholar] [CrossRef] [PubMed]
- Mojica, F.J.; Díez-Villaseñor, C.; García-Martínez, J.; Soria, E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef] [PubMed]
- Lokaj, J.; John, C. Ilya Ilich Metchnikov and Paul Ehrlich: 1908 Nobel Prize winners for their research on immunity. Epidemiol. Mikrobiol. Imunol. Cas. Spol. Epidemiol. Mikrobiol. Ceske Lek. Spol. J. E. Purkyne 2008, 57, 119–124. [Google Scholar]
- Bernards, R. The Nobel Prize in Physiology or Medicine for 2006 for the discovery of RNA interference. Ned. Tijdschr. Geneeskd. 2006, 150, 2849–2853. [Google Scholar] [PubMed]
- Volchenkov, R.; Sprater, F.; Vogelsang, P.; Appel, S. The 2011 Nobel Prize in Physiology or Medicine. Scand. J. Immunol. 2012, 75, 1–4. [Google Scholar] [CrossRef]
- Westermann, L.; Neubauer, B.; Köttgen, M. Nobel Prize 2020 in Chemistry honors CRISPR: A tool for rewriting the code of life. Pflug. Arch. Eur. J. Physiol. 2021, 473, 1–2. [Google Scholar] [CrossRef]
| Animal Group | Effector Cells | Receptors and Effector Molecules |
|---|---|---|
| Mammals | (Wolff and Humeniuk 2013) [105] | (Vandendriessche, Cambier et al., 2021) [19] |
| (Rosental, Appel et al., 2012) [50] | ||
| (Hirayama, Iida et al., 2017) [20] | ||
| (Geffner 2005) [71] | ||
| Osteichthyes | (Potts and Bowman 2017) [106] | (Wei, Zhou et al., 2007) [107] |
| (Moss, Monette et al., 2009) [108] | (Kasahara and Flajnik 2019) [90] | |
| (Tang, Iyer et al., 2017) [109] | (Flajnik and Du Pasquier 2004) [75] | |
| (Flajnik 2018) [65] | ||
| Agnathans | (Han, Das et al., 2015) [110] | (Pancer, Amemiya et al., 2004) [83] |
| (Hirano, Guo et al., 2013) [111] | ||
| (Das, Li et al., 2015) [61] | ||
| (Mayer, Uinuk-ool et al., 2002) [62] | ||
| Tunicates | (Rosental, Kowarsky et al., 2018) [16] | (Voskoboynik, Newman et al., 2013) [59] |
| (Rosental, Raveh et al., 2020) [17] | (Nyholm, Passegue et al., 2006) [112] | |
| (Peronato, Franchi et al., 2020) [25] | ||
| Echinoderms | (Arizza, Giaramita et al., 2007) [113] | (Yakovenko, Donnyo et al., 2021) [32] |
| (Cooper 1980) [48] | (Chou, Lun et al., 2018) [33] | |
| (Lin, Zhang et al., 2001) [56] | ||
| Arthropods | (Muñoz-Chápuli, Carmona et al., 2005) [114] | (Melcarne, Lemaitre et al., 2019) [115] |
| (Lanot, Zachary et al., 2001) [116] | ||
| (Meister and Lagueux 2003) [117] | ||
| (Cattenoz, Sakr et al., 2020) [15] | ||
| (Csordás, Gábor et al., 2021) [14] | ||
| (Cárdenas, Dankert et al., 2004) [78] | ||
| Molluscs | (Franceschi, Cossarizza et al., 1991) [51] | (Schultz, Bu et al., 2018) [76] |
| (Nakayama, Nomoto et al., 1997) [118] | (Loker, Adema et al., 2004) [77] | |
| Sipunculids | (Boiledieu and Valembois 1977) [52] | (Flajnik and Du Pasquier 2004) [75] |
| Platyelminthes | (de Oliveira, Lopes et al., 2018) [119] | (Peiris, Hoyer et al., 2014) [28] |
| (Morita 1991) [11] | ||
| Cnidarians | (Snyder G 2021) [10] | |
| Porifera | (Mukherjee, Ray et al., 2015) [9] | (Pita, Hoeppner et al., 2018) [120] |
| (Yuen, Bayes et al., 2014) [21] | ||
| (Wiens, Korzhev et al., 2005) [22] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mandujano-Tinoco, E.A.; Sultan, E.; Ottolenghi, A.; Gershoni-Yahalom, O.; Rosental, B. Evolution of Cellular Immunity Effector Cells; Perspective on Cytotoxic and Phagocytic Cellular Lineages. Cells 2021, 10, 1853. https://doi.org/10.3390/cells10081853
Mandujano-Tinoco EA, Sultan E, Ottolenghi A, Gershoni-Yahalom O, Rosental B. Evolution of Cellular Immunity Effector Cells; Perspective on Cytotoxic and Phagocytic Cellular Lineages. Cells. 2021; 10(8):1853. https://doi.org/10.3390/cells10081853
Chicago/Turabian StyleMandujano-Tinoco, Edna Ayerim, Eliya Sultan, Aner Ottolenghi, Orly Gershoni-Yahalom, and Benyamin Rosental. 2021. "Evolution of Cellular Immunity Effector Cells; Perspective on Cytotoxic and Phagocytic Cellular Lineages" Cells 10, no. 8: 1853. https://doi.org/10.3390/cells10081853
APA StyleMandujano-Tinoco, E. A., Sultan, E., Ottolenghi, A., Gershoni-Yahalom, O., & Rosental, B. (2021). Evolution of Cellular Immunity Effector Cells; Perspective on Cytotoxic and Phagocytic Cellular Lineages. Cells, 10(8), 1853. https://doi.org/10.3390/cells10081853