Novel, Fully Characterised Bovine Taste Bud Cells of Fungiform Papillae
Abstract
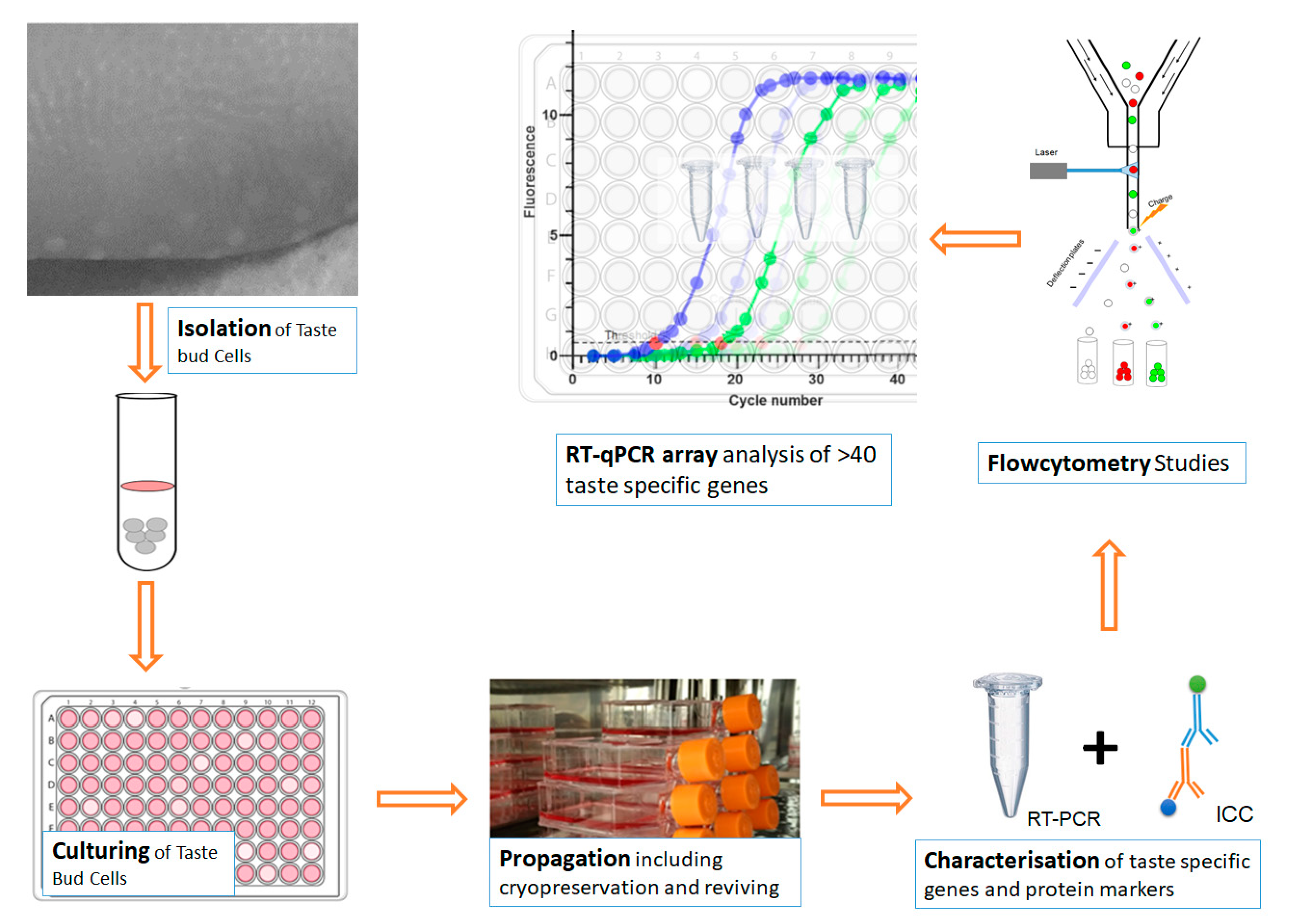
:1. Introduction
2. Materials and Methods
2.1. Isolation and Culturing of BTBCs
2.1.1. Culture Media
2.1.2. Isolation and Culturing of BTBCs
2.1.3. Passaging and Propagation of Cultured Cells
2.1.4. Cryogenic Preservation and Thawing
2.2. Characterisation of BTBCs
2.2.1. Expression of Taste Specific Genes
2.2.2. Primer Design and Specification
2.2.3. Total RNA Extraction
2.2.4. Reverse Transcription PCR (RT-PCR) and Electrophoresis
2.3. Immunocytochemistry
2.3.1. Identification TRC Markers: α-Gustducin and PLCβ2
2.3.2. Identification of Bitter Membrane Receptors: T2R7
2.4. Flow Cytometry
2.4.1. General Flow-Cytometry Procedure
2.4.2. CD73 Expressing Cells
2.4.3. TRPM5-Positive Cells
2.4.4. Bitter Receptors
2.5. Calcium Imaging
2.6. Preliminary Data Mining and Bioinformatics Studies on Bovine α-gustducin
2.6.1. Chromosomal Mapping
2.6.2. Protein Information
2.6.3. Multiple Alignment Sequence and Matrix Identity
2.7. RT-qPCR Analysis of Taste Specific Genes
2.7.1. RT-qPCR Array
2.7.2. Reverse Transcription: First Strand cDNA Synthesis
2.7.3. Quantitative RT-PCR (RT-qPCR)
3. Results
3.1. Bovine Taste Bud Cells (BTBCs): Isolation and In Vitro Culturing
3.2. RT-PCR: Expression of Taste Specific Genes
3.3. Immunocytochemistry
3.4. Flow Cytometry
3.5. Calcium Imaging
3.6. Data Mining and Bioinformatics Studies on Bovine α-Gustducin
3.7. RT-qPCR Analysis of Taste Specific Genes
3.8. RT-qPCR Array Quantification
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Peng, Y.; Gillis-Smith, S.; Jin, H.; Tränkner, D.; Ryba, N.J.P.; Zuker, C.S. Sweet and bitter taste in the brain of awake behaving animals. Nature 2015, 527, 512–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kishi, M.; Emori, Y.; Tsukamoto, Y.; Abe, K. Primary culture of rat taste bud cells that retain molecular markers for taste buds and permit functional expression of foreign genes. Neuroscience 2001, 106, 217–225. [Google Scholar] [CrossRef]
- Ruiz, C.J.; Stone, L.M.; McPheeters, M.; Ogura, T.; Böttger, B.; Lasher, R.S.; Finger, T.E.; Kinnamon, S.C. Mainte-nance of rat taste buds in primary culture. Chem. Senses 2001, 26, 861–873. [Google Scholar] [CrossRef] [Green Version]
- Spielman, A.I.; Nagai, H.; Sunavala, G.; Dasso, M.; Breer, H.; Boekhoff, I.; Huque, T.; Whitney, G.; Brand, J.G. Rapid kinetics of second messenger production in bitter taste. Am. J. Physiol. Content 1996, 270, C926–C931. [Google Scholar] [CrossRef]
