Induced Pluripotent Stem Cells (iPSCs)—Roles in Regenerative Therapies, Disease Modelling and Drug Screening
Abstract
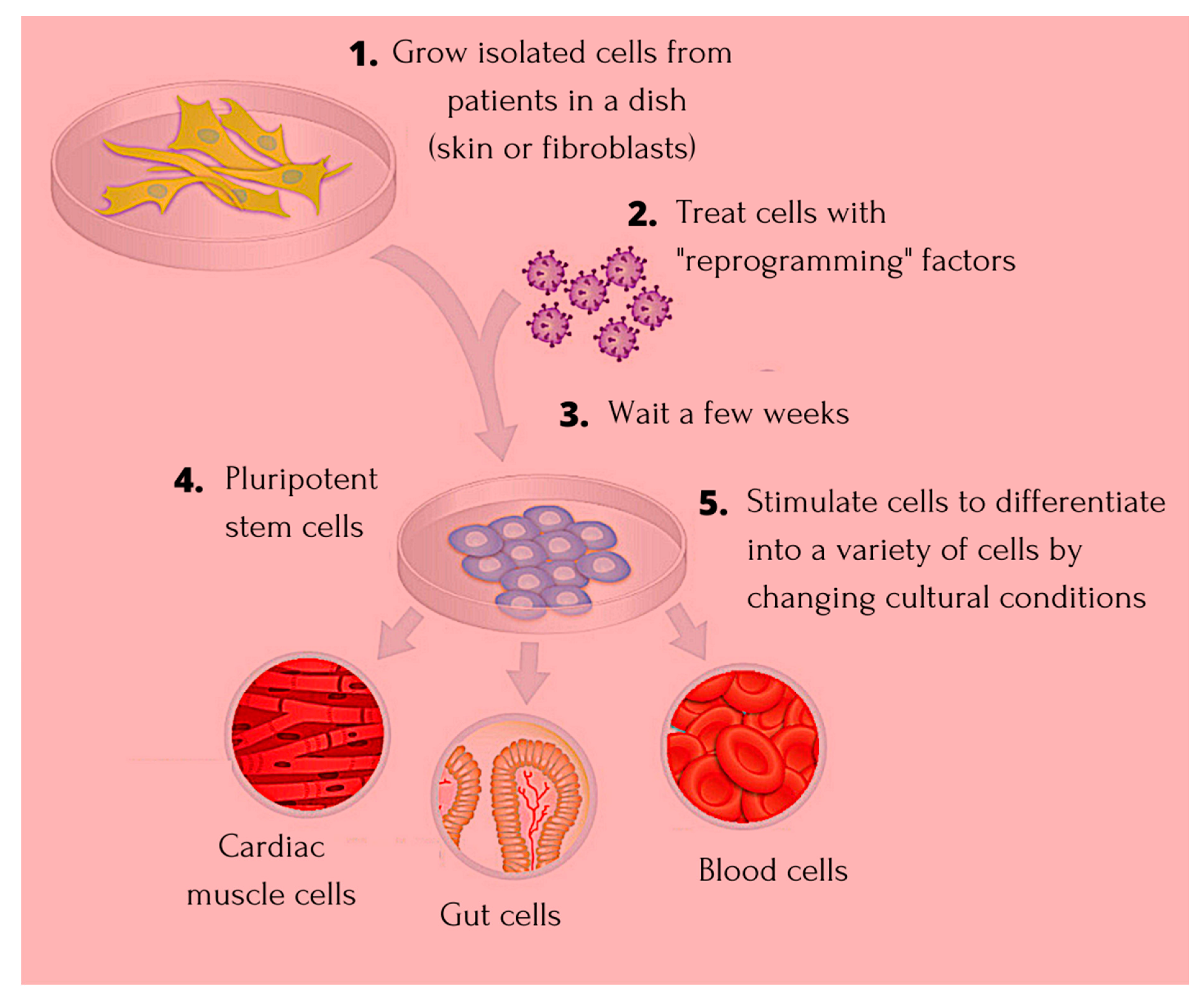
:1. Introduction
1.1. Induced Pluripotent Stem Cells—The Niche Favoring Unique Aspects
1.2. Application of iPSC in Cardiac Disease
1.3. Application of iPSC in Degenerative Diseases
1.4. Application of iPSC in Blood Disorders
1.5. Application of iPSC in Organ Dysfunctions
1.6. Application of iPSCs in Cancer Syndromes
2. Induced Pluripotent Stem Cells: Advantages and Beyond
3. ESCs and iPSCs in Clinical Trials
4. Conclusions
Funding
Conflicts of Interest
References
- Weismann, A. The Germ-Plasm: A Theory of Heredity; Charles Scribner’s Sons: New York, NY, USA, 1893. [Google Scholar]
- Gurdon, J.B.; Byrne, J.A. The first half-century of nuclear transplantation. Proc. Natl. Acad. Sci. USA 2003, 100, 8048–8052. [Google Scholar] [CrossRef] [Green Version]
- Bradley, A.; Evans, M.; Kaufman, M.H.; Robertson, E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 1984, 309, 255–256. [Google Scholar] [CrossRef]
- Cowan, C.A.; Atienza, J.; Melton, D.A.; Eggan, K. Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science 2005, 309, 1369–1373. [Google Scholar] [CrossRef] [Green Version]
- Karagiannis, P.; Takahashi, K.; Saito, M.; Yoshida, Y.; Okita, K.; Watanabe, A.; Inoue, H.; Yamashita, J.K.; Todani, M.; Nakagawa, M.; et al. Induced pluripotent stem cells and their use in human models of disease and development. Physiol. Rev. 2018, 99, 79–114. [Google Scholar] [CrossRef] [PubMed]
- Abbar, A.A.; Ngai, S.C.; Nograles, N.; Alhaji, S.Y.; Abdullah, S. Induced pluripotent stem cells: Reprogramming platforms and applications in cell replacement therapy. BioRes. Open Access 2020, 9, 121–136. [Google Scholar] [CrossRef]
- Nishikawa, S.; Goldstein, R.A.; Nierras, C.R. The promise of human induced pluripotent stem cells for research and therapy. Nat. Rev. Mol. Cell Biol. 2008, 9, 725–729. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Ni, X.; Zhao, Z.-A.; Lei, W.; Hu, S. The application of induced pluripotent stem cells in cardiac disease modelling and drug testing. J. Cardiovasc. Transl. Res. 2018, 11, 366–374. [Google Scholar] [CrossRef]
- Mandai, M.; Watanabe, A.; Kurimoto, Y.; Hirami, Y.; Morinaga, C.; Daimon, T.; Fujihara, M.; Akimaru, H.; Sakai, N.; Shibata, Y.; et al. Autologous induced stem-cell-derived retinal cells for macular degeneration. N. Engl. J. Med. 2017, 376, 1038–1046. [Google Scholar] [CrossRef]
- Sundberg, M.; Bogetofte, H.; Lawson, T.; Jansson, J.; Smith, G.; Astradsson, A.; Moore, M.; Osborn, T.; Cooper, O.; Spealman, R.; et al. Improved cell therapy protocols for Parkinson’s disease based on differentiation efficiency and safety of hESC-, hiPSC-, and non-human primate iPSC-derived dopaminergic neurons. Stem Cells 2013, 31, 1548–1562. [Google Scholar] [CrossRef] [Green Version]
- Sridharan, R.; Tchieu, J.; Mason, M.J.; Yachechko, R.; Kuoy, E.; Horvath, S.; Zhou, Q.; Plath, K. Role of the murine reprogramming factors in the induction of pluripotency. Cell 2009, 136, 364–377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winkler, T.; Cantilena, A.; Metais, J.-Y.; Xu, X.; Nguyen, A.-D.; Borate, B.; Antosiewicz-Bourget, J.E.; Wolfsberg, T.G.; Thomson, J.A.; Dunbar, C.E. No evidence for clonal selection due to lentiviral integration sites in human induced pluripotent stem cells. Stem Cells 2010, 28, 687–694. [Google Scholar] [CrossRef] [Green Version]
- Somers, A.; Jean, J.-C.; Sommer, C.A.; Omari, A.; Ford, C.C.; Mills, J.A.; Ying, L.; Sommer, A.G.; Jean, J.M.; Smith, B.W.; et al. Generation of transgene-free lung disease-specific human induced pluripotent stem cells using a single excisable lentiviral stem cell cassette. Stem Cells 2010, 28, 1728–1740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stadtfeld, M.; Hochedlinger, K. Induced pluripotency: History, mechanisms, and applications. Genes Dev. 2010, 24, 2239–2263. [Google Scholar] [CrossRef] [Green Version]
- Okita, K.; Nakagawa, M.; Hyenjong, H.; Ichisaka, T.; Yamanaka, S. Generation of mouse induced pluripotent stem cells without viral vectors. Science 2008, 322, 949–953. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.S.; Bishop, E.S.; Zhang, R.; Yu, X.; Farina, E.M.; Yan, S.; Zhao, C.; Zeng, Z.; Shu, Y.; Wu, X.; et al. Adenovirus-mediated gene delivery: Potential applications for gene and cell-based therapies in the new era of personalized medicine. Genes Dis. 2017, 4, 43–63. [Google Scholar] [CrossRef]
- Ong, S.-G.; Lee, W.H.; Kodo, K.; Wu, J.C. MicroRNA-mediated regulation of differentiation and trans-differentiation in stem cells. Adv. Drug Deliv. Rev. 2015, 88, 3–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakagawa, M.; Koyanagi, M.; Tanabe, K.; Takahashi, K.; Ichisaka, T.; Aoi, T.; Okita, K.; Mochiduki, Y.; Takizawa, N.; Yamanaka, S. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008, 26, 101–106. [Google Scholar] [CrossRef]
