Genome Editing in Zebrafish by ScCas9 Recognizing NNG PAM
Abstract
:1. Introduction
2. Materials and Methods
2.1. Zebrafish Husbandry
2.2. Plasmid Construction
2.3. crRNA, tracrRNA, sgRNA and mRNA Synthesis
2.4. Sccas9 Expression and Purification
2.5. Guide RNA: Cas9 Ribonucleoprotein (RNP) Complexes Preparation and Zebrafish Microinjection
2.6. Mutations Detection
2.7. Imaging
2.8. Founder and Stable Mutant Line Identification
2.9. Statistical Analysis
3. Results
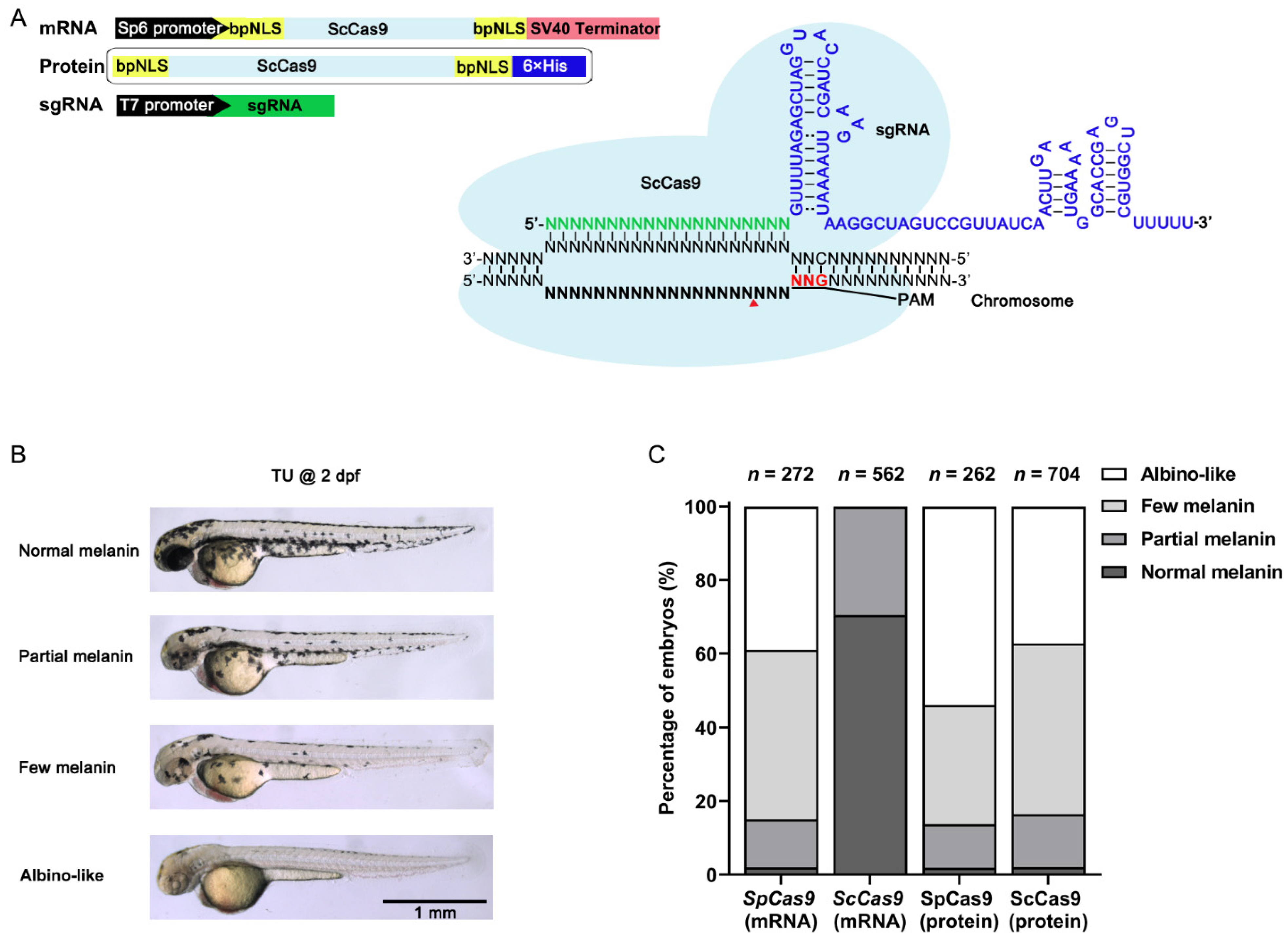
3.1. ScCas9 RNP Complexes Provide Robust Genome Editing in Zebrafish
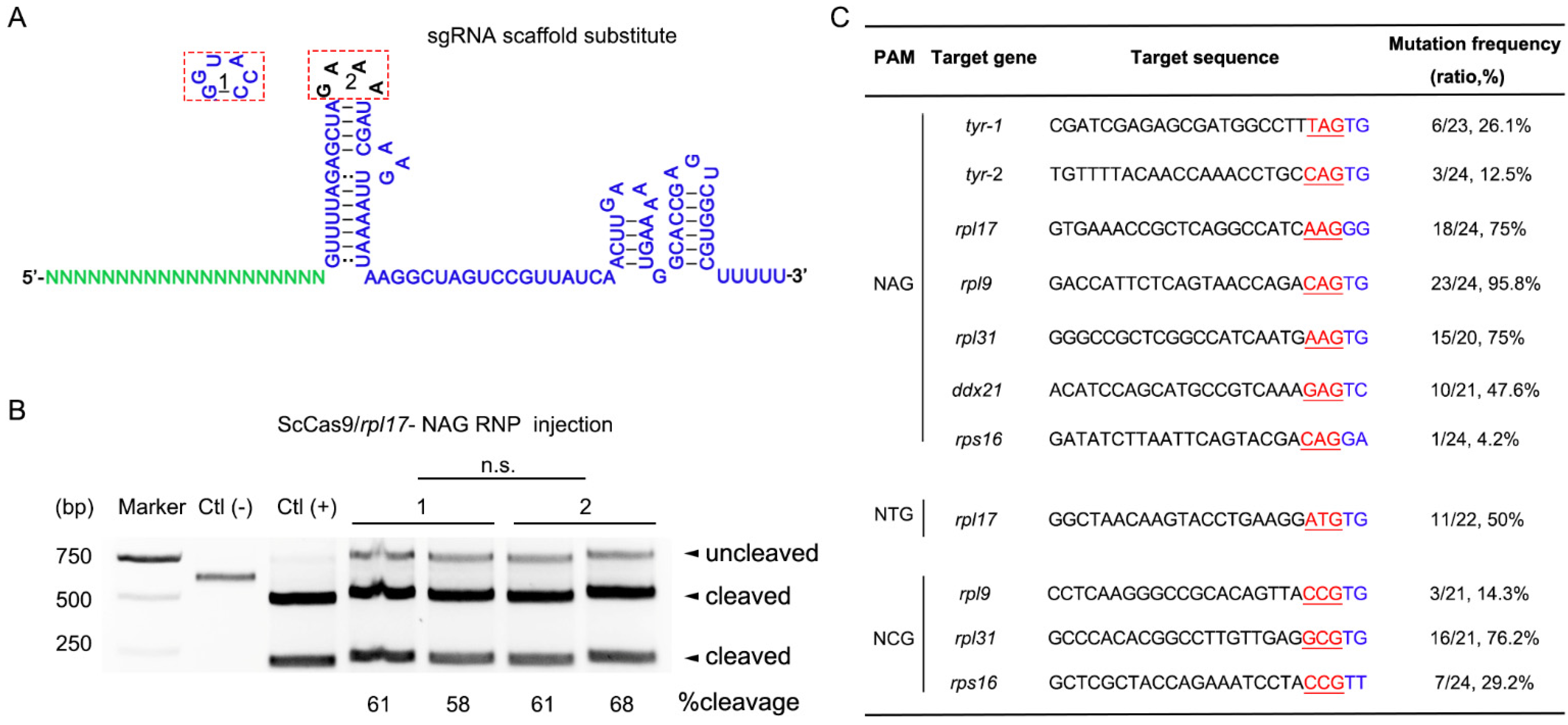
3.2. ScCas9 Can Recognize NNG PAM in Zebrafish Genome
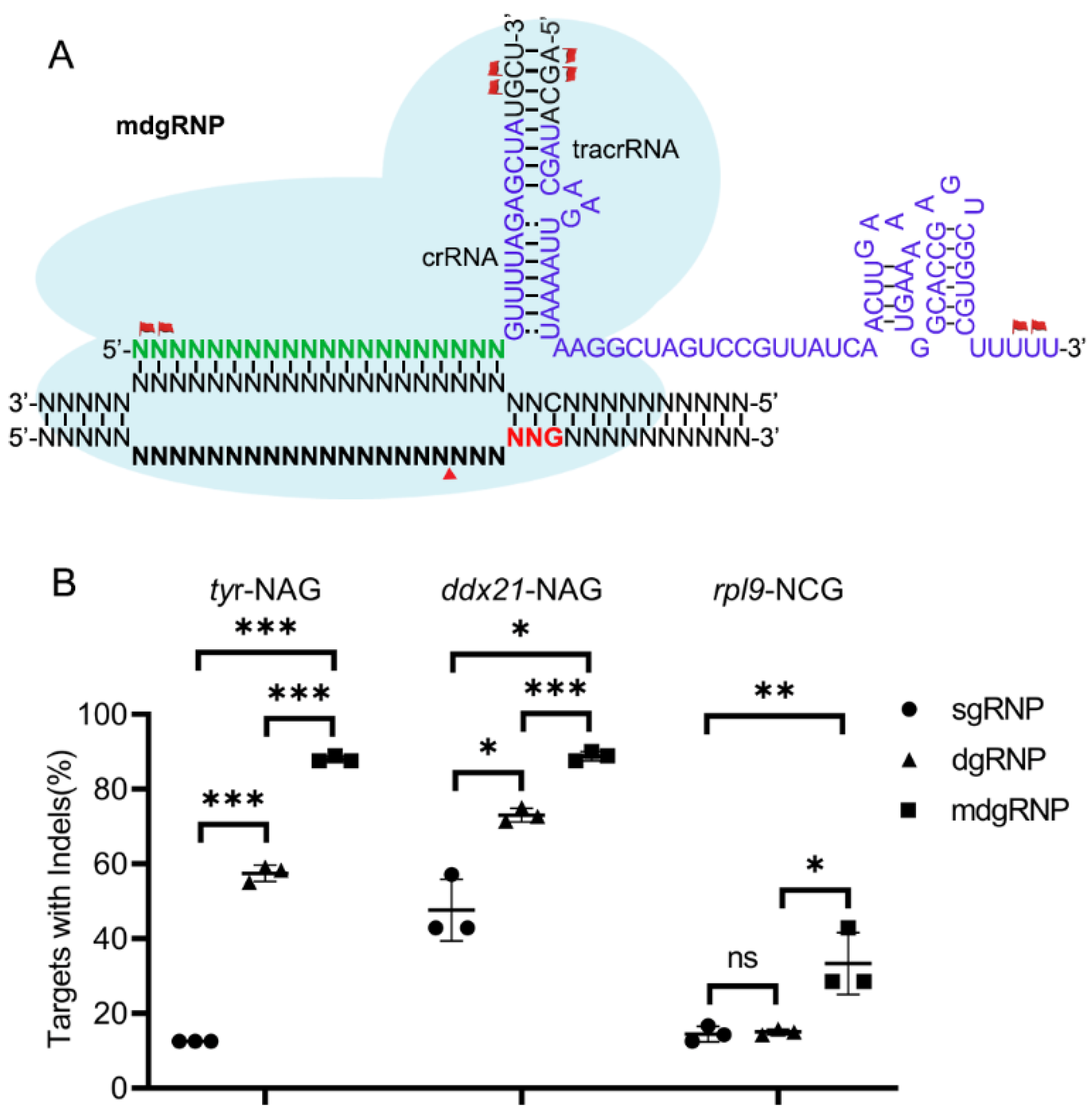
3.3. Chemical Modified dgRNP Can Significantly Improve ScCas9 Activity in Zebrafish Genome
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [Green Version]
- Boel, A.; De Saffel, H.; Steyaert, W.; Callewaert, B.; De Paepe, A.; Coucke, P.J.; Willaert, A. CRISPR/Cas9-mediated homology-directed repair by ssODNs in zebrafish induces complex mutational patterns resulting from genomic integration of repair-template fragments. Dis. Model. Mech. 2018, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, W.; Lu, X.; Liu, Y.; Bai, H.; Li, S.; Lin, S. Precise A*T to G*C base editing in the zebrafish genome. BMC Biol. 2018, 16, 139. [Google Scholar] [CrossRef] [PubMed]
- Esvelt, K.M.; Mali, P.; Braff, J.L.; Moosburner, M.; Yaung, S.J.; Church, G.M. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods 2013, 10, 1116–1121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleinstiver, B.P.; Prew, M.S.; Tsai, S.Q.; Topkar, V.V.; Nguyen, N.T.; Zheng, Z.; Gonzales, A.P.; Li, Z.; Peterson, R.T.; Yeh, J.R.; et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature 2015, 523, 481–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ran, F.A.; Cong, L.; Yan, W.X.; Scott, D.A.; Gootenberg, J.S.; Kriz, A.J.; Zetsche, B.; Shalem, O.; Wu, X.; Makarova, K.S.; et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature 2015, 520, 186–191. [Google Scholar] [CrossRef]
- Hirano, H.; Gootenberg, J.S.; Horii, T.; Abudayyeh, O.O.; Kimura, M.; Hsu, P.D.; Nakane, T.; Ishitani, R.; Hatada, I.; Zhang, F.; et al. Structure and engineering of Francisella novicida Cas9. Cell 2016, 164, 950–961. [Google Scholar] [CrossRef] [Green Version]
- Gao, L.; Cox, D.B.T.; Yan, W.X.; Manteiga, J.C.; Schneider, M.W.; Yamano, T.; Nishimasu, H.; Nureki, O.; Crosetto, N.; Zhang, F. Engineered Cpf1 variants with altered PAM specificities. Nat. Biotechnol. 2017, 35, 789–792. [Google Scholar] [CrossRef] [Green Version]
- Harrington, L.B.; Paez-Espino, D.; Staahl, B.T.; Chen, J.S.; Ma, E.; Kyrpides, N.C.; Doudna, J.A. A thermostable Cas9 with increased lifetime in human plasma. Nat. Commun. 2017, 8, 1424. [Google Scholar] [CrossRef] [Green Version]
- Kim, E.; Koo, T.; Park, S.W.; Kim, D.; Kim, K.; Cho, H.Y.; Song, D.W.; Lee, K.J.; Jung, M.H.; Kim, S.; et al. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat. Commun. 2017, 8, 14500. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Li, J.; He, Y.; Xu, M.; Zhang, J.; Du, W.; Zhao, Y.; Xia, L. Precise gene replacement in rice by RNA transcript-templated homologous recombination. Nat. Biotechnol. 2019, 37, 445–450. [Google Scholar] [CrossRef]
- Hua, K.; Tao, X.; Han, P.; Wang, R.; Zhu, J.K. Genome engineering in rice using Cas9 variants that recognize NG PAM sequences. Mol. Plant 2019, 12, 1003–1014. [Google Scholar] [CrossRef]
