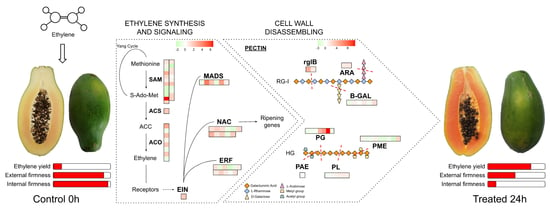
Systems Biology Applied to the Study of Papaya Fruit Ripening: The Influence of Ethylene on Pulp Softening
Abstract
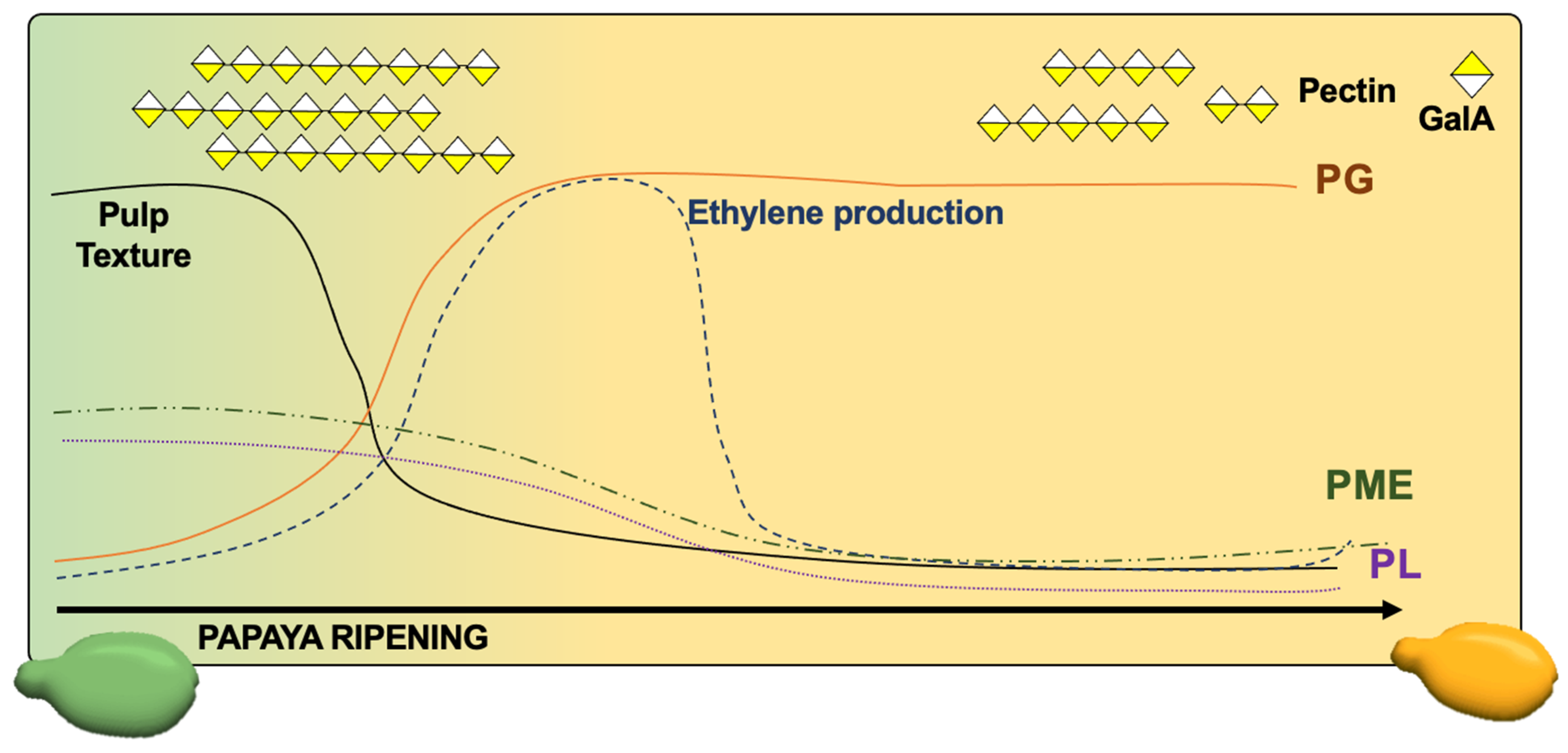
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Experimental Design
2.2. Analysis of the Ripening Parameters
2.3. Total RNA Extraction
2.4. Illumina Sequencing (RNA-Seq)
2.5. Differential Expression Analysis
2.6. Enrichment Analysis
2.7. Co-Expression Analyses
2.8. Differential Expression Analysis through Real-Time PCR
2.9. Statistics
3. Results and Discussion
3.1. Ethylene Treatment Induced Papaya Ripening
3.2. Changes to Transcriptome Profile during Ripening
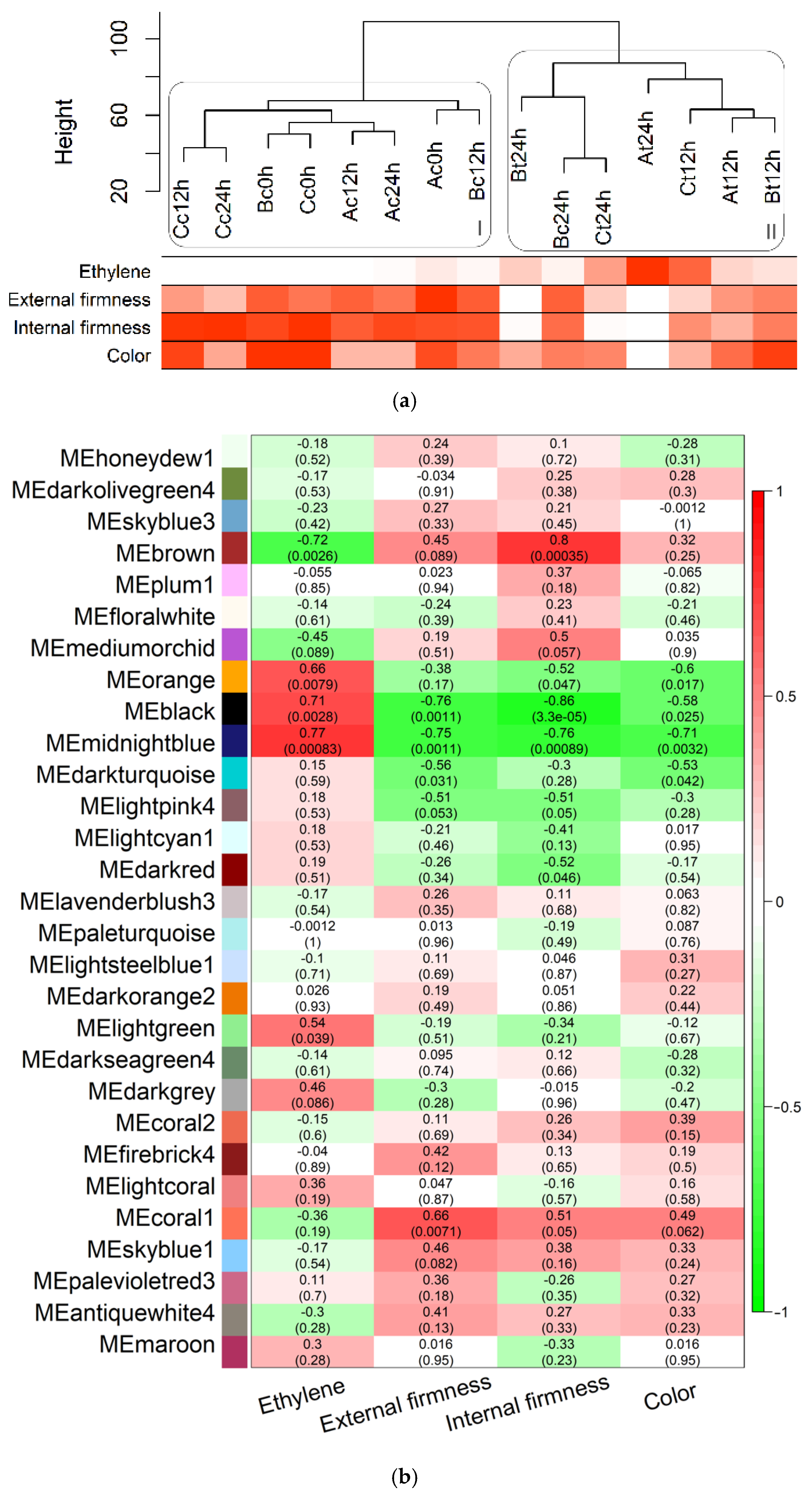
3.3. Correlation among Expression and Phenotypic Data
3.4. Ethylene-Related Genes and Transcription Factors
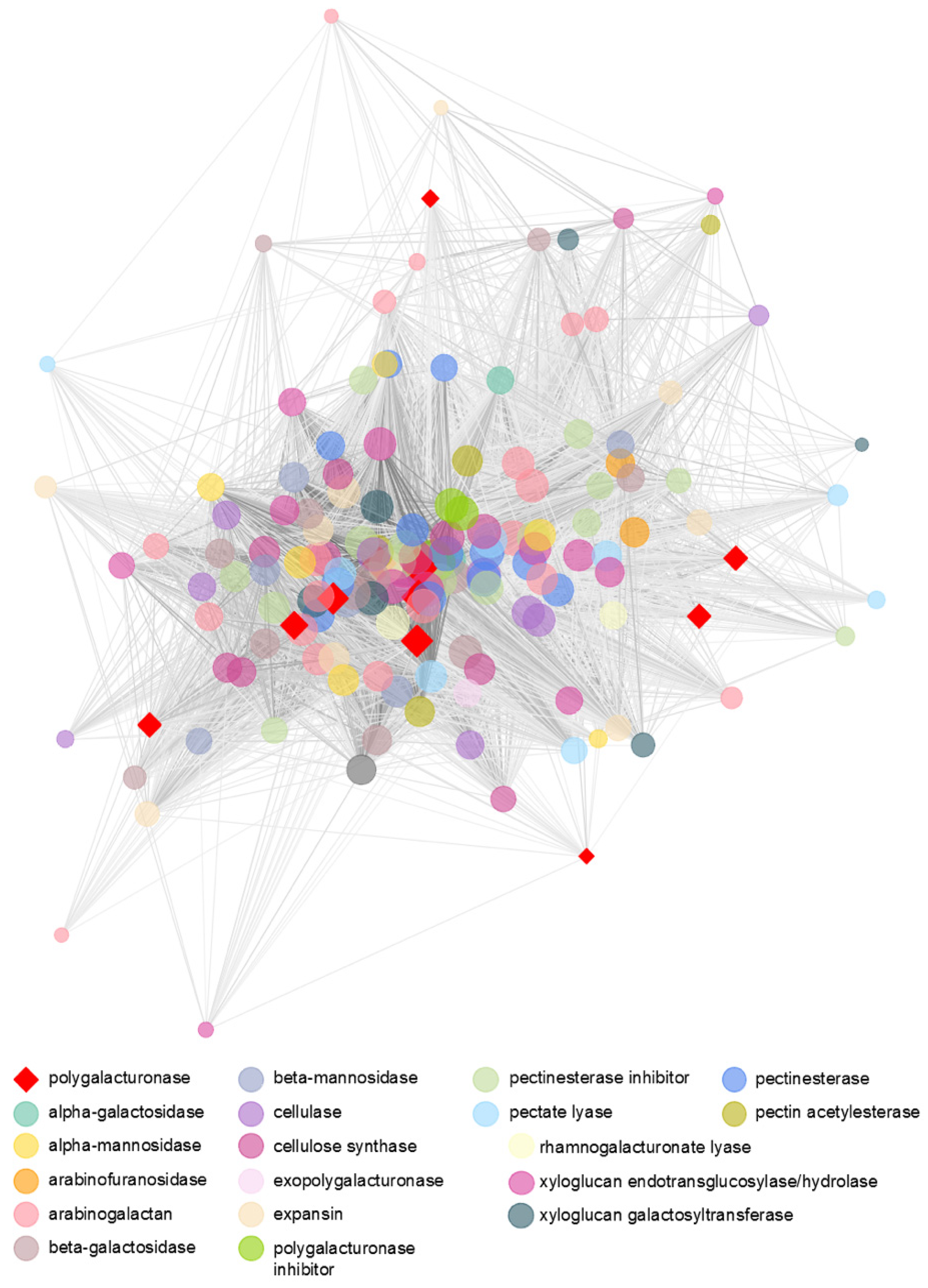
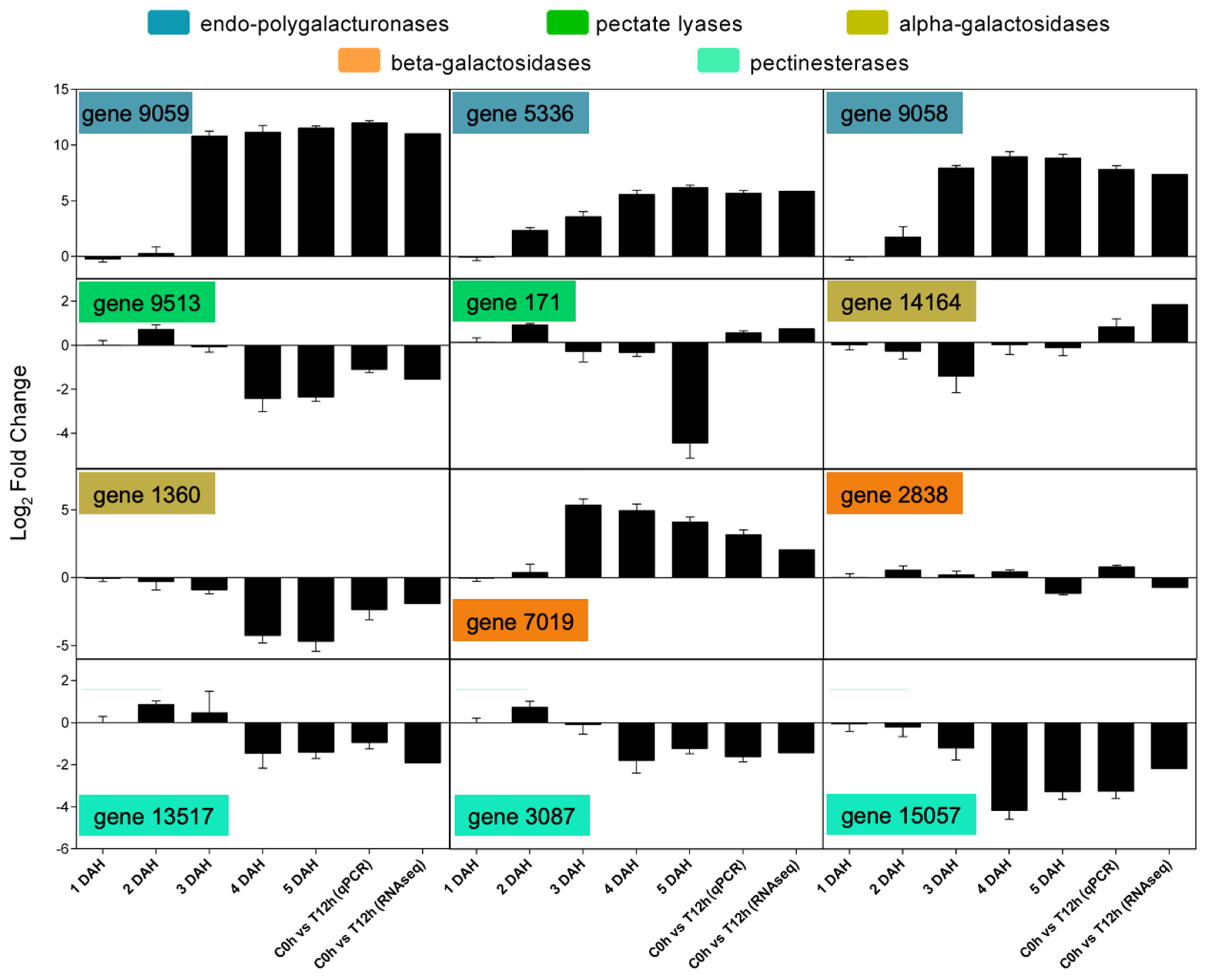
3.5. Transcriptomic Analysis Reveals a Diverse Expression of Papaya Cell-Wall-Related Genes
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fabi, J.P.; Cordenunsi, B.R.; Seymour, G.B.; Lajolo, F.M.; do Nascimento, J.R.O. Molecular Cloning and Characterization of a Ripening-Induced Polygalacturonase Related to Papaya Fruit Softening. Plant Physiol. Biochem. 2009, 47, 1075–1081. [Google Scholar] [CrossRef]