- Huangfu, D.; Osafune, K.; Maehr, R.; Guo, W.; Eijkelenboom, A.; Chen, S.; Muhlestein, W.; Melton, D.A. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 2008, 26, 1269–1275. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-L.; Chang, D.C.; Chang-Lin, S.; Lin, C.-H.; Wu, D.T.S.; Chen, D.T.; Ying, S.-Y. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA 2008, 14, 2115–2124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Wang, X.; Nyberg, S.L. Application of induced pluripotent stem cells in liver diseases. Cell Med. 2014, 7, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [Green Version]
- Zhou, W.; Freed, C.R. Adenoviral gene delivery can reprogram human fibroblasts to induced pluripotent stem cells. Stem Cells 2009, 27, 2667–2674. [Google Scholar] [CrossRef]
- Nishishita, N.; Takenaka, C.; Fusaki, N.; Kawamata, S. Generation of human induced pluripotent stem cells from cord blood cells. J. Stem Cells 2011, 6, 101–108. [Google Scholar]
- Kim, D.; Kim, C.-H.; Moon, J.-I.; Chung, Y.-G.; Chang, M.-Y.; Han, B.-S.; Ko, S.; Yang, E.; Cha, K.Y.; Lanza, R.; et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming factors. Cell Stem Cell 2009, 4, 472–476. [Google Scholar] [CrossRef] [Green Version]
- Yakubov, E.; Rechavi, G.; Rozenblatt, S.; Givol, D. Reprogramming of human fibroblasts to pluripotent stem cells using mRNA of four transcription factors. Biochem. Biophys. Res. Commun. 2010, 394, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Liang, J.; Ni, S.; Zhou, T.; Qing, X.; Li, H.; He, W.; Chen, J.; Li, F.; Zhuang, Q.; et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 2010, 7, 51–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samavarchi-Tehrani, P.; Golipour, A.; David, L.; Sung, H.-K.; Beyer, T.A.; Datti, A.; Woltjen, K.; Nagy, A.; Wrana, J.L. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 2010, 7, 64–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, J.; Nichols, J.; Theunissen, T.W.; Guo, G.; van Oosten, A.L.; Barrandon, O.; Wray, J.; Yamanaka, S.; Chambers, I.; Smith, A. Nanog is the gateway to the pluripotent ground state. Cell 2009, 138, 722–737. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gidekel, S.; Bergman, Y. A unique developmental pattern of Oct-3/4 DNA methylation is controlled by a cis-demodification element. J. Biol. Chem. 2002, 277, 34521–34530. [Google Scholar] [CrossRef] [Green Version]
- Hanna, J.; Wernig, M.; Markoulaki, S.; Sun, C.-W.; Meissner, A.; Cassady, J.P.; Beard, C.; Brambrink, T.; Wu, L.-C.; Townes, T.M.; et al. Treatment of sickle cell anemia mouse model with iPS cells generated from autologous skin. Science 2007, 318, 1920–1923. [Google Scholar] [CrossRef]
- Zou, J.; Sweeney, C.L.; Chou, B.-K.; Choi, U.; Pan, J.; Wang, H.; Dowey, S.N.; Cheng, L.; Malech, H.L. Oxidase-deficient neutrophils from X-linked chronic granulomatous disease iPS cells: Functional correction by zinc finger nuclease-mediated safe harbor targeting. Blood 2011, 117, 5561–5572. [Google Scholar] [CrossRef] [Green Version]
- Olgasi, C.; Talmon, M.; Merlin, S.; Cucci, A.; Richaud-Patin, Y.; Ranaldo, G.; Colangelo, D.; Di Scipio, F.; Berta, G.N.; Borsotti, C.; et al. Patient-specific iPSC-derived endothelial cells provide long-term phenotypic correction of hemophilia A. Stem Cell Rep. 2018, 11, 1391–1406. [Google Scholar] [CrossRef] [Green Version]
- Ramaswamy, S.; Tonnu, N.; Menon, T.; Lewis, B.M.; Green, K.T.; Wampler, D.; Monahan, P.E.; Verma, I.M. Autologous and heterologous cell therapy for hemophilia B towards functional restoration of factor IX. Cell Rep. 2018, 23, 1565–1580. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-W.; Lai, Y.-S.; Westin, E.; Khodadadi-Jamayran, A.; Pawlik, K.M.; Lamb, L.S., Jr.; Goldman, F.D.; Townes, T.M. Modeling human severe combined immunodeficiency and correction by CRISPR/Cas9-enhanced gene targeting. Cell Rep. 2015, 12, 1668–1677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.-J.; Bouhassira, E.E. Zinc-finger nuclease-mediated correction of alpha-thalassemia in iPS cells. Blood 2012, 120, 3906–3914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laperle, A.H.; Sances, S.; Yucer, N.; Dardov, V.J.; Garcia, V.J.; Ho, R.; Fulton, A.N.; Jones, M.R.; Roxas, K.M.; Avalos, P.; et al. iPSC modeling of young-onset Parkinson’s disease reveals a molecular signature of disease and novel therapeutic candidates. Nature 2020, 26, 289–299. [Google Scholar] [CrossRef] [PubMed]
- Ye, T.; Duan, Y.; Tsang, H.W.S.; Xu, H.; Chen, Y.; Cao, H.; Chen, Y.; Fu, A.K.Y.; Ip, N.Y. Efficient manipulation of gene dosage in human iPSCs using CRISPR/Cas9 nickases. Nat. Commun. Biol. 2021, 4, 195. [Google Scholar] [CrossRef]
