Genetic Instability Due to Spindle Anomalies Visualized in Mutants of Dictyostelium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Strains
2.2. Culture Conditions and Sample Preparation for Confocal Microscopy
2.3. Confocal Image Acquisition and Data Processing
3. Results
3.1. Normal Mitosis in D. discoideum
3.2. Spindle Anomalies Caused by Disturbed Centrosome Activities
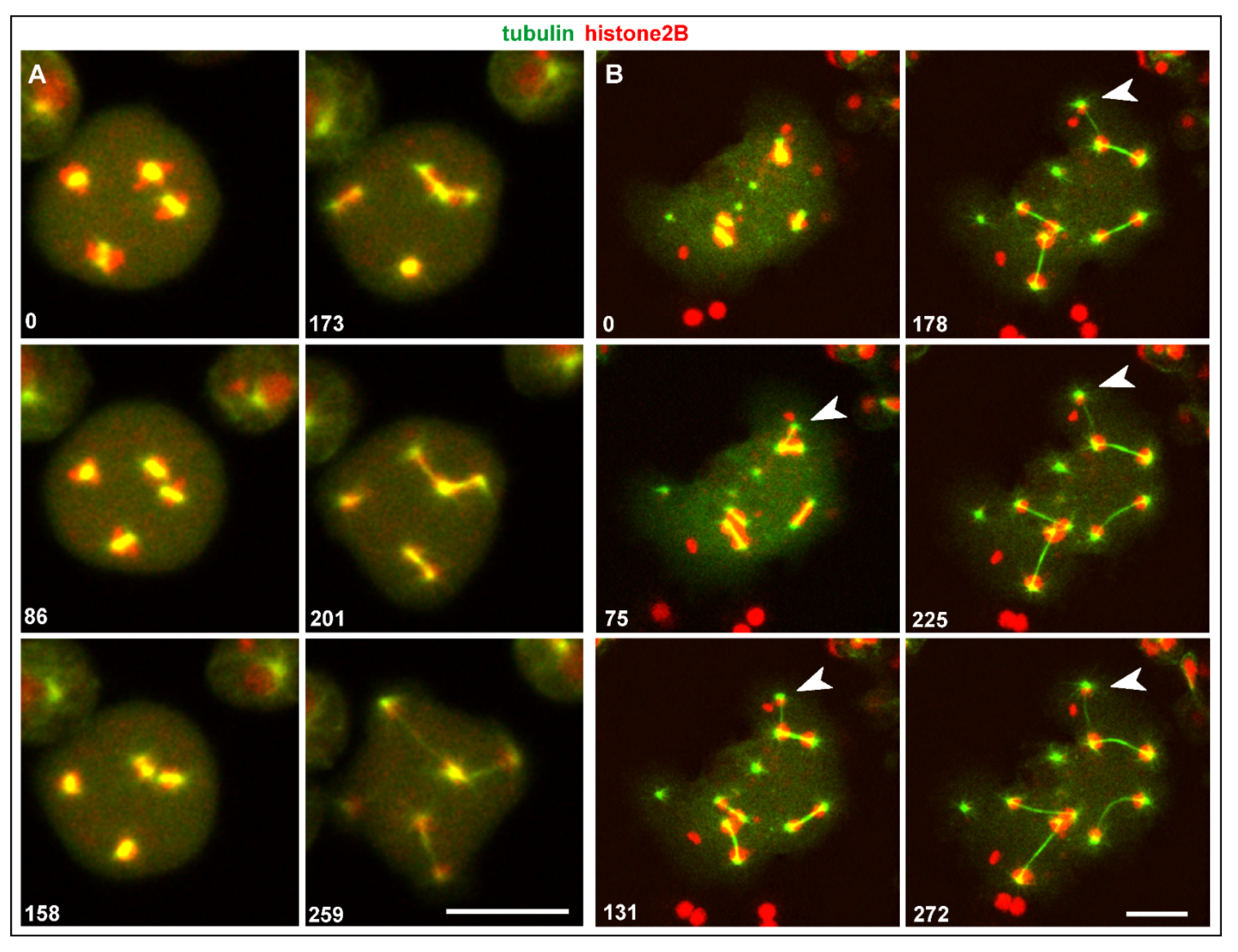
3.3. Spindle Anomalies Resulting in Aberrant Chromosome Segregation
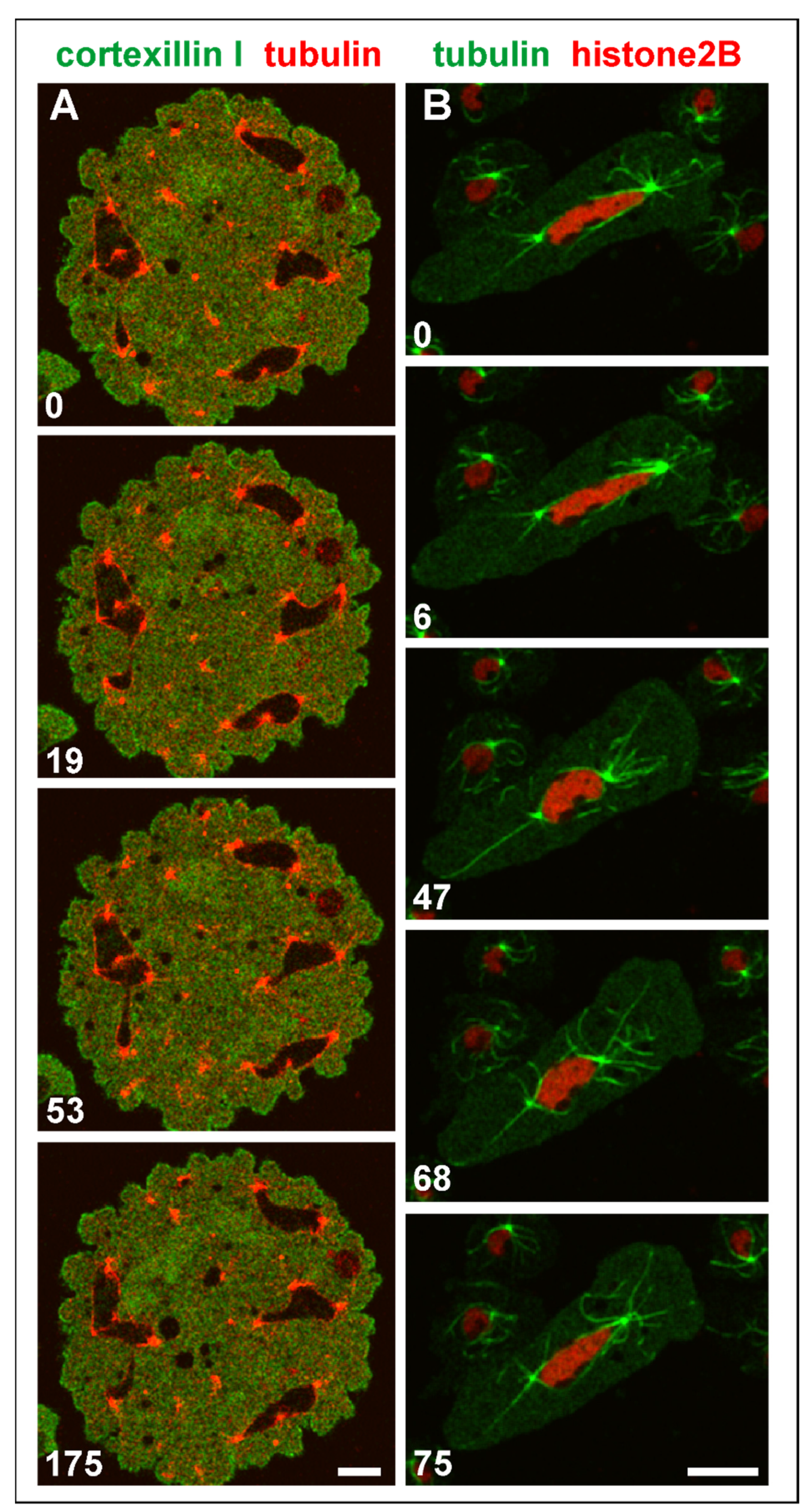
3.4. Single Centrosomes Dividing without Forming a Spindle
3.5. Shape Changes of Multicentric Nuclei in Interphase
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boveri, T. Zur Frage der Entstehung maligner Tumoren; G. Fischer: Jena, Germany, 1914. [Google Scholar]
- Batsios, P.; Gräf, R.; Koonce, M.P.; Larochelle, D.A.; Meyer, I. Nuclear envelope organization in Dictyostelium discoideum. Int. J. Dev. Biol. 2019, 63, 509–519. [Google Scholar] [CrossRef] [Green Version]
- Ueda, M.; Schliwa, M.; Euteneuer, U. Unusual centrosome cycle in Dictyostelium: Correlation of dynamic behavior and structural changes. Mol. Biol. Cell. 1999, 10, 151–160. [Google Scholar] [CrossRef] [Green Version]
- Neujahr, R.; Albrecht, R.; Köhler, J.; Matzner, M.; Schwartz, J.M.; Westphal, M.; Gerisch, G. Microtubule-mediated centrosome motility and the positioning of cleavage furrows in multinucleate myosin II-null cells. J. Cell. Sci. 1998, 111, 1227. [Google Scholar] [CrossRef]
- Gerisch, G.; Faix, J.; Köhler, J.; Müller-Taubenberger, A. Actin-binding proteins required for reliable chromosome segregation in mitosis. Cell Motil. Cytoskelet. 2004, 57, 18–25. [Google Scholar] [CrossRef]
- Müller-Taubenberger, A.; Ishikawa-Ankerhold, H.C.; Kastner, P.M.; Burghardt, E.; Gerisch, G. The STE group kinase SepA controls cleavage furrow formation in Dictyostelium. Cell Motil. Cytoskelet. 2009, 66, 929–939. [Google Scholar] [CrossRef]
- Singh, S.P.; Thomason, P.A.; Lilla, S.; Schaks, M.; Tang, Q.; Goode, B.L.; Machesky, L.M.; Rottner, K.; Insall, R.H. Cell–substrate adhesion drives Scar/WAVE activation and phosphorylation by a Ste20-family kinase, which controls pseudopod lifetime. PLoS Biol. 2020, 18, e3000774. [Google Scholar] [CrossRef]
