Preliminary Evaluation of Proliferation, Wound Healing Properties, Osteogenic and Chondrogenic Potential of Dental Pulp Stem Cells Obtained from Healthy and Periodontitis Affected Teeth
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Size Determination and Sample Collection
2.2. Culture and Expansion of Human Dental Pulp Stem Cells (DPSCs)
2.3. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium Bromide (MTT) Assay for Metabolic Activity of hDPSCs and pDPSCs
2.4. Characterization of hDPSCs and pDPSCs by Flow Cytometry
2.5. Growth Curve Plotting to Assess Proliferative Potential
2.6. Analysis of Cell Migration by Wound Scratch Assay
2.7. Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR) for Quantitative Analysis of Gene Expression Related to Bone and Cartilage Formation
2.8. Osteogenic Differentiation Protocol
2.9. Chondrogenic Differentiation Protocol
2.10. Statistical Analysis
3. Results
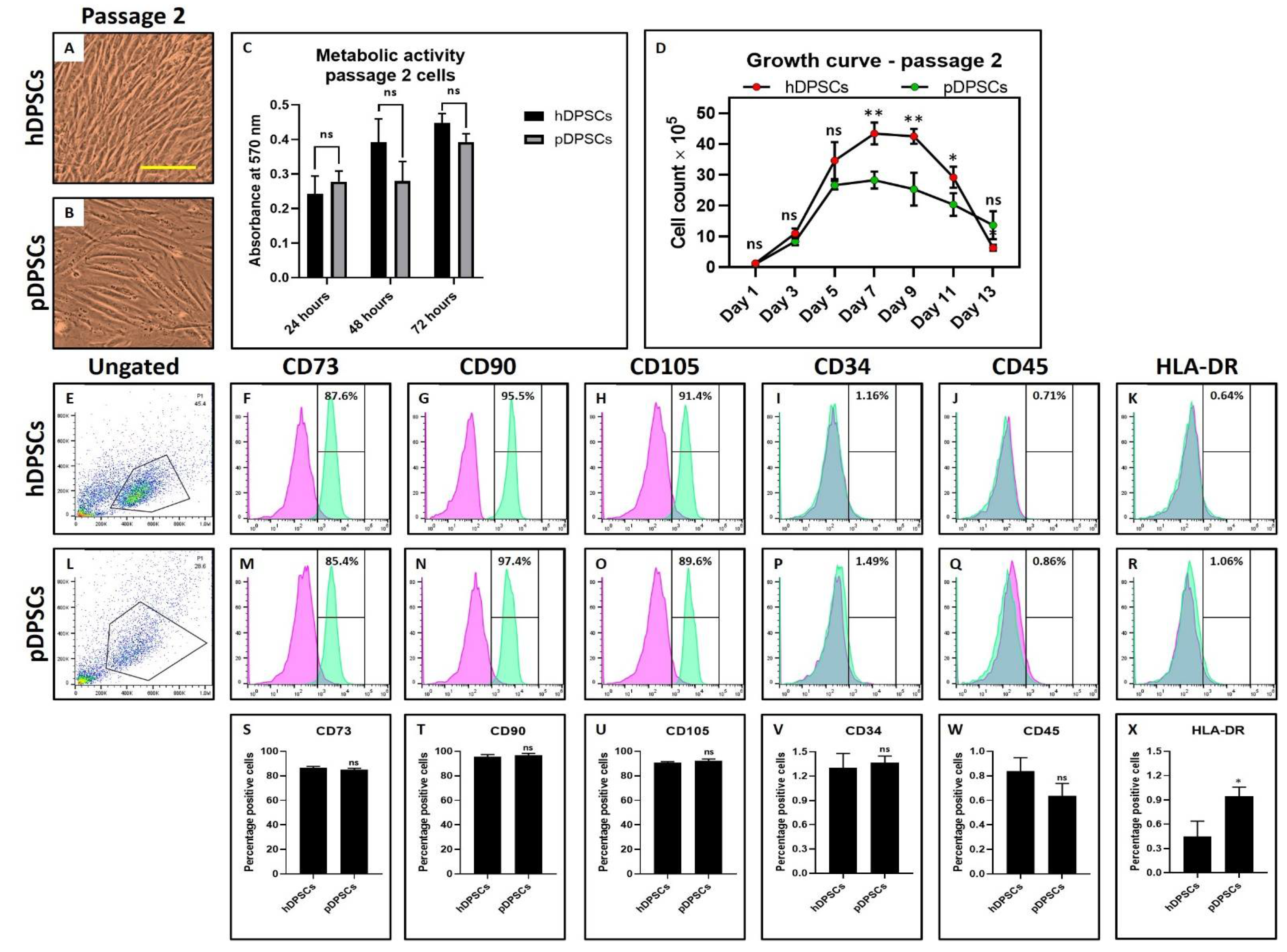
3.1. hDPSCs and pDPSCs Demonstrated No Significant Differences in Morphology, Metabolic Activity and Mesenchymal Stem Cell (MSC) Marker Expression. pDPSCs Demonstrated Slower Growth at Earlier Passage
3.2. pDPSCs Demonstrate Slower Migration and Decreased Chondrogenic Potential. No Significant Difference in Osteogenic Potential
3.3. Mesenchymal Stem Cell (MSC) Marker Expression Affected in pDPSCs at Late Passage. hDPSCs and pDPSCs Show No Significant Differences in Morphology and Metabolic Activity. pDPSCs Demonstrated Slightly Higher Growth at Late Passage
3.4. pDPSCs Demonstrate Increased Migration, Chondrogenic, and Osteogenic Potential at Late Passage
3.5. hDPSCs and pDPSCs Demonstrate Significant Differences in Morphology and Increased Metabolic Activity. Cryopreservation Affected Mesenchymal Stem Cell (MSC) Marker Expression in pDPSCs. pDPSCs Demonstrated Comparable Growth Rate to hDPSCs for Shorter Incubation Time after Cryopreservation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gronthos, S.; Mankani, M.; Brahim, J.; Robey, P.; Shi, S. Postnatal human dental pulp stem cells (DPSCs) in vitro and invivo. Proc. Natl. Acad. Sci. USA 2000, 97, 13625–13630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almushayt, A.; Narayanan, K.; Zaki, A.E.; George, A. Dentin matrix protein 1 induces cytodifferentiation of dental pulp stem cells into odontoblasts. Gene Ther. 2005, 13, 611–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; He, H.; Tang, C.; Zhang, G.; Li, Y.; Wang, R.; Shi, J.; Jin, Y. Differentiation potential of STRO-1+ dental pulp stem cells changes during cell passaging. BMC Cell Biol. 2010, 11, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.-L.; Chang, W.-J.; Lin, C.-Y.; Hsieh, S.-C.; Lee, S.-Y.; Fan, K.-H.; Lin, C.-T.; Huang, H.-M. Static magnetic field increases survival rate of dental pulp stem cells during DMSO-free cryopreservation. Electromagn. Biol. Med. 2014, 34, 302–308. [Google Scholar] [CrossRef] [PubMed]