- Wang, L.; Yi, F.; Fu, L.; Yang, J.; Wang, S.; Wang, Z.; Suzuki, K.; Sun, L.; Xu, X.; Yu, Y.; et al. CRISPR/Cas9-mediated targeted gene correction in amyotrophic lateral sclerosis patient iPSCs. Protein Cell 2017, 8, 365–378. [Google Scholar] [CrossRef]
- Wang, G.; McCain, M.L.; Yang, L.; He, A.; Pasqualini, F.S.; Agarwal, A.; Yuan, H.; Jiang, D.; Zhang, D.; Zangi, L.; et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat. Med. 2014, 20, 616–623. [Google Scholar] [CrossRef]
- Yamamoto, Y.; Makiyama, T.; Harita, T.; Sasaki, K.; Wuriyanghai, Y.; Hayano, M.; Nishiuchi, S.; Kohjitani, H.; Hirose, S.; Chen, J.; et al. Allele-specific ablation rescues electrophysiological abnormalities in a human iPS cell model of long-QT syndrome with a CALM2 mutation. Hum. Mol. Genet. 2017, 26, 1670–1677. [Google Scholar] [CrossRef]
- Firth, A.L.; Menon, T.; Parker, G.S.; Qualls, S.J.; Lewis, B.M.; Ke, E.; Dargitz, C.T.; Wright, R.; Khanna, A.; Gage, F.H.; et al. Functional gene correction for cystic fibrosis in lung epithelial cells generated from patient iPSCs. Cell Rep. 2015, 12, 1385–1390. [Google Scholar] [CrossRef] [Green Version]
- Doi, D.; Magotani, H.; Kikuchi, T.; Ikeda, M.; Hiramatsu, S.; Yoshida, K.; Amano, N.; Nomura, M.; Umekage, M.; Morizane, A.; et al. Pre-clinical study of induced pluripotent stem cell-derived dopaminergic progenitor cells for Parkinson’s disease. Nat. Commun. 2020, 11, 3369. [Google Scholar] [CrossRef] [PubMed]
- Nenasheva, T.; Gerasimova, T.; Serdyuk, Y.; Grigoreva, E.; Kosmiadi, G.; Nikolaev, A.; Dashinimaev, E.; Lyadova, I. Macrophages derived from human induced pluripotent stem cells are low-activated “naïve-like” cells capable of restricting Mycobacteria growth. Front. Immunol. 2020, 11, 1016. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, T.; Shimizu, K.; Matsumoto, R.; Honda, H. Selective elimination of human induced pluripotent stem cells using medium with high concentration of L-alanine. Sci. Rep. 2018, 8, 12427. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, R.; Shimizu, K.; Nagashima, T.; Tanaka, H.; Mizuno, M.; Kikkawa, F.; Hori, M.; Honda, H. Plasma-activated medium selectively eliminates undifferentiated human induced pluripotent stem cells. Regen. Ther. 2016, 5, 55–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burkert, K.; Taheri, H.; Hamad, S.; Oliverio, M.; Peinkofer, G.; Kornfeld, J.-W.; Harnying, W.; Pfannkuche, K.; Hescheler, J.; Berkessel, A.; et al. Salicylic diamines selectively eliminate residual undifferentiated cells from pluripotent stem cell-derived cardiomyocyte preparations. Sci. Rep. 2021, 11, 2391. [Google Scholar] [CrossRef]
- Wu, J.C.; Garg, P.; Yoshida, Y.; Yamanaka, S.; Gepstein, L.; Hulot, J.-S.; Knollmann, B.C.; Schwartz, P.J. Towards precision medicine with human iPSCs for cardiac channelopathies. Circ. Res. 2019, 125, 653–658. [Google Scholar] [CrossRef]
- Schick, R.; Mekies, L.N.; Shemer, Y.; Eisen, B.; Hallas, T.; Jehuda, R.B.; Ben-Ari, M.; Szantai, A.; Willi, L.; Shulman, R.; et al. Functional abnormalities in induced pluripotent stem cell-derived cardiomyocytes generated from titin-mutated patients with dilated cardiomyopathy. PLoS ONE 2018, 13, e0205719. [Google Scholar] [CrossRef]
- Li, S.; Pan, H.; Tan, C.; Sun, Y.; Song, Y.; Zhang, X.; Yang, W.; Wang, X.; Li, D.; Dai, Y.; et al. Mitochondrial dysfunctions contribute to hypertrophic cardiomyopathy in patient iPSC-derived cardiomyocytes with MT-RNR2 mutation. Stem Cell Rep. 2018, 10, 808–821. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Lu, L.; Chen, J.; Wang, K.; Han, J.; Xue, S.; Weng, G. Generation of an induced pluripotential stem cell (iPSC) line from a patient with hypertrophic cardiomyopathy carrying myosin binding protein C (MYBPC3) c.3369-3370 insC mutation. Stem Cell Res. 2020, 50, 102144. [Google Scholar] [CrossRef] [PubMed]
- Gu, M.; Shao, N.-Y.; Sa, S.; Li, D.; Termglinchan, V.; Ameen, M.; Karakikes, I.; Sosa, G.; Grubert, F.; Lee, J.; et al. Patient-specific iPSC-derived endothelial cells uncover pathways that protect against pulmonary hypertension in BMPR2 mutation carriers. Cell Stem Cell 2017, 20, 490–504.e5. [Google Scholar] [CrossRef] [Green Version]
- Kinnear, C.; Chang, W.Y.; Khattak, S.; Hinek, A.; Thompson, T.; Rodrigues, D.C.; Kennedy, K.; Mahmut, N.; Pasceri, P.; Stanford, W.L.; et al. Modeling and rescue of the vascular phenotype of Williams-Beuren syndrome in patient induced pluripotent stem cells. Stem Cells Transl. Med. 2013, 2, 2–15. [Google Scholar] [CrossRef]
- Granata, A.; Serrano, F.; Bernard, W.G.; McNamara, M.; Low, L.; Sastry, P.; Sinha, S. An iPSC-derived vascular model of Marfan syndrome identifies key mediators of smooth muscle cell death. Nat. Genet. 2017, 49, 97–109. [Google Scholar] [CrossRef] [Green Version]