- Romanov, R.; Rogachevskaja, O.; Bystrova, M.; Jiang, P.; Margolskee, R.; Kolesnikov, S. Afferent neurotransmission mediated by hemichannels in mammalian taste cells. EMBO J. 2007, 26, 657–667. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, R.; Shigemura, N.; Sanematsu, K.; Yasumatsu, K.; Ishizuka, S.; Ninomiya, Y. Taste Responsiveness of Fungiform Taste Cells With Action Potentials. J. Neurophysiol. 2006, 96, 3088–3095. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maehashi, K.; Matano, M.; Wang, H.; Vo, L.; Yamamoto, Y.; Huang, L. Bitter peptides activate hTAS2Rs, the human bitter receptors. Biochem. Biophys. Res. Commun. 2008, 365, 851–855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Upadhyaya, J.; Pydi, S.P.; Singh, N.; Aluko, R.E.; Chelikani, P. Bitter taste receptor T2R1 is activated by dipeptides and tripeptides. Biochem. Biophys. Res. Commun. 2010, 398, 331–335. [Google Scholar] [CrossRef]
- Levit, A.; Nowak, S.; Peters, M.; Wiener, A.; Meyerhof, W.; Behrens, M.; Niv, M.Y. The bitter pill: Clinical drugs that activate the human bitter taste receptor TAS2R14. FASEB J. 2013, 28, 1181–1197. [Google Scholar] [CrossRef] [PubMed]
- Behrens, M.; Foerster, S.; Staehler, F.; Raguse, J.-D.; Meyerhof, W. Gustatory Expression Pattern of the Human TAS2R Bitter Receptor Gene Family Reveals a Heterogenous Population of Bitter Responsive Taste Receptor Cells. J. Neurosci. 2007, 27, 12630–12640. [Google Scholar] [CrossRef] [Green Version]
- Farbman, A.I. Fine structure of degenerating taste buds after denervation. Development 1969, 22, 55–68. [Google Scholar] [CrossRef]
- Ozdener, H.; Yee, K.K.; Cao, J.; Brand, J.G.; Teeter, J.H.; Rawson, N.E. Characterization and long-term maintenance of rat taste cells in culture. Chem. Sens. 2006, 31, 279–290. [Google Scholar] [CrossRef] [Green Version]
- Ozdener, M.H.; Rawson, N.E. Culture and maintenance of taste cells in vitro. Vitr. Cell. Dev. Biol.-Anim. 2011, 47, 513–514. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Menon, V.; Thomas, R.; Ghale, A.; Reinhard, C.; Pruszak, J. Flow Cytometry Protocols for Surface and Intracellular Antigen Analyses of Neural Cell Types. J. Vis. Exp. 2014, 52241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, N.; Chakraborty, R.; Bhullar, R.P.; Chelikani, P. Differential expression of bitter taste receptors in non-cancerous breast epithelial and breast cancer cells. Biochem. Biophys. Res. Commun. 2014, 446, 499–503. [Google Scholar] [CrossRef]
- Dando, R.; Dvoryanchikov, G.; Pereira, E.; Chaudhari, N.; Roper, S.D. Adenosine Enhances Sweet Taste through A2B Receptors in the Taste Bud. J. Neurosci. 2012, 32, 322–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pirttimaki, T.; Sims, R.; Saunders, G.; Antonio, S.; Codadu, N.; Parri, H. Astrocyte-Mediated Neuronal Synchronization Properties Revealed by False Gliotransmitter Release. J. Neurosci. 2017, 37, 9859–9870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crowe, J.; El-Tamer, A.; Nagel, D.; Koroleva, A.; Madrid-Wolff, J.; Olarte, O.; Sokolovsky, S.; Estevez-Priego, E.; Ludl, A.; Soriano, J.; et al. Development of two-photon polymerised scaffolds for optical interrogation and neurite guidance of human iPSC-derived cortical neuronal networks. Lab Chip 2020, 20, 1792–1806. [Google Scholar] [CrossRef] [PubMed]