- Tucker, G.; Yin, X.; Zhang, A.; Wang, M.; Zhu, Q.; Liu, X.; Xie, X.; Chen, K.; Grierson, D. Ethylene and Fruit Softening. Food Qual. Saf. 2017, 1, 253–267. [Google Scholar] [CrossRef]
- Fabi, J.P.; Peroni, F.H.G.; Gomez, M.L.P.A. Papaya, Mango and Guava Fruit Metabolism during Ripening: Postharvest Changes Affecting Tropical Fruit Nutritional Content and Quality. Fresh Prod. 2010, 11, 201913937. [Google Scholar]
- Liu, K.; Yuan, C.; Li, H.; Lin, W.; Yang, Y.; Shen, C.; Zheng, X. Genome-Wide Identification and Characterization of Auxin Response Factor (ARF) Family Genes Related to Flower and Fruit Development in Papaya (Carica papaya L.). BMC Genom. 2015, 16, 901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karagiannis, E.; Michailidis, M.; Tanou, G.; Scossa, F.; Sarrou, E.; Stamatakis, G.; Samiotaki, M.; Martens, S.; Fernie, A.R.; Molassiotis, A. Decoding Altitude-Activated Regulatory Mechanisms Occurring during Apple Peel Ripening. Hortic. Res. 2020, 7, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Minas, I.S.; Tanou, G.; Krokida, A.; Karagiannis, E.; Belghazi, M.; Vasilakakis, M.; Papadopoulou, K.K.; Molassiotis, A. Ozone-Induced Inhibition of Kiwifruit Ripening Is Amplified by 1-Methylcyclopropene and Reversed by Exogenous Ethylene. BMC Plant Biol. 2018, 18, 358. [Google Scholar] [CrossRef] [PubMed]
