Microbiota-Gut-Brain Communication in the SARS-CoV-2 Infection
Abstract
:1. SARS-CoV-2
2. General Aspects of Psychiatric Disorders and Neurodegenerative Diseases
3. Inflammation, Psychiatric Disorders, and Neurodegenerative Diseases
4. Microbiota-Gut-Brain Axis, Psychiatric Disorders, and Neurodegenerative Diseases
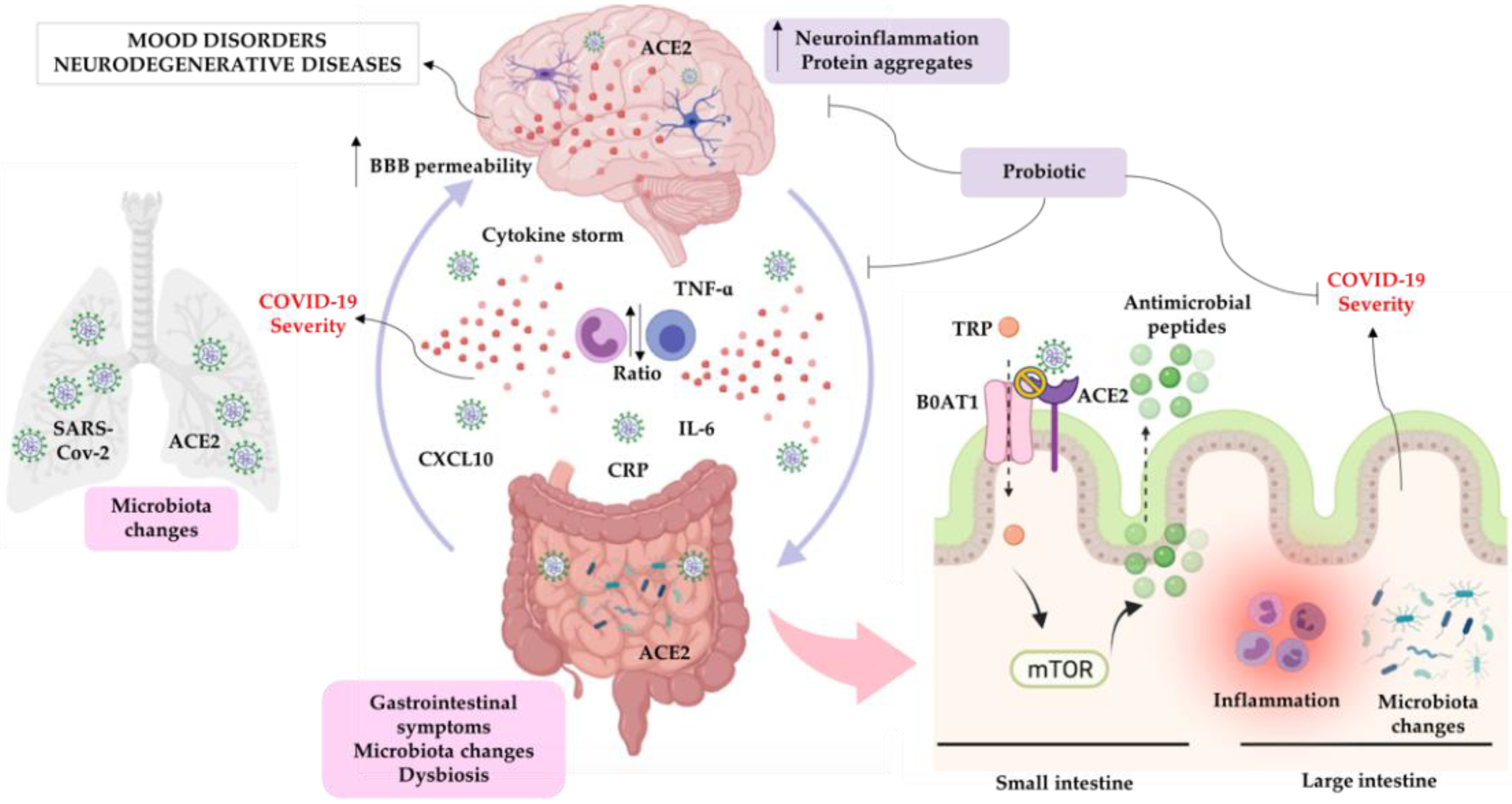
5. SARS-CoV-2 and Inflammation/Neuroinflammation
6. SARS-CoV-2 and Microbiota-Gut-Brain Axis
7. Future Directions and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. WHO Director-General’s Opening Remarks at the Media Briefing on COVID-19—11 March 2020. Available online: https://www.who.int/director-general/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-COVID-19---11-march-2020 (accessed on 30 June 2021).
- WHO. Weekly Epidemiological Update on COVID-19—15 June 2021. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19---15-june-2021 (accessed on 29 June 2021).
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e8. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Madhavan, M.V.; Sehgal, K.; Nair, N.; Mahajan, S.; Sehrawat, T.S.; Bikdeli, B.; Ahluwalia, N.; Ausiello, J.C.; Wan, E.Y.; et al. Extrapulmonary manifestations of COVID-19. Nat. Med. 2020, 26, 1017–1032. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Chen, W.; Zhou, Y.-S.; Lian, J.-Q.; Zhang, Z.; Du, P.; Gong, L.; Zhang, Y.; Cui, H.-Y.; Geng, J.-J.; et al. SARS-CoV-2 invades host cells via a novel route: CD147-spike protein. bioRxiv 2020. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Chen, W.; Zhang, Z.; Deng, Y.; Lian, J.Q.; Du, P.; Wei, D.; Zhang, Y.; Sun, X.X.; Gong, L.; et al. CD147-spike protein is a novel route for SARS-CoV-2 infection to host cells. Signal. Transduct. Target. 2020, 5, 283. [Google Scholar] [CrossRef]
- Ragab, D.; Salah Eldin, H.; Taeimah, M.; Khattab, R.; Salem, R. The COVID-19 Cytokine Storm; What We Know So Far. Front. Immunol. 2020, 11, 1446. [Google Scholar] [CrossRef]
- Sinha, P.; Matthay, M.A.; Calfee, C.S. Is a “cytokine Storm” Relevant to COVID-19? JAMA Intern. Med. 2020, 180, 1152–1154. [Google Scholar] [CrossRef]
- Mangalmurti, N.; Hunter, C.A. Cytokine Storms: Understanding COVID-19. Immunity 2020, 53, 19–25. [Google Scholar] [CrossRef]
- Fernández-Rodríguez, A.; Casas, I.; Culebras, E.; Morilla, E.; Cohen, M.C.; Alberola, J. COVID-19 and post-mortem microbiological studies. Span. J. Leg Med. 2020, 46, 127–138. [Google Scholar] [CrossRef]
- Johnson, K.D.; Harris, C.; Cain, J.K.; Hummer, C.; Goyal, H.; Perisetti, A. Pulmonary and Extra-Pulmonary Clinical Manifestations of COVID-19. Front. Med. 2020, 7, 526. [Google Scholar] [CrossRef]
- Hartung, H.P.; Aktas, O. COVID-19 and management of neuroimmunological disorders. Nat. Rev. Neurol. 2020, 16, 347–348. [Google Scholar] [CrossRef]
- Pezzini, A.; Padovani, A. Lifting the mask on neurological manifestations of COVID-19. Nat. Rev. Neurol. 2020, 16, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Torales, J.; O’Higgins, M.; Castaldelli-Maia, J.M.; Ventriglio, A. The outbreak of COVID-19 coronavirus and its impact on global mental health. Int. J. Soc. Psychiatry 2020, 66, 317–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnamoorthy, Y.; Nagarajan, R.; Saya, G.K.; Menon, V. Prevalence of psychological morbidities among general population, healthcare workers and COVID-19 patients amidst the COVID-19 pandemic: A systematic review and meta-analysis. Psychiatry Res. 2020, 293, 113382. [Google Scholar] [CrossRef] [PubMed]
- Del Brutto, O.H.; Wu, S.; Mera, R.M.; Costa, A.F.; Recalde, B.Y.; Issa, N.P. Cognitive decline among individuals with history of mild symptomatic SARS-CoV-2 infection: A longitudinal prospective study nested to a population cohort. Eur. J. Neurol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Fiani, B.; Covarrubias, C.; Desai, A.; Sekhon, M.; Jarrah, R. A Contemporary Review of Neurological Sequelae of COVID-19. Front. Neurol. 2020, 11, 640. [Google Scholar] [CrossRef]
- Bostanciklioglu, M. Temporal correlation between neurological and gastrointestinal symptoms of sars-cov-2. Inflamm. Bowel Dis. 2020, 26, E89–E91. [Google Scholar]
- Chaves Andrade, M.; Souza de Faria, R.; Avelino Mota Nobre, S. COVID-19: Can the symptomatic SARS-CoV-2 infection affect the homeostasis of the gut-brain-microbiota axis? Med. Hypotheses 2020, 144, 110206. [Google Scholar] [CrossRef] [PubMed]
- Hyman, S.E. A glimmer of light for neuropsychiatric disorders. Nature 2008, 455, 890–893. [Google Scholar] [CrossRef]
- WHO. Depression. Available online: https://www.who.int/news-room/fact-sheets/detail/depression (accessed on 15 April 2020).
- Malhi, G.S.; Mann, J.J. Depression. Lancet 2018, 392, 2299–2312. [Google Scholar] [CrossRef]
- Czarny, P.; Wigner, P.; Galecki, P.; Sliwinski, T. The interplay between inflammation, oxidative stress, DNA damage, DNA repair and mitochondrial dysfunction in depression. Prog. Neuro-Psychopharmacol. Biol. Psychiatry 2018, 80, 309–321. [Google Scholar] [CrossRef]