- Aken, B.L.; Ayling, S.C.; Barrell, D.; Clarke, L.; Curwen, V.; Fairley, S.; Banet, J.F.; Billis, K.; Giron, C.G.; Hourlier, T.; et al. The Ensembl gene annotation system. Database 2016, 2016, baw093. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2016, 45, D200–D203. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2016, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amatori, S.; Persico, G.; Fanelli, M. Real-time quantitative PCR array to study drug-induced changes of gene expression in tumor cell lines. J. Cancer Metastasis Treat. 2017, 3, 90. [Google Scholar] [CrossRef]
- Ozdener, M.H.; Rawson, N.E. Primary culture of mammalian taste epithelium. In Epithelial Cell Culture Protocols. Methods in Molecular Biology (Methods and Protocols), 2nd ed.; Randell, S., Fulcher, M., Eds.; Humana Press: Totowa, NJ, USA, 2013; Volume 945, pp. 95–107. [Google Scholar]
- Calvo, S.S.-C.; Egan, J.M. The endocrinology of taste receptors. Nat. Rev. Endocrinol. 2015, 11, 213–227. [Google Scholar] [CrossRef] [PubMed]
- Nelson, G.; Hoon, M.A.; Chandrashekar, J.; Zhang, Y.; Ryba, N.J.; Zuker, C.S. Mammalian Sweet Taste Receptors. Cell 2001, 106, 381–390. [Google Scholar] [CrossRef] [Green Version]
- Duval, K.; Grover, H.; Han, L.-H.; Mou, Y.; Pegoraro, A.F.; Fredberg, J.; Chen, Z. Modeling Physiological Events in 2D vs. 3D Cell Culture. Physiology 2017, 32, 266–277. [Google Scholar] [CrossRef]
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med Sci. 2016, 12, 910–919. [Google Scholar] [CrossRef]
- Matsumoto, I.; Ohmoto, M.; Abe, K. Functional diversification of taste cells in vertebrates. Semin. Cell Dev. Biol. 2012, 24, 210–214. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, H.M.; Barlow, L.A. Differential expression of a BMP4 reporter allele in anterior fungiform versus posterior circumvallate taste buds of mice. BMC Neurosci. 2010, 11, 129. [Google Scholar] [CrossRef] [Green Version]
- Yee, K.K.; Li, Y.; Redding, K.; Iwatsuki, K.; Margolskee, R.F.; Jiang, P. Lgr5-EGFP Marks Taste Bud Stem/Progenitor Cells in Posterior Tongue. Stem Cells 2013, 31, 992–1000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finger, T.E. Cell Types and Lineages in Taste Buds. Chem. Senses 2005, 30, i54–i55. [Google Scholar] [CrossRef] [Green Version]
- Yang, R.; Dzowo, Y.; Wilson, C.; Russell, R.; Kidd, G.; Salcedo, E.; Lasher, R.; Kinnamon, J.; Finger, T. Three-dimensional reconstructions of mouse circumvallate taste buds using serial blockface scanning electron microscopy: I. Cell types and the apical region of the taste bud. J. Comp. Neurol. 2019, 528, 756–771. [Google Scholar] [CrossRef]