- Park, K.C.; Rivero, F.; Meili, R.; Lee, S.; Apone, F.; Firtel, R.A. Rac regulation of chemotaxis and morphogenesis in Dictyostelium. EMBO J. 2004, 23, 4177–4189. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.; Seastone, D.J.; Harris, E.; Cardelli, J.A.; Knecht, D.A. RacB regulates cytoskeletal function in Dictyostelium spp. Eukaryot. Cell 2003, 2, 474–485. [Google Scholar] [CrossRef] [Green Version]
- Larochelle, D.A.; Vithalani, K.K.; De Lozanne, A. A novel member of the rho family of small GTP-binding proteins is specifically required for cytokinesis. J. Cell Biol. 1996, 133, 1321–1329. [Google Scholar] [CrossRef] [Green Version]
- Litschko, C.; Brühmann, S.; Csiszár, A.; Stephan, T.; Dimchev, V.; Damiano-Guercio, J.; Junemann, A.; Körber, S.; Winterhoff, M.; Nordholz, B.; et al. Functional integrity of the contractile actin cortex is safeguarded by multiple Diaphanous-related formins. Proc. Natl. Acad. Sci. USA 2019, 116, 3594–3603. [Google Scholar] [CrossRef] [Green Version]
- Rivero, F.; Xiong, H. Chapter Two—Rho Signaling in Dictyostelium discoideum. In International Review of Cell and Molecular Biology; Jeon, K.W., Ed.; Academic Press: Cambridge, MA, USA, 2016; Volume 322, pp. 61–181. [Google Scholar]
- Xiong, H.; Rivero, F.; Euteneuer, U.; Mondal, S.; Mana-Capelli, S.; Larochelle, D.; Vogel, A.; Gassen, B.; Noegel, A.A. Dictyostelium Sun-1 connects the centrosome to chromatin and ensures genome stability. Traffic 2008, 9, 708–724. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.; Haase, I.; Simmeth, E.; Gerisch, G.; Müller-Taubenberger, A. A brilliant monomeric red fluorescent protein to visualize cytoskeleton dynamics in Dictyostelium. FEBS Lett. 2004, 577, 227–232. [Google Scholar] [CrossRef] [Green Version]
- Müller-Taubenberger, A.; Lupas, A.N.; Li, H.; Ecke, M.; Simmeth, E.; Gerisch, G. Calreticulin and calnexin in the endoplasmic reticulum are important for phagocytosis. EMBO J. 2001, 20, 6772–6782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Octtaviani, E.; Effler, J.C.; Robinson, D.N. Enlazin, a natural fusion of two classes of canonical cytoskeletal proteins, contributes to cytokinesis dynamics. Mol. Biol. Cell. 2006, 17, 5275–5286. [Google Scholar] [CrossRef]
- Bindl, J.; Molnar, E.S.; Ecke, M.; Prassler, J.; Müller-Taubenberger, A.; Gerisch, G. Unilateral cleavage furrows in multinucleate cells. Cells 2020, 9, 1493. [Google Scholar] [CrossRef]