- Moreno-Mateos, M.A.; Fernandez, J.P.; Rouet, R.; Vejnar, C.E.; Lane, M.A.; Mis, E.; Khokha, M.K.; Doudna, J.A.; Giraldez, A.J. CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat. Commun. 2017, 8, 2024. [Google Scholar] [CrossRef] [Green Version]
- Feng, Y.; Chen, C.; Han, Y.; Chen, Z.; Lu, X.; Liang, F.; Li, S.; Qin, W.; Lin, S. Expanding CRISPR/Cas9 genome editing capacity in zebrafish using SaCas9. G3 Genes Genomes Genet. 2016, 6, 2517–2521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chatterjee, P.; Jakimo, N.; Jacobson, J.M. Minimal PAM specificity of a highly similar SpCas9 ortholog. Sci. Adv. 2018, 4, eaau0766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Meng, X.; Wang, J.; Qin, B.; Wang, K.; Li, J.; Wang, C.; Yu, H. ScCas9 recognizes NNG protospacer adjacent motif in genome editing of rice. Sci. China Life Sci. 2020, 63, 450–452. [Google Scholar] [CrossRef]
- Kimmel, C.B.; Ballard, W.W.; Kimmel, S.R.; Ullmann, B.; Schilling, T.F. Stages of embryonic development of the zebrafish. Dev. Dyn. 1995, 203, 253–310. [Google Scholar] [CrossRef]
- Bassett, A.R.; Tibbit, C.; Ponting, C.P.; Liu, J.L. Highly efficient targeted mutagenesis of drosophila with the CRISPR/Cas9 system. Cell Rep. 2013, 4, 220–228. [Google Scholar] [CrossRef] [Green Version]
- Truett, G.E.; Heeger, P.; Mynatt, R.L.; Truett, A.A.; Walker, J.A.; Warman, M.L. Preparation of PCR-quality mouse genomic DNA with hot sodium hydroxide and tris (HotSHOT). Biotechniques 2000, 29, 52–54. [Google Scholar] [CrossRef] [PubMed]
- Hwang, W.Y.; Fu, Y.; Reyon, D.; Maeder, M.L.; Tsai, S.Q.; Sander, J.D.; Peterson, R.T.; Yeh, J.R.; Joung, J.K. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 227–229. [Google Scholar] [CrossRef]