- Ren, W.; Lewandowski, B.C.; Watson, J.; Aihara, E.; Iwatsuki, K.; Bachmanov, A.; Margolskee, R.; Jiang, P. Single Lgr5- or Lgr6-expressing taste stem/progenitor cells generate taste bud cells ex vivo. Proc. Natl. Acad. Sci. USA 2014, 111, 16401–16406. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perea-Martinez, I.; Nagai, T.; Chaudhari, N. Functional Cell Types in Taste Buds Have Distinct Longevities. PLoS ONE 2013, 8, e53399. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.; Yang, R.; Thomas, S.M.; Kinnamon, J.C. Qualitative and quantitative differences between taste buds of the rat and mouse. BMC Neurosci. 2007, 8, 5. [Google Scholar] [CrossRef] [Green Version]
- Cossarizza, A.; Chang, H.; Radbruch, A.; Akdis, M.; Andrä, I.; Annunziato, F.; Bacher, P.; Barnaba, V.; Battistini, L.; Bauer, W.; et al. Guidelines for the use of flow cytometry and cell sorting in immunological studies. Eur. J. Immunol. 2017, 47, 1584–1797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [Green Version]
- Huang, A.L.; Chen, X.; Hoon, M.A.; Chandrashekar, J.; Guo, W.; Tränkner, D.; Ryba, N.J.P.; Zuker, C.S. The cells and logic for mammalian sour taste detection. Nature 2006, 442, 934–938. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kataoka, S.; Yang, R.; Ishimaru, Y.; Matsunami, H.; Sévigny, J.; Kinnamon, J.C.; Finger, T. The Candidate Sour Taste Receptor, PKD2L1, Is Expressed by Type III Taste Cells in the Mouse. Chem. Senses 2007, 33, 243–254. [Google Scholar] [CrossRef] [Green Version]
- Roche, K.C.; Gracz, A.; Liu, X.F.; Newton, V.; Akiyama, H.; Magness, S.T. SOX9 Maintains Reserve Stem Cells and Preserves Radioresistance in Mouse Small Intestine. Gastroenterology 2015, 149, 1553–1563.e10. [Google Scholar] [CrossRef] [Green Version]
- Smith, N.R.; Swain, J.R.; Davies, P.S.; Gallagher, A.C.; Parappilly, M.S.; Beach, C.Z.; Streeter, P.R.; Williamson, I.A.; Magness, S.T.; Wong, M.H. Monoclonal Antibodies Reveal Dynamic Plasticity Between Lgr5- and Bmi1-Expressing Intestinal Cell Populations. Cell. Mol. Gastroenterol. Hepatol. 2018, 6, 79–96. [Google Scholar] [CrossRef] [Green Version]
- Donati, G.; Watt, F.M. Stem Cell Heterogeneity and Plasticity in Epithelia. Cell Stem Cell 2015, 16, 465–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gaillard, D.; Xu, M.; Liu, F.; Millar, S.; Barlow, L.A. β-Catenin Signaling Biases Multipotent Lingual Epithelial Progenitors to Differentiate and Acquire Specific Taste Cell Fates. PLoS Genet. 2015, 11, e1005208. [Google Scholar] [CrossRef] [PubMed]
- Okubo, T.; Clark, C.; Hogan, B.L. Cell lineage mapping of taste bud cells and keratinocytes in the mouse tongue and soft palate. Stem Cells 2009, 27, 442–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aihara, E.; Mahe, M.M.; Schumacher, M.A.; Matthis, A.L.; Feng, R.; Ren, W.; Noah, T.K.; Matsu-Ura, T.; Moore, S.R.; Hong, C.I.; et al. Characterization of stem/progenitor cell cycle using murine circumvallate papilla taste bud organoid. Sci. Rep. 2015, 5, 17185. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, I.; Ohmoto, M.; Narukawa, M.; Yoshihara, Y.; Abe, K. Skn-1a (Pou2f3) specifies taste receptor cell line-age. Nat. Neurosci. 2011, 14, 685–687. [Google Scholar] [CrossRef] [Green Version]