- Weber, I.; Gerisch, G.; Heizer, C.; Murphy, J.; Badelt, K.; Stock, A.; Schwartz, J.M.; Faix, J. Cytokinesis mediated through the recruitment of cortexillins into the cleavage furrow. EMBO J. 1999, 18, 586–594. [Google Scholar] [CrossRef]
- Effler, J.C.; Kee, Y.-S.; Berk, J.M.; Tran, M.N.; Iglesias, P.A.; Robinson, D.N. Mitosis-specific mechanosensing and contractile-protein redistribution control cell shape. Curr. Biol. 2006, 16, 1962–1967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malchow, D.; Nägele, B.; Schwarz, H.; Gerisch, G. Membrane-bound cyclic AMP phosphodiesterase in chemotactically responding cells of Dictyostelium discoideum. Eur. J. Biochem. 1972, 28, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Gerhardt, M.; Ecke, M.; Walz, M.; Stengl, A.; Beta, C.; Gerisch, G. Actin and PIP3 waves in giant cells reveal the inherent length scale of an excited state. J. Cell. Sci. 2014, 18, 6786–6792. [Google Scholar] [CrossRef] [Green Version]
- Fukui, Y.; Yumura, S.; Yumura, T.K. Chapter 19 Agar-overlay immunofluorescence: High-resolution studies of cytoskeletal components and their changes during chemotaxis. In Methods in Cell Biology; Spudich, J.A., Ed.; Academic Press: Cambridge, MA, USA, 1987; Volume 28, pp. 347–356. [Google Scholar]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Meth. 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Beck, M.; Förster, F.; Ecke, M.; Plitzko, J.M.; Melchior, F.; Gerisch, G.; Baumeister, W.; Medalia, O. Nuclear pore complex structure and dynamics revealed by cryoelectron tomography. Science 2004, 306, 1387. [Google Scholar] [CrossRef] [Green Version]
- Ma, S.; Triviños-Lagos, L.; Gräf, R.; Chisholm, R.L. Dynein intermediate chain mediated Dynein–dynactin interaction is required for interphase microtubule organization and centrosome replication and separation in Dictyostelium. J. Cell Biol. 1999, 147, 1261–1274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gerisch, G.; Ecke, M.; Neujahr, R.; Prassler, J.; Stengl, A.; Hoffmann, M.; Schwarz, U.S.; Neumann, E. Membrane and actin reorganization in electropulse-induced cell fusion. J. Cell. Sci. 2013, 126, 2069–2078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leo, M.; Santino, D.; Tikhonenko, I.; Magidson, V.; Khodjakov, A.; Koonce, M.P. Rules of engagement: Centrosome–nuclear connections in a closed mitotic system. Biol Open 2012, 1, 1111–1117. [Google Scholar] [CrossRef] [Green Version]