- Liang, P.; Lan, F.; Lee, A.S.; Gong, T.; Sanchez-Freire, V.; Wang, Y.; Diecke, S.; Sallam, K.; Knowles, J.W.; Wang, P.J.; et al. Drug screening using a library of human induced pluripotent stem cell-derived cardiomyocytes reveals disease-specific patterns of cardiotoxicity. Circulation 2013, 127, 1677–1691. [Google Scholar] [CrossRef] [Green Version]
- Mathur, A.; Loskill, P.; Shao, K.; Huebsch, N.; Hong, S.; Marcus, S.G.; Marks, N.; Mandegar, M.; Conklin, B.R.; Lee, L.P.; et al. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci. Rep. 2015, 5, 8883. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Blangero, J.; Curran, J.E. Induced pluripotent stem cells in disease modelling and gene identification. Methods Mol. Biol. 2018, 1706, 17–38. [Google Scholar] [CrossRef]
- Wen, Z.; Nguyen, H.N.; Guo, Z.; Lalli, M.A.; Wang, X.; Su, Y.; Kim, N.-S.; Yoon, K.-J.; Shin, J.; Zhang, C.; et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature 2014, 515, 414–418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, I.Y.; Poon, M.-W.; Pang, R.T.; Lian, Q.; Wong, D. Promises of stem cell therapy for retinal degenerative diseases. Graefe’s Arch. Clin. Exp. Ophthalmol. 2011, 249, 1439–1448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimos, J.T.; Rodolfa, K.T.; Niakan, K.K.; Weisenthal, L.M.; Mitsumoto, H.; Chung, W.; Croft, G.F.; Saphier, G.; Leibel, R.; Goland, R.; et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated inti motor neurons. Science 2008, 321, 1218–1221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raya, A.; Rodriguez-Piza, I.; Guenechea, G.; Vassena, R.; Navarro, S.; Barrero, M.J.; Consiglio, A.; Castella, M.; Rio, P.; Sleep, E.; et al. Disease-corrected haematopoietic progenitors from Fanconi anaemia induced pluripotent stem cells. Nature 2009, 460, 53–59. [Google Scholar] [CrossRef] [Green Version]
- Soldner, F.; Hockemeyer, D.; Beard, C.; Gao, Q.; Bell, G.W.; Cook, E.G.; Hargus, G.; Blak, A.; Cooper, O.; Mitalipova, M.; et al. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell 2009, 136, 964–977. [Google Scholar] [CrossRef] [Green Version]
- D’Amour, K.A.; Bang, A.G.; Eliazer, S.; Kelly, O.G.; Agulnick, A.D.; Smart, N.G.; Moorman, M.A.; Kroon, E.; Carpenter, M.K.; Baetge, E.E. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat. Biotechnol. 2006, 24, 1392–1401. [Google Scholar] [CrossRef]
- Kondo, Y.; Toyoda, T.; Inagaki, N.; Osafune, K. iPSC technology-based regenerative therapy for diabetes. J. Diabetes Investig. 2018, 9, 234–243. [Google Scholar] [CrossRef] [Green Version]
- Sahu, S.; Hemlata; Verma, A. Adverse events related to blood transfusion. Indian J. Anaesth. 2014, 58, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.; Shin, K.-H.; Kim, H.H.; Kim, H.-S. Current advances in red blood cell generation using stem cells from diverse sources. Stem Cells Int. 2019, 2019, 9281329. [Google Scholar] [CrossRef] [Green Version]
- Chang, K.-H.; Bonig, H.; Papayannopoulou, T. Generation and characterization of erythroid cells from human embryonic stem cells and induced pluripotent stem cells: An overview. Stem Cells Int. 2011, 2011, 791604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebrahimi, M.; Forouzesh, M.; Raoufi, S.; Ramazii, M.; Ghaedrahmati, F.; Farzaneh, M. Differentiation of human induced pluripotent stem cells into erythroid cells. Stem Cell Res. Ther. 2020, 11, 483. [Google Scholar] [CrossRef] [PubMed]
- Peyrard, T.; Bardiaux, L.; Krause, C.; Kobari, L.; Lapillonne, H.; Andreu, G.; Douay, L. Banking of pluripotent adult stem cells as an unlimited source for red blood cell production: Potential applications for alloimmunized patients and rare blood challenges. Transfus. Med. Rev. 2011, 25, 206–216. [Google Scholar] [CrossRef] [PubMed]
- Papapetrou, E.P. Induced pluripotent stem cells to model blood diseases. Blood 2018, 132, SCI-15. [Google Scholar] [CrossRef]
- Bedel, A.; Pasquet, J.-M.; Lippert, E.; Taillepierre, M.; Lagarde, V.; Dabernat, S.; Dubus, P.; Charaf, L.; Beliveau, F.; de Verneuil, H.; et al. Variable behaviour of iPSCs derived from CML patients for response to TKI and hematopoietic differentiation. PLoS ONE 2013, 8, e71596. [Google Scholar] [CrossRef]