- Levy, M.J.F.; Boulle, F.; Steinbusch, H.W.; van den Hove, D.L.A.; Kenis, G.; Lanfumey, L. Neurotrophic factors and neuroplasticity pathways in the pathophysiology and treatment of depression. Psychopharmacology 2018, 235, 2195–2220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, A.H.; Maletic, V.; Raison, C.L. Inflammation and Its Discontents: The Role of Cytokines in the Pathophysiology of Major Depression. Biol. Psychiatry 2009, 65, 732–741. [Google Scholar] [CrossRef] [Green Version]
- Lyketsos, C.G.; Carrillo, M.C.; Ryan, J.M.; Khachaturian, A.S.; Trzepacz, P.; Amatniek, J.; Cedarbaum, J.; Brashear, R.; Miller, D.S. Neuropsychiatric symptoms in Alzheimer’s disease. Alzheimer’s Dement. 2011, 7, 532–539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poewe, W.; Seppi, K.; Tanner, C.M.; Halliday, G.M.; Brundin, P.; Volkmann, J.; Schrag, A.E.; Lang, A.E. Parkinson disease. Nat. Rev. Dis. Prim. 2017, 3, 1–21. [Google Scholar] [CrossRef]
- Barnham, K.J.; Masters, C.L.; Bush, A.I. Neurodegenerative diseases and oxidatives stress. Nat. Rev. Drug Discov. 2004, 3, 205–214. [Google Scholar] [CrossRef]
- Soto, C. Unfolding the role of protein misfolding in neurodegenerative diseases. Nat. Rev. Neurosci. 2003, 4, 49–60. [Google Scholar] [CrossRef]
- Heneka, M.T.; Carson, M.J.; El Khoury, J.; Landreth, G.E.; Brosseron, F.; Feinstein, D.L.; Jacobs, A.H.; Wyss-Coray, T.; Vitorica, J.; Ransohoff, R.M.; et al. Neuroinflammation in Alzheimer’s disease. Lancet Neurol. 2015, 14, 388–405. [Google Scholar] [CrossRef] [Green Version]
- Hirsch, E.C.; Hunot, S. Neuroinflammation in Parkinson’s disease: A target for neuroprotection? Lancet Neurol. 2009, 8, 382–397. [Google Scholar] [CrossRef]
- Lau, A.; Tymianski, M. Glutamate receptors, neurotoxicity and neurodegeneration. Pflug. Arch. Eur. J. Physiol. 2010, 460, 525–542. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Singh, A.; Ekavali. A review on Alzheimer’s disease pathophysiology and its management: An update. Pharm. Rep. 2015, 67, 195–203. [Google Scholar] [CrossRef]
- Furman, D.; Campisi, J.; Verdin, E.; Carrera-Bastos, P.; Targ, S.; Franceschi, C.; Ferrucci, L.; Gilroy, D.W.; Fasano, A.; Miller, G.W.; et al. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 2019, 25, 1822–1832. [Google Scholar] [CrossRef]
- Nathan, C.; Ding, A. Nonresolving Inflammation. Cell 2010, 140, 871–882. [Google Scholar] [CrossRef] [Green Version]
- Gisterå, A.; Hansson, G.K. The immunology of atherosclerosis. Nat. Rev. Nephrol. 2017, 13, 368–380. [Google Scholar] [CrossRef]
- Wellen, K.E.; Hotamisligil, G.S. Inflammation, stress, and diabetes. J. Clin. Investig. 2005, 115, 1111–1119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taniguchi, K.; Karin, M. NF-B, inflammation, immunity and cancer: Coming of age. Nat. Rev. Immunol. 2018, 18, 309–324. [Google Scholar] [CrossRef]
- Miller, A.H.; Raison, C.L. The role of inflammation in depression: From evolutionary imperative to modern treatment target. Nat. Rev. Immunol. 2016, 16, 22–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perry, V.H.; Cunningham, C.; Holmes, C. Systemic infections and inflammation affect chronic neurodegeneration. Nat. Rev. Immunol. 2007, 7, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Howren, M.B.; Lamkin, D.M.; Suls, J. Associations of depression with c-reactive protein, IL-1, and IL-6: A meta-analysis. Psychosom Med. 2009, 71, 171–186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dowlati, Y.; Herrmann, N.; Swardfager, W.; Liu, H.; Sham, L.; Reim, E.K.; Lanctôt, K.L. A Meta-Analysis of Cytokines in Major Depression. Biol. Psychiatry 2010, 67, 446–457. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.J.; Au, B.; Ollis, L.; Schmitz, N. The association between C-reactive protein, Interleukin-6 and depression among older adults in the community: A systematic review and meta-analysis. Exp. Gerontol. 2018, 102, 109–132. [Google Scholar] [CrossRef]
- Köhler, C.A.; Freitas, T.H.; Maes, M.; de Andrade, N.Q.; Liu, C.S.; Fernandes, B.S.; Stubbs, B.; Solmi, M.; Veronese, N.; Herrmann, N.; et al. Peripheral cytokine and chemokine alterations in depression: A meta-analysis of 82 studies. Acta Psychiatr. Scand. 2017, 135, 373–387. [Google Scholar] [CrossRef] [PubMed]
- Osimo, E.F.; Baxter, L.J.; Lewis, G.; Jones, P.B.; Khandaker, G.M. Prevalence of low-grade inflammation in depression: A systematic review and meta-Analysis of CRP levels. Psychol. Med. 2019, 49, 1958–1970. [Google Scholar] [CrossRef]
- Enache, D.; Pariante, C.M.; Mondelli, V. Markers of central inflammation in major depressive disorder: A systematic review and meta-analysis of studies examining cerebrospinal fluid, positron emission tomography and post-mortem brain tissue. Brain Behav. Immun. 2019, 81, 24–40. [Google Scholar] [CrossRef]
- Lai, K.S.P.; Liu, C.S.; Rau, A.; Lanctôt, K.L.; Köhler, C.A.; Pakosh, M.; Carvalho, A.F.; Herrmann, N. Peripheral inflammatory markers in Alzheimer’s disease: A systematic review and meta-analysis of 175 studies. J. Neurol. Neurosurg. Psychiatry 2017, 88, 876–882. [Google Scholar] [CrossRef] [PubMed]
- Qin, X.Y.; Zhang, S.P.; Cao, C.; Loh, Y.P.; Cheng, Y. Aberrations in peripheral inflammatory cytokine levels in Parkinson disease: A systematic review and meta-analysis. JAMA Neurol. 2016, 73, 1316–1324. [Google Scholar] [CrossRef]