- Fabi, J.P.; do Prado, S.B.R. Fast and Furious: Ethylene-Triggered Changes in the Metabolism of Papaya Fruit during Ripening. Front. Plant Sci. 2019, 10. [Google Scholar] [CrossRef]
- Fabi, J.P.; Lajolo, F.M.; do Nascimento, J.R.O. Cloning and Characterization of Transcripts Differentially Expressed in the Pulp of Ripening Papaya. Sci. Hortic. 2009, 121, 159–165. [Google Scholar] [CrossRef]
- Shinozaki, Y.; Nicolas, P.; Fernandez-Pozo, N.; Ma, Q.; Evanich, D.J.; Shi, Y.; Xu, Y.; Zheng, Y.; Snyder, S.I.; Martin, L.B.B.; et al. High-Resolution Spatiotemporal Transcriptome Mapping of Tomato Fruit Development and Ripening. Nat. Commun. 2018, 9, 364. [Google Scholar] [CrossRef]
- Sánchez-Sevilla, J.F.; Vallarino, J.G.; Osorio, S.; Bombarely, A.; Posé, D.; Merchante, C.; Botella, M.A.; Amaya, I.; Valpuesta, V. Gene Expression Atlas of Fruit Ripening and Transcriptome Assembly from RNA-Seq Data in Octoploid Strawberry (Fragaria × Ananassa). Sci. Rep. 2017, 7, 13737. [Google Scholar] [CrossRef]
- Chen, S.; Li, Y.; Zhao, Y.; Li, G.; Zhang, W.; Wu, Y.; Huang, L. ITRAQ and RNA-Seq Analyses Revealed the Effects of Grafting on Fruit Development and Ripening of Oriental Melon (Cucumis melo L. var. makuwa). Gene 2020, 766, 145142. [Google Scholar] [CrossRef]