- Jao, L.E.; Wente, S.R.; Chen, W. Efficient multiplex biallelic zebrafish genome editing using a CRISPR nuclease system. Proc. Natl. Acad. Sci. USA 2013, 110, 13904–13909. [Google Scholar] [CrossRef] [Green Version]
- Zhao, D.; Jones, J.L.; Gasperini, R.J.; Charlesworth, J.C.; Liu, G.S.; Burdon, K.P. Rapid and efficient cataract gene evaluation in F0 zebrafish using CRISPR-Cas9 ribonucleoprotein complexes. Methods 2021. [Google Scholar] [CrossRef]
- Fonfara, I.; Le Rhun, A.; Chylinski, K.; Makarova, K.S.; Lecrivain, A.L.; Bzdrenga, J.; Koonin, E.V.; Charpentier, E. Phylogeny of Cas9 determines functional exchangeability of dual-RNA and Cas9 among orthologous type II CRISPR-Cas systems. Nucleic Acids Res. 2014, 42, 2577–2590. [Google Scholar] [CrossRef] [PubMed]
- Rahdar, M.; McMahon, M.A.; Prakash, T.P.; Swayze, E.E.; Bennett, C.F.; Cleveland, D.W. Synthetic CRISPR RNA-Cas9-guided genome editing in human cells. Proc. Natl. Acad. Sci. USA 2015, 112, E7110–E7117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hendel, A.; Bak, R.O.; Clark, J.T.; Kennedy, A.B.; Ryan, D.E.; Roy, S.; Steinfeld, I.; Lunstad, B.D.; Kaiser, R.J.; Wilkens, A.B.; et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat. Biotechnol. 2015, 33, 985–989. [Google Scholar] [CrossRef] [PubMed]
- Hoshijima, K.; Jurynec, M.J.; Klatt Shaw, D.; Jacobi, A.M.; Behlke, M.A.; Grunwald, D.J. Highly efficient CRISPR-Cas9-based methods for generating deletion mutations and F0 embryos that lack gene function in zebrafish. Dev. Cell 2019, 51, 645–657.e644. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Xu, Z.; Gosavi, G.; Ren, B.; Cao, Y.; Kuang, Y.; Zhou, C.; Spetz, C.; Yan, F.; Zhou, X.; et al. Targeted base editing in rice with CRISPR/ScCas9 system. Plant Biotechnol. J. 2020, 18, 1645–1647. [Google Scholar] [CrossRef]