- Chen, X.; Hu, Y.; Cao, Z.; Liu, Q.; Cheng, Y. Cerebrospinal fluid inflammatory cytokine aberrations in Alzheimer’s disease, Parkinson’s disease and amyotrophic lateral sclerosis: A systematic review and meta-analysis. Front. Immunol. 2018, 9, 2122. [Google Scholar] [CrossRef] [Green Version]
- Dantzer, R.; O’Connor, J.C.; Freund, G.G.; Johnson, R.W.; Kelley, K.W. From inflammation to sickness and depression: When the immune system subjugates the brain. Nat. Rev. Neurosci. 2008, 9, 46–56. [Google Scholar] [CrossRef] [Green Version]
- Kaufmann, F.N.; Costa, A.P.; Ghisleni, G.; Diaz, A.P.; Rodrigues, A.L.S.; Peluffo, H.; Kaster, M.P. NLRP3 inflammasome-driven pathways in depression: Clinical and preclinical findings. Brain Behav. Immun. 2017, 64, 367–383. [Google Scholar] [CrossRef] [PubMed]
- Haruwaka, K.; Ikegami, A.; Tachibana, Y.; Ohno, N.; Konishi, H.; Hashimoto, A.; Matsumoto, M.; Kato, D.; Ono, R.; Kiyama, H.; et al. Dual microglia effects on blood brain barrier permeability induced by systemic inflammation. Nat. Commun. 2019, 10, 5816. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kealy, J.; Greene, C.; Campbell, M. Blood-brain barrier regulation in psychiatric disorders. Neurosci. Lett. 2020, 726, 133664. [Google Scholar] [CrossRef] [Green Version]
- Najjar, S.; Pearlman, D.M.; Devinsky, O.; Najjar, A.; Zagzag, D. Neurovascular unit dysfunction with blood-brain barrier hyperpermeability contributes to major depressive disorder: A review of clinical and experimental evidence. J. Neuroinflamm. 2013, 10, 142. [Google Scholar] [CrossRef] [Green Version]
- Sweeney, M.D.; Sagare, A.P.; Zlokovic, B.V. Blood–brain barrier breakdown in Alzheimer disease and other neurodegenerative disorders. Nat. Rev. Neurol. 2018, 14, 133–150. [Google Scholar] [CrossRef]
- Bowman, G.L.; Kaye, J.A.; Moore, M.; Waichunas, D.; Carlson, N.E.; Quinn, J.F. Blood-brain barrier impairment in Alzheimer disease: Stability and functional significance. Neurology 2007, 68, 1809–1814. [Google Scholar] [CrossRef] [Green Version]
- Berk, M.; Williams, L.J.; Jacka, F.N.; O’Neil, A.; Pasco, J.A.; Moylan, S.; Allen, N.B.; Stuart, A.L.; Hayley, A.C.; Byrne, M.L.; et al. So depression is an inflammatory disease, but where does the inflammation come from? BMC Med. 2013, 11, 200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gubert, C.; Kong, G.; Renoir, T.; Hannan, A.J. Exercise, diet and stress as modulators of gut microbiota: Implications for neurodegenerative diseases. Neurobiol. Dis. 2020, 134, 104621. [Google Scholar] [CrossRef]
- Kamada, N.; Seo, S.U.; Chen, G.Y.; Núñez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 2013, 13, 321–335. [Google Scholar] [CrossRef] [PubMed]
- Maslowski, K.M.; Vieira, A.T.; Ng, A.; Kranich, J.; Sierro, F.; Di, Y.; Schilter, H.C.; Rolph, M.S.; MacKay, F.; Artis, D.; et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 2009, 461, 1282–1286. [Google Scholar] [CrossRef]
- Morais, L.H.; Schreiber, H.L.; Mazmanian, S.K. The gut microbiota–brain axis in behaviour and brain disorders. Nat. Rev. Microbiol. 2021, 19, 241–255. [Google Scholar] [CrossRef] [PubMed]
- Sanada, K.; Nakajima, S.; Kurokawa, S.; Barceló-Soler, A.; Ikuse, D.; Hirata, A.; Yoshizawa, A.; Tomizawa, Y.; Salas-Valero, M.; Noda, Y.; et al. Gut microbiota and majore depressive disorder: A systematic review and meta-analysis. J. Affect. Disord. 2020, 266, 1–13. [Google Scholar] [CrossRef]
- Foster, J.A.; McVey Neufeld, K.A. Gut-brain axis: How the microbiome influences anxiety and depression. Trends Neurosci. 2013, 36, 305–312. [Google Scholar] [CrossRef]
- Mayer, E.A. Gut feelings: The emerging biology of gut-brain communication. Nat. Rev. Neurosci. 2011, 12, 453–466. [Google Scholar] [CrossRef] [PubMed]
- Cryan, J.F.; O’riordan, K.J.; Cowan, C.S.M.; Sandhu, K.V.; Bastiaanssen, T.F.S.; Boehme, M.; Codagnone, M.G.; Cussotto, S.; Fulling, C.; Golubeva, A.V.; et al. The microbiota-gut-brain axis. Physiol. Rev. 2019, 99, 1877–2013. [Google Scholar] [CrossRef] [PubMed]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut microbiota in health and disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [Green Version]
- Stilling, R.M.; Dinan, T.G.; Cryan, J.F. Microbial genes, brain & behaviour—Epigenetic regulation of the gut-brain axis. GenesBrain Behav. 2014, 13, 69–86. [Google Scholar]
- Carabotti, M.; Scirocco, A.; Maselli, M.A.; Severi, C. The gut-brain axis: Interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 2015, 28, 203–209. [Google Scholar]
- Rao, M.; Gershon, M.D. The bowel and beyond: The enteric nervous system in neurological disorders. Nat. Rev. Gastroenterol. Hepatol. 2016, 13, 517–528. [Google Scholar] [CrossRef] [Green Version]
- Breit, S.; Kupferberg, A.; Rogler, G.; Hasler, G. Vagus nerve as modulator of the brain-gut axis in psychiatric and inflammatory disorders. Front. Psychiatry 2018, 9, 44. [Google Scholar] [CrossRef] [Green Version]
- Mittal, R.; Debs, L.H.; Patel, A.P.; Nguyen, D.; Patel, K.; O’Connor, G.; Grati, M.; Mittal, J.; Yan, D.; Eshraghi, A.A.; et al. Neurotransmitters: The Critical Modulators Regulating Gut–Brain Axis. J. Cell Physiol. 2017, 232, 2359–2372. [Google Scholar] [CrossRef] [Green Version]
- Strandwitz, P. Neurotransmitter modulation by the gut microbiota. Brain Res. 2018, 1693, 128–133. [Google Scholar] [CrossRef]