- Miura, H.; Barlow, L.A. Taste bud regeneration and the search for taste progenitor cells. Arch Ital. Biol. 2010, 148, 107–118. [Google Scholar]
- He, W.; Danilova, V.; Zou, S.; Hellekant, G.; Max, M.; Margolskee, R.F.; Damak, S. Partial rescue of taste responses of α-gustducin null mice by transgenic expression of α-transducin. Chem. Senses 2002, 27, 719–727. [Google Scholar] [CrossRef] [Green Version]
- Danilova, V.; Damak, S.; Margolskee, R.F.; Hellekant, G. Taste responses to sweet stimuli in α-gustducin knockout and wild-type mice. Chem. Senses 2006, 31, 573–580. [Google Scholar] [CrossRef] [Green Version]
- He, W.; Yasumatsu, K.; Varadarajan, V.; Yamada, A.; Lem, J.; Ninomiya, Y.; Damak, S. Umami taste responses are mediated by α-transducin and α-gustducin. J. Neurosci. 2004, 24, 7674–7680. [Google Scholar] [CrossRef] [PubMed]
- Ruiz, C.J.; Wray, K.; Delay, E.; Margolskee, R.F.; Kinnamon, S.C. Behavioral evidence for a role of α-gustducin in glutamate taste. Chem. Senses 2003, 28, 573–579. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Hoon, M.A.; Chandrashekar, J.; Mueller, K.L.; Cook, B.; Wu, D.; Zuker, C.S.; Ryba, N.J. Coding of Sweet, Bitter, and Umami Tastes: Different Receptor Cells Sharing Similar Signaling Pathways. Cell 2003, 112, 293–301. [Google Scholar] [CrossRef] [Green Version]
- Pérez, C.; Huang, L.; Rong, M.; Kozak, J.; Preuss, A.; Zhang, H.; Max, M.; Margolskee, R. A transient receptor potential channel expressed in taste receptor cells. Nat. Neurosci. 2002, 5, 1169–1176. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Zhao, Z.; Margolskee, R.; Liman, E. The Transduction Channel TRPM5 Is Gated by Intracellular Calcium in Taste Cells. J. Neurosci. 2007, 27, 5777–5786. [Google Scholar] [CrossRef]
- Nishida, K.; Kubota, T.; Matsumoto, S.; Kato, J.; Watanabe, Y.; Yamamoto, A.; Furui, M.; Ohishi, A.; Nagasawa, K. Expression of Prostatic Acid Phosphatase in Rat Circumvallate Papillae. PLoS ONE 2016, 11, e0158401. [Google Scholar] [CrossRef]
- Ramsköld, D.; Wang, E.T.; Burge, C.B.; Sandberg, R. An Abundance of Ubiquitously Expressed Genes Revealed by Tissue Transcriptome Sequence Data. PLoS Comput. Biol. 2009, 5, e1000598. [Google Scholar] [CrossRef] [PubMed]
- Saimi, Y.; Kung, C. Calmodulin as an Ion Channel Subunit. Annu. Rev. Physiol. 2002, 64, 289–311. [Google Scholar] [CrossRef] [PubMed]
- Nagano, O.; Murakami, D.; Hartmann, D.; De Strooper, B.; Saftig, P.; Iwatsubo, T.; Nakajima, M.; Shinohara, M.; Saya, H. Cell–matrix interaction via CD44 is independently regulated by different metalloproteinases activated in response to extracellular Ca2+ influx and PKC activation. J. Cell Biol. 2004, 165, 893–902. [Google Scholar] [CrossRef]