- Kumano, K.; Arai, S.; Hosoi, M.; Taoka, K.; Takayama, N.; Otsu, M.; Nagae, G.; Ueda, K.; Nakazaki, K.; Kamikubo, Y.; et al. Generation of induced pluripotent stem cells from primary chronic myelogenous leukemia patient samples. Blood 2012, 119, 6234–6242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kotini, A.G.; Chang, C.-J.; Boussaad, I.; Delrow, J.J.; Dolezal, E.K.; Nagulapally, A.B.; Perna, F.; Fishbein, G.A.; Klimek, V.M.; Hawkins, R.D.; et al. Functional analysis of a chromosomal deletion associated with myelodysplastic syndromes using isogenic human induced pluripotent stem cells. Nat. Biotechnol. 2015, 33, 646–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, C.-J.; Kotini, A.G.; Olszewska, M.; Georgomanoli, M.; Teruya-Feldstein, J.; Sperber, H.; Sanchez, R.; DeVita, R.; Martins, T.J.; Abdel-Wahab, O.; et al. Dissecting the contributions of cooperating gene mutations to cancer phenotypes and drug responses with patient-derived iPSCs. Stem Cell Rep. 2018, 10, 1610–1624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boulad, F.; Wang, X.; Qu, J.; Taylor, C.; Ferro, L.; Karponi, G.; Bartido, S.; Giardina, P.; Heller, G.; Prockop, S.E.; et al. Safe mobilization of CD34+ cells in adults with β-thalassemia and validation of effective globin gene transfer for clinical investigation. Blood 2014, 123, 1483–1486. [Google Scholar] [CrossRef] [Green Version]
- Papapetrou, E.P.; Schambach, A. Gene insertion into genomic safe harbors for human gene therapy. Mol. Ther. 2016, 24, 678–684. [Google Scholar] [CrossRef] [Green Version]
- Papapetrou, E.P.; Lee, G.; Malani, N.; Setty, M.; Riviere, I.; Tirunagari, L.M.S.; Kadota, K.; Roth, S.L.; Giardina, P.; Viale, A.; et al. Genomic safe harbors permit high β-globin transgene expression in thalassemia induced pluripotent stem cells. Nat. Biotechnol. 2011, 29, 73–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flynn, R.; Grundmann, A.; Renz, P.; Hanseler, W.; James, W.S.; Cowley, S.A.; Moore, M.D. CRISPR-mediated genotypic and phenotypic correction of a chronic granulomatous disease mutation in human iPS cells. Gene Ther. 2015, 43, 838–848. [Google Scholar] [CrossRef] [Green Version]
- Ding, Q.; Cowan, C.A. Liver in a dish. Cell Res. 2013, 23, 1242–1243. [Google Scholar] [CrossRef] [Green Version]
- Fox, I.J.; Duncan, S.A. Engineering liver tissue from induced pluripotent stem cells: A first step in generating new organs for transplantation? Hepatology 2013, 58, 2198–2201. [Google Scholar] [CrossRef] [Green Version]
- Willenbring H and Soto-Gutierrez, A. Transplantable liver organoids made from only three ingredients. Cell Stem Cell 2013, 13, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Takebe, T.; Sekine, K.; Enomura, M.; Koike, H.; Kimura, M.; Ogaeri, T.; Zhang, R.-R.; Ueno, Y.; Zheng, Y.-W.; Koike, N.; et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 2013, 499, 481–484. [Google Scholar] [CrossRef]
- Olgasi, C.; Cucci, A.; Follenzi, A. iPSC-derived liver organoids: A journey from drug screening, to disease modelling, arriving to regenerative medicine. Int. J. Mol. Sci. 2020, 21, 6215. [Google Scholar] [CrossRef] [PubMed]
- Tasnim, F.; Xing, J.; Huang, X.; Mo, S.; Wei, X.; Tan, M.-H.; Yu, H. Generation of mature Kupffer cells from human induced pluripotent stem cells. Biomaterials 2019, 192, 377–391. [Google Scholar] [CrossRef]
- Akram, K.M.; Patel, N.; Spiteri, M.A.; Forsyth, N.R. Lung regeneration: Endogenous and exogenous stem cell mediated therapeutic approaches. Int. J. Mol. Sci. 2016, 17, 128. [Google Scholar] [CrossRef] [Green Version]
- Warburton, D.; El-Hashash, A.; Carraro, G.; Tiozzo, C.; Sala, F.; Rogers, O.; De Langhe, S.; Kemp, P.J.; Riccardi, D.; Torday, J.; et al. Lung organogenesis. Curr. Top. Dev. Biol. 2010, 90, 73–158. [Google Scholar] [CrossRef]
- Mou, H.; Zhao, R.; Sherwood, R.; Ahfeldt, T.; Lapey, A.; Wain, J.; Sicillan, L.; Izvolsky, K.; Lau, F.H.; Musunuru, K.; et al. Generation of multipotent lung and airway progenitors from mouse ESCs and patient-specific cystic fibrosis iPSCs. Cell Stem Cell 2012, 10, 385–397. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.X.L.; Islam, M.N.; O’Neill, J.; Hu, Z.; Yang, Y.-G.; Chen, Y.-W.; Mumau, M.; Green, M.D.; Vunjak-Novakovic, G.; Bhattacharya, J.; et al. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 2014, 32, 84–91. [Google Scholar] [CrossRef] [Green Version]
- Samuel, R.M.; Majd, H.; Richter, M.N.; Ghazizadeh, Z.; Zekavat, S.M.; Navickas, A.; Ramirez, J.T.; Asgharian, H.; Simoneau, C.R.; Bonser, L.R.; et al. Androgen signaling regulates SARS-CoV-2 receptor levels and is associated with severe COVID-19 symptoms in men. Cell Stem Cell 2020, 27, 876–889. [Google Scholar] [CrossRef]
- Katsura, H.; Sontake, V.; Tata, A.; Kobayashi, Y.; Edwards, C.E.; Heaton, B.E.; Konkimalla, A.; Asakura, T.; Mikami, Y.; Fritch, E.J.; et al. Human lung stem cell-based alveolospheres provide insights into SARS-CoV-2 mediated interferon responses and pneumocyte dysfunction. Cell Stem Cell 2020, 27, 890–904. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Hume, A.J.; Abo, K.M.; Werder, R.B.; Villacorta-Martin, C.; Alysandratos, K.-D.; Beermann, M.L.; Simone-Roach, C.; Lindstrom-Vautrin, J.; Olejnik, J.; et al. SARS-CoV-2 infection of pluripotent stem cell-derived human lung alveolar type 2 cells elicits a rapid epithelial-intrinsic inflammatory response. Cell Stem Cell 2020, 27, 962–973.e7. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Duan, X.; Yang, L.; Nilsson-Payant, B.E.; Wang, P.; Duan, F.; Tang, X.; Yaron, T.M.; Zhang, T.; Uhl, S.; et al. Identification of SARS-CoV-2 inhibitors using lung and colonic organoids. Nature 2020, 589, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Xu, A.; Tu, J.; Liu, M.; Gingold, J.A.; Zhao, R.; Lee, D.F. Modeling osteosarcoma using Li-Fraumeni syndrome patient-derived induced pluripotent stem cells. JOVE 2018, 136, 57664. [Google Scholar] [CrossRef]