- Gioventù, S.; Andriolo, G.; Bonino, F.; Frasca, S.; Lazzari, L.; Montelatici, E.; Santoro, F.; Rebulla, P. A novel method for banking dental pulp stem cells. Transfus. Apher. Sci. 2012, 47, 199–206. [Google Scholar] [CrossRef]
- Graziano, A.; D’Aquino, R.; Laino, G.; Papaccio, G. Dental pulp stem cells: A promising tool for bone regeneration. Stem Cell Rev. 2008, 4, 21–26. [Google Scholar] [CrossRef] [Green Version]
- Alongi, D.J.; Yamaza, T.; Song, Y.; Fouad, A.F.; Romberg, E.E.; Shi, S.; Tuan, R.S.; Huang, G.T.-J. Stem/progenitor cells from inflamed human dental pulp retain tissue regeneration potential. Regen. Med. 2010, 5, 617–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.; Pan, J.; Wright, J.T.; Bencharit, S.; Zhang, S.; Everett, E.T.; Teixeira, F.B.; Preisser, J.S. Putative Stem Cells in Human Dental Pulp with Irreversible Pulpitis: An Exploratory Study. J. Endod. 2010, 36, 820–825. [Google Scholar] [CrossRef] [Green Version]
- Pereira, L.O.; Rubini, M.R.; Silva, J.R.; Oliveira, D.M.; Silva, I.C.R.; Poças-Fonseca, M.J. Comparison of stem cell properties of cells isolated from normal and inflamed dental pulps. Int. Endod. J. 2012, 45, 1080–1090. [Google Scholar] [CrossRef]
- Yazid, F.B.; Gnanasegaran, N.; Kunasekaran, W.; Govindasamy, V.; Musa, S. Comparison of immunodulatory properties of dental pulp stem cells derived from healthy and inflamed teeth. Clin. Oral Investig. 2014, 18, 2103–2112. [Google Scholar] [CrossRef]
- Tomasello, L.; Mauceri, R.; Coppola, A.; Pitrone, M.; Pizzo, G.; Campisi, G.; Pizzolanti, G.; Giordano, C. Mesenchymal stem cells derived from inflamed dental pulpal and gingival tissue: A potential application for bone formation. Stem Cell Res. Ther. 2017, 8, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, H.-H.; Chen, B.; Zhu, Q.-L.; Kong, H.; Li, Q.-H.; Gao, L.-N.; Xiao, M.; Chen, F.-M.; Yu, Q. Investigation of dental pulp stem cells isolated from discarded human teeth extracted due to aggressive periodontitis. Biomaterials 2014, 35, 9459–9472. [Google Scholar] [CrossRef]
- Di Tinco, R.; Bertani, G.; Pisciotta, A.; Bertoni, L.; Bertacchini, J.; Colombari, B. Evaluation of Antimicrobial Effect of Air-Polishing Treatments and Their Influence on Human Dental Pulp Stem Cells Seeded on Titanium Disks. Int. J. Mol. Sci. 2021, 22, 865. [Google Scholar] [CrossRef] [PubMed]
- Park, S.; Kim, J.E.; Han, J.; Jeong, S.; Lim, J.W.; Lee, M.C.; Son, H.; Kim, H.B.; Choung, Y.-H.; Seonwoo, H.; et al. 3D-Printed Poly(ε-Caprolactone)/Hydroxyapatite Scaffolds Modified with Alkaline Hydrolysis Enhance Osteogenesis In Vitro. Polymers 2021, 13, 257. [Google Scholar] [CrossRef] [PubMed]