- Konturek, P.C.; Brzozowski, T.; Konturek, S.J. Stress and the gut: Pathophysiology, clinical consequences, diagnostic approach and treatment options. J. Physiol. Pharm. 2011, 62, 591–599. [Google Scholar]
- Vodička, M.; Ergang, P.; Hrnčíř, T.; Mikulecká, A.; Kvapilová, P.; Vagnerová, K.; Šestáková, B.; Fajstová, A.; Hermanová, P.; Hudcovic, T.; et al. Microbiota affects the expression of genes involved in HPA axis regulation and local metabolism of glucocorticoids in chronic psychosocial stress. Brain Behav. Immun. 2018, 73, 615–624. [Google Scholar] [CrossRef]
- Sudo, N.; Chida, Y.; Aiba, Y.; Sonoda, J.; Oyama, N.; Yu, X.N.; Kubo, C.; Koga, Y. Postnatal microbial colonization programs the hypothalamic-pituitary-adrenal system for stress response in mice. J. Physiol. 2004, 558, 263–275. [Google Scholar] [CrossRef]
- Yoo, B.B.; Mazmanian, S.K. The Enteric Network: Interactions between the Immune and Nervous Systems of the Gut. Immunity 2017, 46, 910–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Powell, N.; Walker, M.M.; Talley, N.J. The mucosal immune system: Master regulator of bidirectional gut-brain communications. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 143–159. [Google Scholar] [CrossRef]
- Belkaid, Y.; Hand, T.W. Role of the microbiota in immunity and inflammation. Cell 2014, 157, 121–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dalile, B.; Van Oudenhove, L.; Vervliet, B.; Verbeke, K. The role of short-chain fatty acids in microbiota-gut-brain communication. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 461–478. [Google Scholar] [CrossRef]
- Maqsood, R.; Stone, T.W. The Gut-Brain Axis, BDNF, NMDA and CNS Disorders. Neurochem. Res. 2016, 41, 2819–2835. [Google Scholar] [CrossRef] [PubMed]
- Savignac, H.M.; Corona, G.; Mills, H.; Chen, L.; Spencer, J.P.E.; Tzortzis, G.; Burnet, P.W.J. Prebiotic feeding elevates central brain derived neurotrophic factor, N-methyl-d-aspartate receptor subunits and d-serine. Neurochem. Int. 2013, 63, 756–764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonaz, B.; Bazin, T.; Pellissier, S. The vagus nerve at the interface of the microbiota-gut-brain axis. Front. Neurosci. 2018, 12, 49. [Google Scholar] [CrossRef] [Green Version]
- Obrenovich, M. Leaky Gut, Leaky Brain? Microorganisms 2018, 6, 107. [Google Scholar] [CrossRef] [Green Version]
- Camilleri, M. Leaky gut: Mechanisms, measurement and clinical implications in humans. Gut 2019, 68, 1516–1526. [Google Scholar] [CrossRef]
- Kelly, J.R.; Keane, V.O.; Cryan, J.F.; Clarke, G.; Dinan, T.G. Mood and Microbes: Gut to Brain Communication in Depression. Gastroenterol. Clin. North. Am. 2019, 48, 389–405. [Google Scholar] [CrossRef]
- Manosso, L.M.; Lin, J.; Carlessi, A.S.; Recco, K.C.C.; Quevedo, J.; Gonçalves, C.L.; Réus, G.Z. Sex-related patterns of the gut-microbiota-brain axis in the neuropsychiatric conditions. Brain Res. Bull. 2021, 171, 196–208. [Google Scholar] [CrossRef]
- Fond, G.; Loundou, A.; Hamdani, N.; Boukouaci, W.; Dargel, A.; Oliveira, J.; Roger, M.; Tamouza, R.; Leboyer, M.; Boyer, L. Anxiety and depression comorbidities in irritable bowel syndrome (IBS): A systematic review and meta-analysis. Eur. Arch. Psychiatry Clin. Neurosci. 2014, 264, 651–660. [Google Scholar] [CrossRef]
- Mikocka-Walus, A.; Knowles, S.R.; Keefer, L.; Graff, L. Controversies Revisited: A Systematic Review of the Comorbidity of Depression and Anxiety with Inflammatory Bowel Diseases. Inflamm. Bowel Dis. 2016, 22, 752–762. [Google Scholar] [CrossRef] [PubMed]
- Maes, M.; Kubera, M.; Leunis, J.-C. The gut-brain barrier in major depression: Intestinal mucosal dysfunction with an increased translocation of LPS from gram negative enterobacteria (leaky gut) plays a role in the inflammatory pathophysiology of depression. Neuroendocr. Lett. 2008, 29, 117–124. [Google Scholar]
- Kelly, J.R.; Borre, Y.; O’ Brien, C.; Patterson, E.; El Aidy, S.; Deane, J.; Kennedy, P.J.; Beers, S.; Scott, K.; Moloney, G.; et al. Transferring the blues: Depression-associated gut microbiota induces neurobehavioural changes in the rat. J. Psychiatr. Res. 2016, 82, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Ling, Z.; Zhang, Y.; Mao, H.; Ma, Z.; Yin, Y.; Wang, W.; Tang, W.; Tan, Z.; Shi, J.; et al. Altered fecal microbiota composition in patients with major depressive disorder. Brain Behav. Immun. 2015, 48, 186–194. [Google Scholar] [CrossRef] [Green Version]
- Naseribafrouei, A.; Hestad, K.; Avershina, E.; Sekelja, M.; Linløkken, A.; Wilson, R.; Rudi, K. Correlation between the human fecal microbiota and depression. Neurogastroenterol. Motil 2014, 26, 1155–1162. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.T.; Walsh, R.F.L.; Sheehan, A.E. Prebiotics and probiotics for depression and anxiety: A systematic review and meta-analysis of controlled clinical trials. Neurosci. Biobehav. Rev. 2019, 102, 13–23. [Google Scholar] [CrossRef]
- Houser, M.C.; Tansey, M.G. The gut-brain axis: Is intestinal inflammation a silent driver of Parkinson’s disease pathogenesis? NPJ Park Dis. 2017, 3, 3. [Google Scholar] [CrossRef]