- Mulero-Navarro, S.; Sevilla, A.; Roman, A.C.; Lee, D.-F.; D’Souza, S.L.; Pardo, S.; Riess, I.; Su, J.; Cohen, N.; Schaniel, C.; et al. Myeloid dysregulation in a human induced pluripotent stem cell model of PTPN11–associated juvenile myelomonocytic leukemia. Cell Rep. 2015, 13, 504–515. [Google Scholar] [CrossRef] [Green Version]
- Portier, L.; Desterke, C.; Chaker, D.; Oudrhiri, N.; Asgarova, A.; Dkhissi, F.; Turhan, A.G.; Bennaceur-Griscelli, A.; Griscelli, F. iPSC-derived hereditary breast cancer model reveals the BRCA1-deleted tumor niche as a new culprit in disease progression. Int. J. Mol. Sci. 2021, 22, 1227. [Google Scholar] [CrossRef]
- Chao, H.-M.; Chern, E. Patient-derived induced pluripotent stem cells for models of cancer and cancer stem cell research. J. Formos. Med. Assoc. 2018, 117, 1046–1057. [Google Scholar] [CrossRef]
- Reyal, F.; Guyader, C.; Decraene, C.; Lucchesi, C.; Auger, N.; Assayag, F.; De Plater, L.; Gentien, D.; Poupon, M.-F.; Cottu, P.; et al. Molecular profiling of patient-derived breast cancer xenografts. Breast Cancer Res. 2012, 14, R11. [Google Scholar] [CrossRef]
- Krumbach, R.; Schuler, J.; Hofmann, M.; Giesemann, T.; Fiebig, H.-H.; Beckers, T. Primary resistance to cetuximab in a panel of patient-derived tumour xenograft models: Activation of MET as one mechanism for drug resistance. Eur. J. Cancer 2011, 47, 1231–1243. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Trotman, L.C.; Shaffer, D.; Lin, H.-K.; Dotan, Z.A.; Niki, M.; Koutcher, J.A.; Scher, H.I.; Ludwig, T.; Gerald, W.; et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature 2005, 436, 725–730. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Pegram, M.D.; Wu, J.C. Induced pluripotent stem cells as a novel cancer vaccine. Expert Opin. Biol. Ther. 2019, 19, 1191–1197. [Google Scholar] [CrossRef] [PubMed]
- Medvedev, S.P.; Shevchenko, A.I.; Zakian, A.M. Induced pluripotent stem cells: Problems and advantages when applying them in regenerative medicine. Acta Nat. 2010, 2, 18–28. [Google Scholar] [CrossRef] [Green Version]
- Honda, Y.; Li, J.; Hino, A.; Tsujimoto, S.; Lee, J.-K. High-throughput drug screening system based on human induced pluripotent stem cell-derived atrial myocytes~A novel platform to detect cardiac toxicity for atrial arrhythmias. Front. Pharmacol. 2021, 12, 680618. [Google Scholar] [CrossRef]
- Djidrovski, I.; Georgiou, M.; Hughes, G.L.; Patterson, E.I.; Casas-Sanchez, A.; Pennington, S.H.; Biagini, G.A.; Moya-Molina, M.; van den Bor, J.; Smit, M.J.; et al. SARS-CoV-2 infects an upper airway model derived from induced pluripotent stem cells. Stem Cells 2021. [CrossRef]
- Yamanaka, S. Pluripotent stem cell-based cell therapy–Promises and challenges. Cell Stem Cell 2020, 27, 523–531. [Google Scholar] [CrossRef] [PubMed]
- Pang, L. Toxicity testing in the era of induced pluripotent stem cells: A perspective regarding the use of patient-specific induced pluripotent stem cell-derived cardiomyocytes for cardiac safety evaluation. Curr. Opin. Toxicol. 2020, 23–24, 50–55. [Google Scholar] [CrossRef]
- Induced Pluripotent Stem Cells for Disease Research; ClinicalTrials.gov Identifier: NCT04476225; University of California: San Francisco, CA, USA. Available online: https://clinicaltrials.gov/ct2/show/NCT04476225 (accessed on 10 May 2021).
- Deinsberger, J.; Reisinger, D.; Weber, B. Global trends in clinical trials involving pluripotent stem cells: A systematic multi-database analysis. NPJ Regener. Med. 2020, 5, 15. [Google Scholar] [CrossRef] [PubMed]
- Help Therapeutics. Treating Heart Failure with hPSC-CMs (HEAL-CHF). Epicardial Injection of Allogeneic Human Pluripotent Stem Cell-derived Cardiomyocytes to Treat Severe Chronic Heart Failure; ClinicalTrials.gov Identifier: NCT03763136. Available online: https://clinicaltrials.gov/ct2/show/NCT03763136 (accessed on 10 May 2021).
- Chinese Academy of Sciences. Safety and Efficacy Study of Human ESC-derived Neural Precursor Cells in the Treatment of Parkinson’s Disease; A Phase I/II, Open-Label Study to Assess the Safety and Efficacy of Striatum Transplantation of Human Embryonic Stem Cells-derived Neural Precursor Cells in Patients with Parkinson’s Disease; ClinicalTrials.gov Identifier: NCT03119636. Available online: https://clinicaltrials.gov/ct2/show/NCT03119636 (accessed on 10 May 2021).