- Deshpande, A.B.; Anamika, K.; Jha, V.; Chidley, H.G.; Oak, P.S.; Kadoo, N.Y.; Pujari, K.H.; Giri, A.P.; Gupta, V.S. Transcriptional Transitions in Alphonso Mango (Mangifera indica L.) during Fruit Development and Ripening Explain Its Distinct Aroma and Shelf Life Characteristics. Sci. Rep. 2017, 7, 8711. [Google Scholar] [CrossRef]
- Pei, M.; Gu, C.; Zhang, S. Genome-Wide Identification and Expression Analysis of Genes Associated with Peach (Prunus persica) Fruit Ripening. Sci. Hortic. 2019, 246, 317–327. [Google Scholar] [CrossRef]
- Li, T.; Yun, Z.; Wu, Q.; Qu, H.; Duan, X.; Jiang, Y. Combination of Transcriptomic, Proteomic, and Metabolomic Analysis Reveals the Ripening Mechanism of Banana Pulp. Biomolecules 2019, 9, 523. [Google Scholar] [CrossRef] [Green Version]
- Fu, C.-C.; Chen, H.-J.; Gao, H.-Y.; Wang, S.-L.; Wang, N.; Jin, J.-C.; Lu, Y.; Yu, Z.-L.; Ma, Q.; Han, Y.-C. Papaya CpMADS4 and CpNAC3 Co-Operatively Regulate Ethylene Signal Genes CpERF9 and CpEIL5 during Fruit Ripening. Postharvest Biol. Technol. 2021, 175, 111485. [Google Scholar] [CrossRef]
- Zhou, D.; Shen, Y.; Zhou, P.; Fatima, M.; Lin, J.; Yue, J.; Zhang, X.; Chen, L.-Y.; Ming, R. Papaya CpbHLH1/2 Regulate Carotenoid Biosynthesis-Related Genes during Papaya Fruit Ripening. Hortic. Res. 2019, 6, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daagema, A.A.; Orafa, P.N.; Igbua, F.Z. Nutritional Potentials and Uses of Pawpaw (Carica papaya): A Review. Eur. J. Nutr. Food Saf. 2020, 52–66. [Google Scholar] [CrossRef]