- Westfall, S.; Lomis, N.; Kahouli, I.; Dia, S.Y.; Singh, S.P.; Prakash, S. Microbiome, probiotics and neurodegenerative diseases: Deciphering the gut brain axis. Cell Mol. Life Sci. 2017, 74, 3769–3787. [Google Scholar] [CrossRef] [PubMed]
- Adams-Carr, K.L.; Bestwick, J.P.; Shribman, S.; Lees, A.; Schrag, A.; Noyce, A.J. Constipation preceding Parkinson’s disease: A systematic review and meta-analysis. J. Neurol. Neurosurg. Psychiatry 2016, 87, 710–716. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.H.; Lin, C.L.; Kao, C.H. Irritable bowel syndrome is associated with an increased risk of dementia: A nationwide population-based study. PLoS ONE 2016, 11, e0144589. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, H.E.; Bai, Y.M.; Tsai, S.J.; Su, T.P.; Chen, T.J.; Wang, Y.P.; Chen, M.H. Inflammatory bowel disease is associated with higher dementia risk: A nationwide longitudinal study. Gut 2021, 70, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Barichella, M.; Severgnini, M.; Cilia, R.; Cassani, E.; Bolliri, C.; Caronni, S.; Ferri, V.; Cancello, R.; Ceccarani, C.; Faierman, S.; et al. Unraveling gut microbiota in Parkinson’s disease and atypical parkinsonism. Mov. Disord. 2019, 34, 396–405. [Google Scholar] [CrossRef]
- Vogt, N.M.; Kerby, R.L.; Dill-McFarland, K.A.; Harding, S.J.; Merluzzi, A.P.; Johnson, S.C.; Carlsson, C.M.; Asthana, S.; Zetterberg, H.; Blennow, K.; et al. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 2017, 7, 13537. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Dong, X.; Chen, M.; Zou, Z. Efficacy of probiotics on cognition, and biomarkers of inflammation and oxidative stress in adults with Alzheimer’s disease or mild cognitive impairment-A meta-analysis of randomized controlled trials. Aging 2020, 12, 4010–4039. [Google Scholar]
- Tan, A.H.; Lim, S.Y.; Chong, K.K.; A Manap, M.A.A.; Hor, J.W.; Lim, J.L.; Low, S.C.; Chong, C.W.; Mahadeva, S.; Lang, A.E. Probiotics for Constipation in Parkinson Disease: A Randomized Placebo-Controlled Study. Neurology 2021, 96, e772–e782. [Google Scholar]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef]
- Hussman, J.P. Cellular and Molecular Pathways of COVID-19 and Potential Points of Therapeutic Intervention. Front. Pharm. 2020, 11, 1169. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Tay, M.Z.; Poh, C.M.; Rénia, L.; MacAry, P.A.; Ng, L.F.P. The trinity of COVID-19: Immunity, inflammation and intervention. Nat. Rev. Immunol. 2020, 20, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Hume, A.J.; Abo, K.M.; Werder, R.B.; Villacorta-Martin, C.; Alysandratos, K.D.; Beermann, M.L.; Simone-Roach, C.; Lindstrom-Vautrin, J.; Olejnik, J.; et al. SARS-CoV-2 Infection of Pluripotent Stem Cell-Derived Human Lung Alveolar Type 2 Cells Elicits a Rapid Epithelial-Intrinsic Inflammatory Response. Cell Stem Cell 2020, 27, 962–973.e7. [Google Scholar] [CrossRef]
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front. Immunol. 2020, 11, 827. [Google Scholar] [CrossRef]
- Chen, G.; Wu, D.; Guo, W.; Cao, Y.; Huang, D.; Wang, H.; Wang, T.; Zhang, X.; Chen, H.; Yu, H.; et al. Clinical and immunological features of severe and moderate coronavirus disease 2019. J. Clin. Investig. 2020, 130, 2620–2629. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Li, S.; Liu, J.; Liang, B.; Wang, X.; Wang, H.; Li, W.; Tong, Q.; Yi, J.; Zhao, L.; et al. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine 2020, 55, 102763. [Google Scholar] [CrossRef]
- Chen, X.; Zhao, B.; Qu, Y.; Chen, Y.; Xiong, J.; Feng, Y.; Men, D.; Huang, Q.; Liu, Y.; Yang, B.; et al. Detectable Serum Severe Acute Respiratory Syndrome Coronavirus 2 Viral Load (RNAemia) Is Closely Correlated with Drastically Elevated Interleukin 6 Level in Critically Ill Patients with Coronavirus Disease 2019. Clin. Infect. Dis. 2020, 71, 1937–1942. [Google Scholar] [CrossRef]
- Leisman, D.E.; Ronner, L.; Pinotti, R.; Taylor, M.D.; Sinha, P.; Calfee, C.S.; Hirayama, A.V.; Mastroiani, F.; Turtle, C.J.; Harhay, M.O.; et al. Cytokine elevation in severe and critical COVID-19: A rapid systematic review, meta-analysis, and comparison with other inflammatory syndromes. Lancet Respir. Med. 2020, 8, 1233–1244. [Google Scholar] [CrossRef]
- Henry, B.M.; De Oliveira, M.H.S.; Benoit, S.; Plebani, M.; Lippi, G. Hematologic, biochemical and immune biomarker abnormalities associated with severe illness and mortality in coronavirus disease 2019 (COVID-19): A meta-analysis. Clin. Chem Lab. Med. 2020, 58, 1021–1028. [Google Scholar] [CrossRef] [Green Version]
- Florindo, H.F.; Kleiner, R.; Vaskovich-Koubi, D.; Acúrcio, R.C.; Carreira, B.; Yeini, E.; Tiram, G.; Liubomirski, Y.; Satchi-Fainaro, R. Immune-mediated approaches against COVID-19. Nat. Nanotechnol. 2020, 15, 630–645. [Google Scholar] [CrossRef]
- Xia, H.; Lazartigues, E. Angiotensin-converting enzyme 2 in the brain: Properties and future directions. J. Neurochem. 2008, 107, 1482–1494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zubair, A.S.; McAlpine, L.S.; Gardin, T.; Farhadian, S.; Kuruvilla, D.E.; Spudich, S. Neuropathogenesis and neurologic manifestations of the coronaviruses in the age of coronavirus disease 2019: A review. JAMA Neurol. 2020, 77, 1018–1027. [Google Scholar] [CrossRef]