- Chinese Academy of Sciences. Mesenchymal Stem Cells (MSCs)—Like Cell Transplantation in Women with Primary Ovarian Insufficiency (MSCLCTWPOI). Safety Study of Human Embryonic Stem Cell Derived Mesenchymal Stem Cell (MSC)—Like Cells Transplantation in Women with Primary Ovarian Insufficiency (POI); ClinicalTrials.gov Identifier: NCT03877471. Available online: https://clinicaltrials.gov/ct2/show/NCT03877471 (accessed on 10 May 2021).
- Fate Therapeutics. FT500 as Monotherapy and in Combination with Immune Checkpoint Inhibitors in Subjects with Advanced Solid Tumors (Phase 1). ClinicalTrials.gov Identifier: NCT03841110. Available online: https://clinicaltrials.gov/ct2/show/NCT03841110 (accessed on 10 May 2021).
- Human iPSC for Repair of Vasodegenerative Vessels in Diabetic Retinopathy (iPSC); ClinicalTrials.gov Identifier: NCT03403699; University of Alabama: Birmingham, AL, USA. Available online: https://clinicaltrials.gov/ct2/show/NCT03403699 (accessed on 10 May 2021).
- Sun, X. Thalassemia Treatment Based on the Stem Cell Technology. ClinicalTrials.gov Identifier: NCT03222453. Available online: https://clinicaltrials.gov/ct2/show/NCT03222453 (accessed on 10 May 2021).
- Hadassah Medical Organization. Development of iPS from Donated Somatic Cells of Patients with Neurological Diseases. Derivation of Induced Pluripotent Stem Cells from Somatic Cells Donated by Patients with Neurological Diseases for the Study of the Pathogenesis of the Disorders and Development of Novel Therapies; ClinicalTrials.gov Identifier: NCT00874783. Available online: https://clinicaltrials.gov/ct2/show/NCT00874783 (accessed on 10 May 2021).
- Memorial Sloan Kettering Cancer Center. Generation of Heart Muscle Cells from Blood or Skin Cells of Breast Cancer Patients. Generation of Induced Pluripotent Stem Cell Derived Cardiomyocytes from Patients Exposed to Trastuzumab Therapy for Breast Cancer; ClinicalTrials.gov Identifier: NCT02772367. Available online: https://clinicaltrials.gov/ct2/show/NCT02772367 (accessed on 10 May 2021).
- Moorfields Eye Hospital NHS Foundation Trust. A Study Of Implantation Of Retinal Pigment Epithelium in Subjects with Acute Wet Age Related Macular Degeneration. Phase 1, Open-Label, Safety and Feasibility Study of Implantation of PF-05206388 (Human Embryonic Stem Cell Derived Retinal Pigment Epithelium (RPE) Living Tissue Equivalent) in Subjects with Acute Wet Age Related Macular Degeneration and Recent Rapid Vision Decline; ClinicalTrials.gov Identifier: NCT01691261. Available online: https://clinicaltrials.gov/ct2/show/NCT01691261 (accessed on 10 May 2021).


| Vector | Cell Type | Genes | Efficiency | Reference |
|---|---|---|---|---|
| Retrovirus | Skin fibroblasts | OCT4, SOX2, KLF4 | 0.001% | [18] |
| Fibroblasts | OCT4, SOX2 and Valproic acid | 0.001% | [19] | |
| Skin cancer cell line | miR-302 | Unknown | [20] | |
| Lentivirus | Embryonic fibroblasts | OCT4, SOX2, NANOG, LIN28 | 0.01% | [21] |
| Fibroblasts | OCT4, SOX2, KLF4, c-MYC | 0.01% | [22] | |
| Adenovirus | Embryonic fibroblasts | OCT4, SOX2, KLF4, c-MYC | 0.0002% | [23] |
| Sendai virus | Cord blood CD34+ cells | OCT4, SOX2, KLF4, c-MYC | 0.2% | [24] |
| Recombinant protein | Fibroblasts | OCT4, SOX2, KLF4, c-MYC | 0.001% | [25] |
| mRNA | Fibroblasts | OCT4, SOX2, NANOG, LIN28 | 0.05% | [26] |
| Disorder | iPSC Characteristic | Therapy | Reference |
|---|---|---|---|
| Chronic Granulomatous Disease (CGD)—Preclinical | CGD-iPS-cells which transformed to neutrophils lacked production of reactive oxygen species (ROS) | The zinc finger nuclease -mediated functional correction of the causative CYBB gene in the neutrophils restored ROS production. | [32] |
| Hemophilia A (HA)—Preclinical mice model | The HA-iPSC derived endothelial cells lacked F8 gene expression and secretory protein | Lentiviral-based vector with F8 transgene and driven by endothelial-specific promoter was used, and the derived endothelial cells exhibited restored F8 gene expression | [33] |
| Hemophilia B (HB)—Preclinical hemophilic mice model | HB-iPSC derived hepatocyte-like cell lacked secretion of clotting factor IX | The CRISPR/Cas9 gene editing system was used to correct the cDNA in the HB-iPSCs and the resultant hepatocyte-like cells exhibited restored synthesis ability for clotting factor IX. | [34] |
| Severe combined immunodeficiency (SCID)—Preclinical | SCID-iPSCs with JAK3 deficiency exhibited lack of early T cell development | Gene editing by CRISPR/Cas9—enhanced gene targeting was used to correct the JAK3 mutation, which restored normal T cell development along with production of mature T cells with a broad T cell receptor repertoire. | [35] |
| Thalassemia—Preclinical | The iPSC-derived erythroid cells from homozygous alpha thalassemia exhibited lack of expression of the alpha globin gene | Zinc finger nuclease-mediated insertion of the globin transgene was done in the safe harbor site; AAVS1 on human chromosome 19 for correction of alpha-thalassemia major hydrops fetalis. The homozygous insertion corrected the imbalance of the globin chain in the erythroid cells. | [36] |
| Young-onset Parkinson’s disease (YOPD)—Preclinical | YOPD-iPSCs were differentiated to midbrain dopaminergic neural culture that exhibited increased accumulation of soluble α-synuclein protein and phosphorylated protein kinase Cα, and reduced abundance of the lysosomal membrane protein LAMP1 | Activation of lysosomal-specific pathway by phorbol ester PEP005 reduced α-synuclein, and phosphorylated protein kinase Cα levels, and increasing LAMP1 levels. | [37] |
| Parkinson’s disease (PD)—Proof-of-concept rodent study | Human iPSC-derived midbrain dopaminergic neurons were subjected to sorting to enrich the ventral midbrain (VM) neurons and improve efficacy and safety of cell therapy | Sorting using NCAM(+)/CD29(low) enriched VM dopaminergic neurons better. Further, PiPSC-derived NCAM(+)/CD29(low) DA neurons were able to restore motor function of 6-hydroxydopamine (6-OHDA) lesioned rats 16 weeks after transplantation. | [10] |