- Papapanou, P.N.; Sanz, M.; Buduneli, N.; Dietrich, T.; Feres, M.; Fine, D.H. Periodontitis: Consensus report of workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Clin. Periodontol. 2018, 45, S162–S170. [Google Scholar] [CrossRef]
- Patil, V.R.; Kharat, A.H.; Kulkarni, D.G.; Kheur, S.M.; Bhonde, R.R. Long term explant culture for harvesting homogeneous population of human dental pulp stem cells. Cell Biol. Int. 2018, 42, 1602–1610. [Google Scholar] [CrossRef]
- Cappiello, F.; Casciaro, B.; Mangoni, M.L. A Novel In Vitro Wound Healing Assay to Evaluate Cell Migration. J. Vis. Exp. 2018, e56825. [Google Scholar] [CrossRef]
- Kwok, V.; Caton, J.G. Commentary: Prognosis Revisited: A System for Assigning Periodontal Prognosis. J. Periodontol. 2007, 78, 2063–2071. [Google Scholar] [CrossRef]
- Svärdström, G.; Wennström, J.L. Periodontal Treatment Decisions for Molars: An Analysis of Influencing Factors and Long-Term Outcome. J. Periodontol. 2000, 71, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Lozano, F.J.; Bueno, C.R.; Insausti, C.L.; Meseguer-Olmo, L.; Ramírez, M.C.; Blanquer, M.B.; Marín, N.; Martinez, S.; Moraleda, J.M. Mesenchymal stem cells derived from dental tissues. Int. Endod. J. 2011, 44, 800–806. [Google Scholar] [CrossRef] [Green Version]
- Karamzadeh, R.; Eslaminejad, M.B.; Aflatoonian, R. Isolation, Characterization and Comparative Differentiation of Human Dental Pulp Stem Cells Derived from Permanent Teeth by Using Two Different Methods. J. Vis. Exp. 2012, e4372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raja, V.; Emmadi, P.; Namasivayam, A.; Thyegarajan, R.; Rajaraman, V. The periodontal—endodontic continuum: A review. J. Conserv. Dent. 2008, 11, 54–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heasman, P.A. An endodontic conundrum: The association between pulpal infection and periodontal disease. Br. Dent. J. 2014, 216, 275–279. [Google Scholar] [CrossRef] [PubMed]
- Adriaens, P.A.; De Boever, J.A.; Loesche, W.J. Bacterial Invasion in Root Cementum and Radicular Dentin of Periodontally Diseased Teeth in Humans. J. Periodontol. 1988, 59, 222–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boostani, H.R.; Fatemi, K.; Disfani, R.; Zare, R.; Moeintaghavi, A.; Ali, S.A. Influence of moderate to severe chronic periodontitis on dental pulp. J. Indian Soc. Periodontol. 2012, 16, 558–561. [Google Scholar] [CrossRef] [PubMed]
- Widbiller, M.; Eidt, A.; Wölflick, M.; Lindner, S.R.; Schweikl, H.; Hiller, K.-A.; Buchalla, W.; Galler, K.M. Interactive effects of LPS and dentine matrix proteins on human dental pulp stem cells. Int. Endod. J. 2018, 51, 877–888. [Google Scholar] [CrossRef]