- Ribeiro, D.E.; Oliveira-Giacomelli, Á.; Glaser, T.; Arnaud-Sampaio, V.F.; Andrejew, R.; Dieckmann, L.; Baranova, J.; Lameu, C.; Ratajczak, M.Z.; Ulrich, H. Hyperactivation of P2X7 receptors as a culprit of COVID-19 neuropathology. Mol. Psychiatry 2021, 26, 1044–1059. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Li, W.; Bao, J.; Peng, Q.; Wen, D.; Wang, J.; Sun, B. The expression of SARS-CoV-2 receptor ACE2 and CD147, and protease TMPRSS2 in human and mouse brain cells and mouse brain tissues. Biochem. Biophys. Res. Commun. 2020, 533, 867–871. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Meng, S.; Wang, L.; Wang, Y.; Chen, T.; Wang, C. Suppression of oxLDL-induced MMP-9 and EMMPRIN expression by berberine via inhibition of NF-κB activation in human THP-1 macrophages. Anat. Rec. 2012, 295, 78–86. [Google Scholar] [CrossRef]
- Yurchenko, V.; Constant, S.; Eisenmesser, E.; Bukrinsky, M. Cyclophilin-CD147 interactions: A new target for anti-inflammatory therapeutics. Clin. Exp. Immunol. 2010, 160, 305–317. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.C.; Bai, W.Z.; Hashikawa, T. The neuroinvasive potential of SARS-CoV2 may play a role in the respiratory failure of COVID-19 patients. J. Med. Virol. 2020, 92, 552–555. [Google Scholar] [CrossRef]
- Desforges, M.; Le Coupanec, A.; Dubeau, P.; Bourgouin, A.; Lajoie, L.; Dubé, M.; Talbot, P.J. Human coronaviruses and other respiratory viruses: Underestimated opportunistic pathogens of the central nervous system? Viruses 2019, 12, 14. [Google Scholar] [CrossRef] [Green Version]
- Najjar, S.; Pahlajani, S.; De Sanctis, V.; Stern, J.N.H.; Najjar, A.; Chong, D. Neurovascular Unit Dysfunction and Blood–Brain Barrier Hyperpermeability Contribute to Schizophrenia Neurobiology: A Theoretical Integration of Clinical and Experimental Evidence. Front. Psychiatry 2017, 8, 83. [Google Scholar] [CrossRef] [PubMed]
- Mao, L.; Jin, H.; Wang, M.; Hu, Y.; Chen, S.; He, Q.; Chang, J.; Hong, C.; Zhou, Y.; Wang, D.; et al. Neurologic Manifestations of Hospitalized Patients with Coronavirus Disease 2019 in Wuhan, China. JAMA Neurol. 2020, 77, 683–690. [Google Scholar] [CrossRef] [Green Version]
- John, G.R.; Lee, S.C.; Brosnan, C.F. Cytokines: Powerful regulators of glial cell activation. Neuroscientist 2003, 9, 10–22. [Google Scholar] [CrossRef]
- Dantzer, R. Neuroimmune interactions: From the brain to the immune system and vice versa. Physiol. Rev. 2018, 98, 477–504. [Google Scholar] [CrossRef] [PubMed]
- Shigemoto-Mogami, Y.; Hoshikawa, K.; Sato, K. Activated microglia disrupt the blood-brain barrier and induce chemokines and cytokines in a rat in vitro model. Front. Cell Neurosci. 2018, 12, 494. [Google Scholar] [CrossRef] [PubMed]
- Colombo, E.; Farina, C. Astrocytes: Key Regulators of Neuroinflammation. Trends Immunol. 2016, 37, 608–620. [Google Scholar] [CrossRef]
- Glass, C.K.; Saijo, K.; Winner, B.; Marchetto, M.C.; Gage, F.H. Mechanisms Underlying Inflammation in Neurodegeneration. Cell 2010, 140, 918–934. [Google Scholar] [CrossRef] [Green Version]
- Leng, F.; Edison, P. Neuroinflammation and microglial activation in Alzheimer disease: Where do we go from here? Nat. Rev. Neurol. 2021, 17, 157–172. [Google Scholar] [CrossRef]
- Tsivgoulis, G.; Palaiodimou, L.; Katsanos, A.H.; Caso, V.; Köhrmann, M.; Molina, C.; Cordonnier, C.; Fischer, U.; Kelly, P.; Sharma, V.K.; et al. Neurological manifestations and implications of COVID-19 pandemic. Adv. Neurol. Disord. 2020, 13, 1756286420932036. [Google Scholar] [CrossRef]
- Deng, J.; Zhou, F.; Hou, W.; Silver, Z.; Wong, C.Y.; Chang, O.; Huang, E.; Zuo, Q.K. The prevalence of depression, anxiety, and sleep disturbances in COVID-19 patients: A meta-analysis. Ann. N. Y. Acad Sci. 2021, 1486, 90–111. [Google Scholar] [CrossRef]
- Heneka, M.T.; Golenbock, D.; Latz, E.; Morgan, D.; Brown, R. Immediate and long-term consequences of COVID-19 infections for the development of neurological disease. Alzheimer’s Res. 2020, 12, 69. [Google Scholar] [CrossRef] [PubMed]
- Cilia, R.; Bonvegna, S.; Straccia, G.; Andreasi, N.G.; Elia, A.E.; Romito, L.M.; Devigili, G.; Cereda, E.; Eleopra, R. Effects of COVID-19 on Parkinson’s Disease Clinical Features: A Community-Based Case-Control Study. Mov. Disord. 2020, 35, 1287–1292. [Google Scholar] [CrossRef]
- Kappelmann, N.; Dantzer, R.; Khandaker, G.M. Interleukin-6 as potential mediator of long-term neuropsychiatric symptoms of COVID-19. Psychoneuroendocrinology 2021, 131, 105295. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Lian, J.S.; Hu, J.H.; Gao, J.; Zheng, L.; Zhang, Y.M.; Hao, S.R.; Jia, H.Y.; Cai, H.; Zhang, X.L.; et al. Epidemiological, clinical and virological characteristics of 74 cases of coronavirus-infected disease 2019 (COVID-19) with gastrointestinal symptoms. Gut 2020, 69, 1002–1009. [Google Scholar] [CrossRef] [Green Version]
- Galanopoulos, M.; Gkeros, F.; Doukatas, A.; Karianakis, G.; Pontas, C.; Tsoukalas, N.; Viazis, N.; Liatsos, C.; Mantzaris, G.J. COVID-19 pandemic: Pathophysiology and manifestations from the gastrointestinal tract. World J. Gastroenterol. 2020, 26, 4579–4588. [Google Scholar] [CrossRef]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Guan, W.; Ni, Z.; Hu, Y.; Liang, W.; Ou, C.; He, J.; Liu, L.; Shan, H.; Lei, C.; Hui, D.S.C.; et al. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl J. Med. 2020, 382, 1708–1720. [Google Scholar] [CrossRef]