- Lu, S.G.; Zhao, F.L.; Herness, S. Physiological phenotyping of cholecystokinin-responsive rat taste receptor cells. Neurosci. Lett. 2003, 351, 157–160. [Google Scholar] [CrossRef]
- Yoshida, R.; Shin, M.; Yasumatsu, K.; Takai, S.; Inoue, M.; Shigemura, N.; Takiguchi, S.; Nakamura, S.; Ninomiya, Y. The Role of Cholecystokinin in Peripheral Taste Signaling in Mice. Front. Physiol. 2017, 8, 866. [Google Scholar] [CrossRef] [Green Version]
- Palmer, R.K.; Long, D.; Brennan, F.; Buber, T.; Bryant, R.; Salemme, F.R. A High Throughput In Vivo Assay for Taste Quality and Palatability. PLoS ONE 2013, 8, e72391. [Google Scholar] [CrossRef] [Green Version]
- Mohamed-Ahmed, A.; Soto, J.; Ernest, T.; Tuleu, C. Non-human tools for the evaluation of bitter taste in the design and development of medicines: A systematic review. Drug Discov. Today 2016, 21, 1170–1180. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Gabitto, M.; Peng, Y.; Ryba, N.J.P.; Zuker, C.S. A Gustotopic Map of Taste Qualities in the Mammalian Brain. Science 2011, 333, 1262–1266. [Google Scholar] [CrossRef] [Green Version]
- Anand, V.; Kataria, M.; Kukkar, V.; Saharan, V.; Choudhury, P. The latest trends in the taste assessment of pharmaceuticals. Drug Discov. Today 2007, 12, 257–265. [Google Scholar] [CrossRef] [PubMed]
- Spector, A.C.; Kopka, S.L. Rats Fail to Discriminate Quinine from Denatonium: Implications for the Neural Coding of Bitter-Tasting Compounds. J. Neurosci. 2002, 22, 1937–1941. [Google Scholar] [CrossRef] [Green Version]
- Hettinger, T.P.; Formaker, B.K.; Frank, M.E. Cycloheximide: No ordinary bitter stimulus. Behav. Brain Res. 2007, 180, 4–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boughter, J.D.; John, S.J.S.; Noel, D.T.; Ndubuizu, O.; Smith, D.V. A brief-access test for bitter taste in mice. Chem. Senses 2002, 27, 133–142. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Gillis-Smith, S.; Peng, Y.; Zhang, J.; Chen, X.; Salzman, C.D.; Ryba, N.J.P.; Zuker, C.S. The coding of valence and identity in the mammalian taste system. Nature 2018, 558, 127–131. [Google Scholar] [CrossRef]








Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ftuwi, H.; Parri, R.; Mohammed, A.R. Novel, Fully Characterised Bovine Taste Bud Cells of Fungiform Papillae. Cells 2021, 10, 2285. https://doi.org/10.3390/cells10092285
Ftuwi H, Parri R, Mohammed AR. Novel, Fully Characterised Bovine Taste Bud Cells of Fungiform Papillae. Cells. 2021; 10(9):2285. https://doi.org/10.3390/cells10092285
Chicago/Turabian StyleFtuwi, Habtom, Rheinallt Parri, and Afzal R. Mohammed. 2021. "Novel, Fully Characterised Bovine Taste Bud Cells of Fungiform Papillae" Cells 10, no. 9: 2285. https://doi.org/10.3390/cells10092285
APA StyleFtuwi, H., Parri, R., & Mohammed, A. R. (2021). Novel, Fully Characterised Bovine Taste Bud Cells of Fungiform Papillae. Cells, 10(9), 2285. https://doi.org/10.3390/cells10092285