- do Prado, S.B.; Ferreira, G.F.; Harazono, Y.; Shiga, T.M.; Raz, A.; Carpita, N.C.; Fabi, J.P. Ripening-Induced Chemical Modifications of Papaya Pectin Inhibit Cancer Cell Proliferation. Sci. Rep. 2017, 7, 16564. [Google Scholar] [CrossRef] [PubMed]
- Ming, R.; Hou, S.; Feng, Y.; Yu, Q.; Dionne-Laporte, A.; Saw, J.H.; Senin, P.; Wang, W.; Ly, B.V.; Lewis, K.L.T.; et al. The Draft Genome of the Transgenic Tropical Fruit Tree Papaya (Carica papaya Linnaeus). Nature 2008, 452, 991–996. [Google Scholar] [CrossRef] [Green Version]
- Manrique, G.D.; Lajolo, F.M. Cell-Wall Polysaccharide Modifications during Postharvest Ripening of Papaya Fruit (Carica papaya). Postharvest Biol. Technol. 2004, 33, 11–26. [Google Scholar] [CrossRef]
- Shiga, T.M.; Fabi, J.P.; do Nascimento, J.R.O.; Petkowicz, C.L.d.O.; Vriesmann, L.C.; Lajolo, F.M.; Cordenunsi, B.R. Changes in Cell Wall Composition Associated to the Softening of Ripening Papaya: Evidence of Extensive Solubilization of Large Molecular Mass Galactouronides. J. Agric. Food Chem. 2009, 57, 7064–7071. [Google Scholar] [CrossRef]
- do Prado, S.B.; Melfi, P.R.; Castro-Alves, V.C.; Broetto, S.G.; Araújo, E.S.; do Nascimento, J.R.; Fabi, J.P. Physiological Degradation of Pectin in Papaya Cell Walls: Release of Long Chains Galacturonans Derived from Insoluble Fractions during Postharvest Fruit Ripening. Front. Plant Sci. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.H.; Lu, B.G.; Feng, L.; Yang, F.Y.; Geng, J.J.; Ming, R.; Chen, X.J. Isolation of Ripening-Related Genes from Ethylene/1-MCP Treated Papaya through RNA-Seq. BMC Genom. 2017, 18, 671. [Google Scholar] [CrossRef] [PubMed]
- Fabi, J.P.; Cordenunsi, B.R.; de Mattos Barreto, G.P.; Mercadante, A.Z.; Lajolo, F.M.; Oliveira do Nascimento, J.R. Papaya Fruit Ripening: Response to Ethylene and 1-Methylcyclopropene (1-MCP). J. Agric. Food Chem. 2007, 55, 6118–6123. [Google Scholar] [CrossRef]
- Zhbannikov, I.Y.; Hunter, S.S.; Foster, J.A.; Settles, M.L. SeqyClean: A pipeline for high-throughput sequence data pre-processing. In Proceedings of the 8th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, Boston, MS, USA, 20 August 2017; pp. 407–416. [Google Scholar]
- FastQC: A Quality Control Tool for High Throughput Sequence Data; Version 2.6.0a; Software for Sequence Data; Babraham Bioinformatics: Cambridge, UK, 2010.
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2014. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [Green Version]
- High-Throughput Functional Annotation and Data Mining with the Blast2GO Suite—Nucleic Acids Research—Oxford Academic. Available online: https://academic.oup.com/nar/article/36/10/3420/2410320 (accessed on 30 April 2021).