- Gräf, R.; Euteneuer, U.; Ho, T.-H.; Rehberg, M. Regulated expression of the centrosomal protein DdCP224 affects microtubule dynamics and reveals mechanisms for the control of supernumerary centrosome number. Mol. Biol. Cell. 2003, 14, 4067–4074. [Google Scholar] [CrossRef] [Green Version]
- Lénárt, P.; Bacher, C.P.; Daigle, N.; Hand, A.R.; Eils, R.; Terasaki, M.; Ellenberg, J. A contractile nuclear actin network drives chromosome congression in oocytes. Nature 2005, 436, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Mogessie, B.; Schuh, M. Actin protects mammalian eggs against chromosome segregation errors. Science 2017, 357, eaal1647. [Google Scholar] [CrossRef] [Green Version]
- Farina, F.; Gaillard, J.; Guérin, C.; Couté, Y.; Sillibourne, J.; Blanchoin, L.; Théry, M. The centrosome is an actin-organizing centre. Nat. Cell Biol. 2016, 18, 65–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fey, P.; Dodson, R.J.; Basu, S.; Chisholm, R.L. One stop shop for everything Dictyostelium: DictyBase and the Dicty Stock Center in 2012. In Methods in Molecular Biology; Eichinger, L., Rivero, F., Eds.; Humana Press: Totowa, NJ, USA, 2013; pp. 59–92. [Google Scholar]







| Strain | GFP Label | RFP Label | References |
|---|---|---|---|
| AX2-214 | NUP43-GFP | mRFPM-α-tubulin | [13,14] |
| AX2-214 | Calnexin-GFP | mRFPM-α-tubulin | [14,15] |
| RacE-null | pDRH-HygR:GFP- α-tubulin | mRFP1-histone2B | [16,17] |
| RacB-null | pDRH-HygR:GFP- α-tubulin | mRFP1-histone2B | [16,17] |
| Septase-null | GFP-α-tubulin | mRFP1-histone2B | [4,17] |
| Septase-null | GFP-cortexillin I | pDRH-HygR:RFP- α-tubulin | [18,19] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ecke, M.; Prassler, J.; Gerisch, G. Genetic Instability Due to Spindle Anomalies Visualized in Mutants of Dictyostelium. Cells 2021, 10, 2240. https://doi.org/10.3390/cells10092240
Ecke M, Prassler J, Gerisch G. Genetic Instability Due to Spindle Anomalies Visualized in Mutants of Dictyostelium. Cells. 2021; 10(9):2240. https://doi.org/10.3390/cells10092240
Chicago/Turabian StyleEcke, Mary, Jana Prassler, and Günther Gerisch. 2021. "Genetic Instability Due to Spindle Anomalies Visualized in Mutants of Dictyostelium" Cells 10, no. 9: 2240. https://doi.org/10.3390/cells10092240