| Alzheimer’s disease (AD)—Proof-of-concept preclinical study | AD patient-derived iPSCs were carriers of three copies of the amyloid precursor protein (APP) gene | Gene editing by CRISPR/Cas9 system enables generation of iPS-cell lines with monoallelic, biallelic, or triallelic knockout of APP. The corticol neurons generated from isogenically corrected iPSCs were found to exhibit gene-dosage correlation dependent disease-phenotype correlation. | [38] |
| Amyotrophic lateral sclerosis (ALS)—Preclinical | ALS-iPSCs from fibroblasts exhibited SOD1+/A272C and FUS+/G1566A mutations | The CRISPR/Cas-9 nickases was used to correct the mutation and the gene corrected ALS-iPSCs (FUS+/+ and SOD1+/+) exhibited all pluripotency markers including OCT4, NANOG, and SOX2. | [39] |
| Barth syndrome (BTHS)—Proof-of-concept preclinical study | The BTHS-iPSC-derived cardiomyocytes exhibited abnormalities associated with mutations in the TAZ gene. Further, the cardiomyocytes assembled sparsely, and exhibited irregular sarcomeres. | The CRISPR/Cas9 system was used to introduce TAZ gene mutation in healthy donor iPSC-derived cardiomyocytes to identify relationship. Further, administration of antioxidant mitoTEMPO in the BTHS-iPCs-derived cardiomyocytes exhibited suppression of excess ROS production and normalization of the sarcomere organization and contractility. | [40] |
| Long QT syndrome (LQTS)—Preclinical | The LQT15-iPSC with CALM2-N98S mutation were differentiated into cardiomyocytes exhibited significantly lower beating rates, prolonged AP durations, and impaired inactivation of LTCC currents | The CRISPR/Cas9 system was used to correct the mutation in CALM2 and the resultant gene corrected iPSC-derived cardiomyocytes showed reversal in electrophysiological abnormalities with successfully recapitulating the disease phenotype. | [41] |
| Cystic fibrosis (CF)—Preclinical | The CF-iPSCs were positive for the CFTR mutation involving homozygous deletion of F508 | The CRISPR/Cas9 system was used to correct the CFTR mutation, in combination with a completely excisable selection system. The gene correct iPSCs successfully differentiated to mature airway epithelial cells and recovered normal CFTR expression. | [42] |
| ID Number | Disease | Cell Type | Title | Intervention | Country |
|---|---|---|---|---|---|
| NCT03763136 [108] | Heart disease | hiPSCs-derived cardiomyocytes | Epicardial Injection of Allogeneic Human Pluripotent Stem Cell-derived Cardiomyocytes to Treat Severe Chronic Heart Failure | Injection into the myocardium | China |
| NCT03119636 [109] | Parkinson’s disease | hESC-derived neural precursor cells | A Phase I/II, Open-Label Study to Assess the Safety and Efficacy of Striatum Transplantation of Human Embryonic Stem Cells-derived Neural Precursor Cells in Patients with Parkinson’s Disease | Stereotaxic intra-striatal injection | China |
| NCT03877471 [110] | Primary ovarian insufficiency | hESC-derived mesenchymal stem cells-like cells | Safety Study of Human Embryonic Stem Cell Derived Mesenchymal Stem Cell (MSC)-Like Cells Transplantation in Women with Primary Ovarian Insufficiency (POI) | Injection into ovaries | China |
| NCT03841110 [111] | Advanced solid tumors | iPSC-derived NK cell cancer immunotherapy | FT500 as monotherapy and in combination with immune checkpoint inhibitors in subjects with advanced solid tumors (Phase 1) | USA | |
| NCT03403699 [112] | Diabetic Retinopathy | iPSC-derived mesoderm cells | Human iPSC for Repair of Vasodegenerative Vessels in Diabetic Retinopathy (iPSC) | injection into the vitreous cavity of diabetic rodents and primate eyes | USA |
| NCT03222453 [113] | Beta thalassemia | Hematopoetic stem cells from beta-thalassemia iPSCs | Thalassemia Treatment Based on the Stem Cell Technology | China | |
| NCT00874783 [114] | Neurodegenerative Disorders | iPS cells from cell cultures from skin biopsies or the patient’s hair | Derivation of Induced Pluripotent Stem Cells from Somatic Cells Donated by Patients with Neurological Diseases for the Study of the Pathogenesis of the Disorders and Development of Novel Therapies | Israel | |
| NCT02772367 [115] | Breast cancer | iPSC-derived cardiomyocytes | Generation of Induced Pluripotent Stem Cell Derived Cardiomyocytes from Patients Exposed to Trastuzumab Therapy for Breast Cancer | USA | |
| NCT01691261 [116] | Acute Wet Age-Related Macular Degeneration | hESC-derived retinal pigment epithelium | Phase 1, open-label, safety and feasibility study of implantation of PF-05206388 (Human embryonic stem cell derived retinal pigment epithelium (RPE) living tissue equivalent) in subjects with acute wet age-related macular degeneration and recent rapid vision decline | Intraocular use of retinal pigment epithelium living tissue | United Kingdom (UK) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aboul-Soud, M.A.M.; Alzahrani, A.J.; Mahmoud, A. Induced Pluripotent Stem Cells (iPSCs)—Roles in Regenerative Therapies, Disease Modelling and Drug Screening. Cells 2021, 10, 2319. https://doi.org/10.3390/cells10092319
Aboul-Soud MAM, Alzahrani AJ, Mahmoud A. Induced Pluripotent Stem Cells (iPSCs)—Roles in Regenerative Therapies, Disease Modelling and Drug Screening. Cells. 2021; 10(9):2319. https://doi.org/10.3390/cells10092319
Chicago/Turabian StyleAboul-Soud, Mourad A. M., Alhusain J. Alzahrani, and Amer Mahmoud. 2021. "Induced Pluripotent Stem Cells (iPSCs)—Roles in Regenerative Therapies, Disease Modelling and Drug Screening" Cells 10, no. 9: 2319. https://doi.org/10.3390/cells10092319
APA StyleAboul-Soud, M. A. M., Alzahrani, A. J., & Mahmoud, A. (2021). Induced Pluripotent Stem Cells (iPSCs)—Roles in Regenerative Therapies, Disease Modelling and Drug Screening. Cells, 10(9), 2319. https://doi.org/10.3390/cells10092319