- Langfelder, P.; Horvath, S. WGCNA: An R Package for Weighted Correlation Network Analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Shannon, P. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Fabi, J.P.; Broetto, S.G.; Silva, S.L.; Zhong, S.; Lajolo, F.M.; do Nascimento, J.R.O. Analysis of Papaya Cell Wall-Related Genes during Fruit Ripening Indicates a Central Role of Polygalacturonases during Pulp Softening. PLoS ONE 2014, 9, e105685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabi, J.P.; Seymour, G.B.; Graham, N.S.; Broadley, M.R.; May, S.T.; Lajolo, F.M.; Cordenunsi, B.R.; Oliveira do Nascimento, J.R. Analysis of Ripening-Related Gene Expression in Papaya Using an Arabidopsis-Based Microarray. BMC Plant Biol. 2012, 12, 242. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.H.; Yang, F.Y.; Lu, B.G.; Zhao, W.W.; Jiang, T.; Feng, L.; Chen, X.J.; Ming, R. Exploring the Differential Mechanisms of Carotenoid Biosynthesis in the Yellow Peel and Red Flesh of Papaya. BMC Genom. 2019, 20, 49. [Google Scholar] [CrossRef] [Green Version]
- Saini, R.K.; Zamany, A.J.; Keum, Y.-S. Ripening improves the content of carotenoid, α-tocopherol, and polyunsaturated fatty acids in tomato (Solanum lycopersicum L.) fruits. 3 Biotech 2017, 7, 43. [Google Scholar] [CrossRef] [Green Version]
- Pino, J.A. Odour-Active Compounds in Papaya Fruit Cv. Red Maradol. Food Chem. 2014, 146, 120–126. [Google Scholar] [CrossRef]
- Gomez, M.; Lajolo, F.; Cordenunsi, B. Evolution of Soluble Sugars during Ripening of Papaya Fruit and Its Relation to Sweet Taste. J. Food Sci. 2002, 67, 442–447. [Google Scholar] [CrossRef]
- Gayosso-García Sancho, L.E.; Yahia, E.M.; González-Aguilar, G.A. Identification and Quantification of Phenols, Carotenoids, and Vitamin C from Papaya (Carica papaya L., cv. Maradol) Fruit Determined by HPLC-DAD-MS/MS-ESI. Food Res. Int. 2011, 44, 1284–1291. [Google Scholar] [CrossRef]
- Binder, B.M. Ethylene Signaling in Plants. J. Biol. Chem. 2020, 295, 7710–7725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zhu, X.; Mao, J.; Zou, Y.; Fu, D.; Chen, W.; Lu, W. Isolation and Characterization of Ethylene Response Factor Family Genes during Development, Ethylene Regulation and Stress Treatments in Papaya Fruit. Plant Physiol. Biochem. 2013, 70, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Sharoni, A.M.; Kikuchi, S. Roles of NAC Transcription Factors in the Regulation of Biotic and Abiotic Stress Responses in Plants. Front. Microbiol. 2013, 4. [Google Scholar] [CrossRef] [Green Version]
- Zhong, R.; Lee, C.; Zhou, J.; McCarthy, R.L.; Ye, Z.-H. A Battery of Transcription Factors Involved in the Regulation of Secondary Cell Wall Biosynthesis in Arabidopsis. Plant Cell 2008, 20, 2763–2782. [Google Scholar] [CrossRef] [Green Version]
- Lü, P.; Yu, S.; Zhu, N.; Chen, Y.-R.; Zhou, B.; Pan, Y.; Tzeng, D.; Fabi, J.P.; Argyris, J.; Garcia-Mas, J.; et al. Genome Encode Analyses Reveal the Basis of Convergent Evolution of Fleshy Fruit Ripening. Nat. Plants 2018, 4, 784–791. [Google Scholar] [CrossRef] [Green Version]
- Ochoa-Jiménez, V.-A.; Berumen-Varela, G.; Burgara-Estrella, A.; Orozco-Avitia, J.-A.; Ojeda-Contreras, Á.-J.; Trillo-Hernández, E.-A.; Rivera-Domínguez, M.; Troncoso-Rojas, R.; Báez-Sañudo, R.; Datsenka, T.; et al. Functional Analysis of Tomato Rhamnogalacturonan Lyase Gene Solyc11g011300 during Fruit Development and Ripening. J. Plant Physiol. 2018, 231, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Parmigiani, G.; Johnson, W.E. ComBat-Seq: Batch Effect Adjustment for RNA-Seq Count Data. NAR Genom. Bioinforma. 2020, 2, lqaa078. [Google Scholar] [CrossRef]