- Cheung, K.S.; Hung, I.F.N.; Chan, P.P.Y.; Lung, K.C.; Tso, E.; Liu, R.; Ng, Y.Y.; Chu, M.Y.; Chung, T.W.H.; Tam, A.R.; et al. Gastrointestinal Manifestations of SARS-CoV-2 Infection and Virus Load in Fecal Samples from a Hong Kong Cohort: Systematic Review and Meta-analysis. Gastroenterology 2020, 159, 81–95. [Google Scholar] [CrossRef]
- Pan, L.; Mu, M.; Yang, P.; Sun, Y.; Wang, R.; Yan, J.; Li, P.; Hu, B.; Wang, J.; Hu, C.; et al. Clinical characteristics of COVID-19 patients with digestive symptoms in Hubei, China: A descriptive, cross-sectional, multicenter study. Am. J. Gastroenterol. 2020, 115, 766–773. [Google Scholar] [CrossRef]
- Zuo, T.; Liu, Q.; Zhang, F.; Lui, G.C.Y.; Tso, E.Y.K.; Yeoh, Y.K.; Chen, Z.; Boon, S.S.; Chan, F.K.L.; Chan, P.K.S.; et al. Depicting SARS-CoV-2 faecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut 2021, 70, 276–284. [Google Scholar]
- Xu, K.; Cai, H.; Shen, Y.; Ni, Q.; Chen, Y.; Hu, S.; Li, J.; Wang, H.; Yu, L.; Huang, H.; et al. Management of COVID-19: The Zhejiang experience. Zhejiang Da Xue Xue Bao Yi Xue Ban 2020, 49, 147–157. [Google Scholar]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology 2020, 159, 944–955.e8. [Google Scholar] [CrossRef]
- Geva-Zatorsky, N.; Sefik, E.; Kua, L.; Pasman, L.; Tan, T.G.; Ortiz-Lopez, A.; Yanortsang, T.B.; Yang, L.; Jupp, R.; Mathis, D.; et al. Mining the Human Gut Microbiota for Immunomodulatory Organisms. Cell 2017, 168, 928–943.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miquel, S.; Martín, R.; Rossi, O.; Bermúdez-Humarán, L.G.; Chatel, J.M.; Sokol, H.; Thomas, M.; Wells, J.M.; Langella, P. Faecalibacterium prausnitzii and human intestinal health. Curr. Opin. Microbiol. 2013, 16, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Kaakoush, N.O. Insights into the role of Erysipelotrichaceae in the human host. Front. Cell Infect. Microbiol. 2015, 5, 84. [Google Scholar] [CrossRef] [Green Version]
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C.; et al. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin. Infect. Dis. 2020, 71, 2669–2678. [Google Scholar] [CrossRef]
- Ramanan, P.; Barreto, J.N.; Osmon, D.R.; Tosh, P.K. Rothia bacteremia: A 10-year experience at Mayo Clinic, Rochester, Minnesota. J. Clin. Microbiol. 2014, 52, 3184–3189. [Google Scholar] [CrossRef] [Green Version]
- Yeoh, Y.K.; Zuo, T.; Lui, G.C.Y.; Zhang, F.; Liu, Q.; Li, A.Y.L.; Chung, A.C.K.; Cheung, C.P.; Tso, E.Y.K.; Fung, K.S.C.; et al. Gut microbiota composition reflects disease severity and dysfunctional immune responses in patients with COVID-19. Gut 2021, 70, 698–706. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhu, J.; Hu, Q.; Rao, X. Morganella morganii, a non-negligent opportunistic pathogen. Int. J. Infect. Dis. 2016, 50, 10–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shinagawa, N.; Taniguchi, M.; Hirata, K.; Furuhata, T.; Fukuhara, K.; Mizugucwi, T.; Osanai, H.; Yanai, Y.; Hata, F.; Kihara, C.; et al. Bacteria isolated from surgical infections and its susceptibilities to antimicrobial agents—Special references to bacteria isolated between April 2010 and March 2011. Jpn J. Antibiot. 2014, 67, 293–334. [Google Scholar]
- Hwang, N.; Eom, T.; Gupta, S.K.; Jeong, S.Y.; Jeong, D.Y.; Kim, Y.S.; Lee, J.H.; Sadowsky, M.J.; Unno, T. Genes and gut bacteria involved in luminal butyrate reduction caused by diet and loperamide. Genes 2017, 8, 350. [Google Scholar] [CrossRef] [Green Version]
- Kläring, K.; Just, S.; Lagkouvardos, I.; Hanske, L.; Haller, D.; Blaut, M.; Wenning, M.; Clavel, T. Murimonas intestini gen. nov., sp. nov., an acetateproducing bacterium of the family Lachnospiraceae isolated from the mouse gut. Int. J. Syst Evol. Microbiol. 2015, 65, 870–878. [Google Scholar] [CrossRef]
- Schroeder, B.O.; Bäckhed, F. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 2016, 22, 1079–1089. [Google Scholar] [CrossRef]
- Fan, J.; Li, X.; Gao, Y.; Zhou, J.; Wang, S.; Huang, B.; Wu, J.; Cao, Q.; Chen, Y.; Wang, Z.; et al. The lung tissue microbiota features of 20 deceased patients with COVID-19. J. Clean Prod. 2020, 81, e64–e67. [Google Scholar] [CrossRef] [PubMed]
- Doi, Y.; Murray, G.L.; Peleg, A.Y. Acinetobacter baumannii: Evolution of antimicrobial resistance-treatment options. Semin. Respir. Crit. Care Med. 2015, 36, 85–98. [Google Scholar]
- Di Stadio, A.; Costantini, C.; Renga, G.; Pariano, M.; Ricci, G.; Romani, L. The microbiota/host immune system interaction in the nose to protect from COVID-19. Life 2020, 10, 345. [Google Scholar] [CrossRef]
- Hashimoto, T.; Perlot, T.; Rehman, A.; Trichereau, J.; Ishiguro, H.; Paolino, M.; Sigl, V.; Hanada, T.; Hanada, R.; Lipinski, S.; et al. ACE2 links amino acid malnutrition to microbial ecology and intestinal inflammation. Nature 2012, 487, 477–481. [Google Scholar] [CrossRef] [PubMed]
- Perlot, T.; Penninger, J.M. ACE2—From the renin-angiotensin system to gut microbiota and malnutrition. Microbes Infect. 2013, 15, 866–873. [Google Scholar] [CrossRef]
- Mönkemüller, K.; Fry, L.C.; Rickes, S. Covid-19, Coronavirus, SARS-CoV-2 and the small bowel. Rev. Esp. Enferm. Dig. 2020, 2020, 383–388. [Google Scholar]