- Liu, Y.; Gao, Y.; Zhan, X.; Cui, L.; Xu, S.; Ma, D.; Yue, J.; Wu, B.; Gao, J. TLR4 Activation by Lipopolysaccharide and Streptococcus mutans Induces Differential Regulation of Proliferation and Migration in Human Dental Pulp Stem Cells. J. Endod. 2014, 40, 1375–1381. [Google Scholar] [CrossRef] [PubMed]
- Petrou, P.; Gothelf, Y.; Argov, Z.; Gotkine, M.; Levy, Y.S.; Kassis, I. Safety and Clinical Effects of Mesenchymal Stem Cells Secreting Neurotrophic Factor Transplantation in Patients With Amyotrophic Lateral Sclerosis: Results of Phase 1/2 and 2a Clinical Trials. JAMA Neurol. 2016, 73, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Zhao, K.; Liu, Q. The clinical application of mesenchymal stromal cells in hematopoietic stem cell transplantation. J. Hematol. Oncol. 2016, 9, 46. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Carrasco, R.; Sánchez-Abarca, L.I.; Nieto-Gómez, C.; Martín García, E.; Sánchez-Guijo, F.; Argüeso, P. Subconjunctival injection of mesenchymal stromal cells protects the cornea in an experimental model of GVHD. Ocul Surf. 2019, 17, 285–294. [Google Scholar] [CrossRef]
- Zhou, T.; Yuan, Z.; Weng, J.; Pei, D.; Du, X.; He, C.; Lai, P. Challenges and advances in clinical applications of mesenchymal stromal cells. J. Hematol. Oncol. 2021, 14, 1–24. [Google Scholar] [CrossRef] [PubMed]





| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| RUNX2 | 5′-GTG CCT AGG CGC ATT TCA-3′ | 5′-GCT CTT CTT ACT GAG AGT GGA AGG-3′ |
| OCN | 5′-GGC GCT ACC TGT ATC AAT GG-3′ | 5′-TCA GCC AAC TCG TCA CAG TC-3′ |
| SOX9 | 5′-GCC GAA AGC GGG CTC GAA AC-3′ | 5′-AAA AGT GGG GGC GCT TGC ACC-3′ |
| ACAN | 5′-GCG AGT TGT CAT GGT CTG AA-3′ | 5′-TTC TTG GAG AAG GGA GTC CA-3′ |
| ACTB | 5′-AGA GCT ACG AGC TGC CTG AC-3′ | 5′-AGC ACT GTG TTG GCG TAC AG-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fageeh, H.N. Preliminary Evaluation of Proliferation, Wound Healing Properties, Osteogenic and Chondrogenic Potential of Dental Pulp Stem Cells Obtained from Healthy and Periodontitis Affected Teeth. Cells 2021, 10, 2118. https://doi.org/10.3390/cells10082118
Fageeh HN. Preliminary Evaluation of Proliferation, Wound Healing Properties, Osteogenic and Chondrogenic Potential of Dental Pulp Stem Cells Obtained from Healthy and Periodontitis Affected Teeth. Cells. 2021; 10(8):2118. https://doi.org/10.3390/cells10082118
Chicago/Turabian StyleFageeh, Hytham N. 2021. "Preliminary Evaluation of Proliferation, Wound Healing Properties, Osteogenic and Chondrogenic Potential of Dental Pulp Stem Cells Obtained from Healthy and Periodontitis Affected Teeth" Cells 10, no. 8: 2118. https://doi.org/10.3390/cells10082118