- Chen, Y.; Grimplet, J.; David, K.; Castellarin, S.D.; Terol, J.; Wong, D.C.J.; Luo, Z.; Schaffer, R.; Celton, J.-M.; Talon, M.; et al. Ethylene Receptors and Related Proteins in Climacteric and Non-Climacteric Fruits. Plant Sci. 2018, 276, 63–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ke, X.; Wang, H.; Li, Y.; Zhu, B.; Zang, Y.; He, Y.; Cao, J.; Zhu, Z.; Yu, Y. Genome-Wide Identification and Analysis of Polygalacturonase Genes in Solanum Lycopersicum. Int. J. Mol. Sci. 2018, 19, 2290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chourasia, A.; Sane, V.A.; Nath, P. Differential Expression of Pectate Lyase during Ethylene-Induced Postharvest Softening of Mango (Mangifera indica var. Dashehari). Physiol. Plant. 2006, 128, 546–555. [Google Scholar] [CrossRef]
- Dautt-Castro, M.; Ochoa-Leyva, A.; Contreras-Vergara, C.A.; Pacheco-Sanchez, M.A.; Casas-Flores, S.; Sanchez-Flores, A.; Kuhn, D.N.; Islas-Osuna, M.A. Mango (Mangifera indica L.) Cv. Kent Fruit Mesocarp de Novo Transcriptome Assembly Identifies Gene Families Important for Ripening. Front. Plant Sci. 2015, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villarreal, N.M.; Marina, M.; Nardi, C.F.; Civello, P.M.; Martínez, G.A. Novel Insights of Ethylene Role in Strawberry Cell Wall Metabolism. Plant Sci. 2016, 252, 1–11. [Google Scholar] [CrossRef]
- Posé, S.; Kirby, A.R.; Paniagua, C.; Waldron, K.W.; Morris, V.J.; Quesada, M.A.; Mercado, J.A. The Nanostructural Characterization of Strawberry Pectins in Pectate Lyase or Polygalacturonase Silenced Fruits Elucidates Their Role in Softening. Carbohydr. Polym. 2015, 132, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Paniagua, C.; Santiago-Doménech, N.; Kirby, A.R.; Gunning, A.P.; Morris, V.J.; Quesada, M.A.; Matas, A.J.; Mercado, J.A. Structural Changes in Cell Wall Pectins during Strawberry Fruit Development. Plant Physiol. Biochem. 2017, 118, 55–63. [Google Scholar] [CrossRef] [PubMed]
- Goulao, L.; Oliveira, C. Cell Wall Modifications during Fruit Ripening: When a Fruit Is Not the Fruit. Trends Food Sci. Technol. 2008, 19, 4–25. [Google Scholar] [CrossRef] [Green Version]
- Brummell, D.A. Cell Wall Metabolism during Maturation, Ripening and Senescence of Peach Fruit. J. Exp. Bot. 2004, 55, 2029–2039. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dheilly, E.; Gall, S.L.; Guillou, M.-C.; Renou, J.-P.; Bonnin, E.; Orsel, M.; Lahaye, M. Cell Wall Dynamics during Apple Development and Storage Involves Hemicellulose Modifications and Related Expressed Genes. BMC Plant Biol. 2016, 16, 201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Yeats, T.H.; Uluisik, S.; Rose, J.K.C.; Seymour, G.B. Fruit Softening: Revisiting the Role of Pectin. Trends Plant Sci. 2018, 23, 302–310. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soares, C.G.; do Prado, S.B.R.; Andrade, S.C.S.; Fabi, J.P. Systems Biology Applied to the Study of Papaya Fruit Ripening: The Influence of Ethylene on Pulp Softening. Cells 2021, 10, 2339. https://doi.org/10.3390/cells10092339
Soares CG, do Prado SBR, Andrade SCS, Fabi JP. Systems Biology Applied to the Study of Papaya Fruit Ripening: The Influence of Ethylene on Pulp Softening. Cells. 2021; 10(9):2339. https://doi.org/10.3390/cells10092339
Chicago/Turabian StyleSoares, Caroline Giacomelli, Samira Bernardino Ramos do Prado, Sónia C. S. Andrade, and João Paulo Fabi. 2021. "Systems Biology Applied to the Study of Papaya Fruit Ripening: The Influence of Ethylene on Pulp Softening" Cells 10, no. 9: 2339. https://doi.org/10.3390/cells10092339
APA StyleSoares, C. G., do Prado, S. B. R., Andrade, S. C. S., & Fabi, J. P. (2021). Systems Biology Applied to the Study of Papaya Fruit Ripening: The Influence of Ethylene on Pulp Softening. Cells, 10(9), 2339. https://doi.org/10.3390/cells10092339