- Li, B.; Yang, J.; Zhao, F.; Zhi, L.; Wang, X.; Liu, L.; Bi, Z.; Zhao, Y. Prevalence and impact of cardiovascular metabolic diseases on COVID-19 in China. Clin. Res. Cardiol. 2020, 109, 531–538. [Google Scholar] [CrossRef]
- Wang, B.; Li, R.; Lu, Z.; Huang, Y. Does comorbidity increase the risk of patients with covid-19: Evidence from meta-analysis. Aging 2020, 12, 6049–6057. [Google Scholar] [CrossRef] [PubMed]
- Demeulemeester, F.; de Punder, K.; van Heijningen, M.; van Doesburg, F. Obesity as a Risk Factor for Severe COVID-19 and Complications: A Review. Cells 2021, 10, 933. [Google Scholar] [CrossRef] [PubMed]
- Musso, G.; Gambino, R.; Cassader, M. Obesity, diabetes, and gut microbiota: The hygiene hypothesis expanded? Diabetes Care 2010, 33, 2277–2284. [Google Scholar] [CrossRef] [Green Version]
- Ortega, M.A.; Fraile-Martínez, O.; Naya, I.; García-Honduvilla, N.; Álvarez-Mon, M.; Buján, J.; Asúnsolo, Á.; de la Torre, B. Type 2 diabetes mellitus associated with obesity (Diabesity). The central role of gut microbiota and its translational applications. Nutrients 2020, 12, 2749. [Google Scholar] [CrossRef]
- Tang, W.H.W.; Kitai, T.; Hazen, S.L. Gut microbiota in cardiovascular health and disease. Circ. Res. 2017, 120, 1183–1196. [Google Scholar] [CrossRef] [Green Version]
- Kazemian, N.; Mahmoudi, M.; Halperin, F.; Wu, J.C.; Pakpour, S. Gut microbiota and cardiovascular disease: Opportunities and challenges. Microbiome 2020, 8, 36. [Google Scholar] [CrossRef] [Green Version]
- Mak, J.W.Y.; Chan, F.K.L.; Ng, S.C. Probiotics and COVID-19: One size does not fit all. Lancet Gastroenterol. Hepatol. 2020, 5, 644–645. [Google Scholar] [CrossRef]
- Akour, A. Probiotics and COVID-19: Is there any link? Lett. Appl. Microbiol. 2020, 71, 229–234. [Google Scholar] [CrossRef]
- Bottari, B.; Castellone, V.; Neviani, E. Probiotics and Covid-19. Int. J. Food Sci. Nutr. 2021, 72, 293–299. [Google Scholar] [CrossRef]
- Sundararaman, A.; Ray, M.; Ravindra, P.V.; Halami, P.M. Role of probiotics to combat viral infections with emphasis on COVID-19. Appl. Microbiol. Biotechnol. 2020, 104, 8089–8104. [Google Scholar] [CrossRef] [PubMed]
- Mahooti, M.; Miri, S.M.; Abdolalipour, E.; Ghaemi, A. The immunomodulatory effects of probiotics on respiratory viral infections: A hint for COVID-19 treatment? Microb. Pathog. 2020, 148, 104452. [Google Scholar] [CrossRef] [PubMed]
- Hao, Q.; Dong, B.R.; Wu, T. Probiotics for preventing acute upper respiratory tract infections. Cochrane Database Syst. Rev. 2015, 2015, CD006895. [Google Scholar] [CrossRef]
- West, N.P.; Horn, P.L.; Pyne, D.B.; Gebski, V.J.; Lahtinen, S.J.; Fricker, P.A.; Cripps, A.W. Probiotic supplementation for respiratory and gastrointestinal illness symptoms in healthy physically active individuals. Clin. Nutr. 2014, 33, 581–587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Li, X.; Ge, T.; Xiao, Y.; Liao, Y.; Cui, Y.; Zhang, Y.; Ho, W.; Yu, G.; Zhang, T. Probiotics for prevention and treatment of respiratory tract infections in children: A systematic review and meta-analysis of randomized controlled trials. Medicine 2016, 95, e4509. [Google Scholar] [CrossRef] [PubMed]
- Gao, Q.Y.; Chen, Y.X.; Fang, J.Y. 2019 Novel coronavirus infection and gastrointestinal tract. J. Dig. Dis. 2020, 21, 125–126. [Google Scholar] [CrossRef] [Green Version]
- Serra, D.; Almeida, L.M.; Dinis, T.C.P. The Impact of Chronic Intestinal Inflammation on Brain Disorders: The Microbiota-Gut-Brain Axis. Mol. Neurobiol. 2019, 56, 6941–6951. [Google Scholar] [CrossRef] [PubMed]
- Mazza, M.G.; De Lorenzo, R.; Conte, C.; Poletti, S.; Vai, B.; Bollettini, I.; Melloni, E.M.T.; Furlan, R.; Ciceri, F.; Rovere-Querini, P.; et al. Anxiety and depression in COVID-19 survivors: Role of inflammatory and clinical predictors. Brain Behav. Immun. 2020, 89, 594–600. [Google Scholar] [CrossRef] [PubMed]
- O’Mahony, S.M.; Clarke, G.; Borre, Y.E.; Dinan, T.G.; Cryan, J.F. Serotonin, tryptophan metabolism and the brain-gut-microbiome axis. Behav. Brain Res. 2015, 277, 32–48. [Google Scholar] [CrossRef]
- Fernstrom, J.D.; Wurtman, R.J. Brain serotonin content: Physiological dependence on plasma tryptophan levels. Science 1971, 173, 149–152. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Manosso, L.M.; Arent, C.O.; Borba, L.A.; Ceretta, L.B.; Quevedo, J.; Réus, G.Z. Microbiota-Gut-Brain Communication in the SARS-CoV-2 Infection. Cells 2021, 10, 1993. https://doi.org/10.3390/cells10081993
Manosso LM, Arent CO, Borba LA, Ceretta LB, Quevedo J, Réus GZ. Microbiota-Gut-Brain Communication in the SARS-CoV-2 Infection. Cells. 2021; 10(8):1993. https://doi.org/10.3390/cells10081993
Chicago/Turabian StyleManosso, Luana M., Camila O. Arent, Laura A. Borba, Luciane B. Ceretta, João Quevedo, and Gislaine Z. Réus. 2021. "Microbiota-Gut-Brain Communication in the SARS-CoV-2 Infection" Cells 10, no. 8: 1993. https://doi.org/10.3390/cells10081993
APA StyleManosso, L. M., Arent, C. O., Borba, L. A., Ceretta, L. B., Quevedo, J., & Réus, G. Z. (2021). Microbiota-Gut-Brain Communication in the SARS-CoV-2 Infection. Cells, 10(8), 1993. https://doi.org/10.3390/cells10081993