- Zhang, L.; Rube, H.T.; Vakulskas, C.A.; Behlke, M.A.; Bussemaker, H.J.; Pufall, M.A. Systematic in vitro profiling of off-target affinity, cleavage and efficiency for CRISPR enzymes. Nucleic Acids Res. 2020, 48, 5037–5053. [Google Scholar] [CrossRef]
- Chatterjee, P.; Jakimo, N.; Lee, J.; Amrani, N.; Rodriguez, T.; Koseki, S.R.T.; Tysinger, E.; Qing, R.; Hao, S.; Sontheimer, E.J.; et al. An engineered ScCas9 with broad PAM range and high specificity and activity. Nat. Biotechnol. 2020, 38, 1154–1158. [Google Scholar] [CrossRef]
- Hu, J.H.; Miller, S.M.; Geurts, M.H.; Tang, W.; Chen, L.; Sun, N.; Zeina, C.M.; Gao, X.; Rees, H.A.; Lin, Z.; et al. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature 2018, 556, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Nishimasu, H.; Shi, X.; Ishiguro, S.; Gao, L.; Hirano, S.; Okazaki, S.; Noda, T.; Abudayyeh, O.O.; Gootenberg, J.S.; Mori, H.; et al. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science 2018, 361, 1259–1262. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Liang, F.; Dong, Z.; Li, S.; Ye, J.; Qin, W. Genome Editing in Zebrafish by ScCas9 Recognizing NNG PAM. Cells 2021, 10, 2099. https://doi.org/10.3390/cells10082099
Liu Y, Liang F, Dong Z, Li S, Ye J, Qin W. Genome Editing in Zebrafish by ScCas9 Recognizing NNG PAM. Cells. 2021; 10(8):2099. https://doi.org/10.3390/cells10082099
Chicago/Turabian StyleLiu, Yunxing, Fang Liang, Zijiong Dong, Song Li, Jianmin Ye, and Wei Qin. 2021. "Genome Editing in Zebrafish by ScCas9 Recognizing NNG PAM" Cells 10, no. 8: 2099. https://doi.org/10.3390/cells10082099
APA StyleLiu, Y., Liang, F., Dong, Z., Li, S., Ye, J., & Qin, W. (2021). Genome Editing in Zebrafish by ScCas9 Recognizing NNG PAM. Cells, 10(8), 2099. https://doi.org/10.3390/cells10082099





