Towards Understanding the Pathogenicity of DROSHA Mutations in Oncohematology
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection: Selection of Variants
2.2. Sequence-Based Prediction of Deleterious Nonsynonymous Variants
2.3. Structure-Based Prediction of Variants’ Effect on Protein Stability
2.4. Prediction of Changes in Vibrational Entropy and Normal Mode Analysis
2.5. Protein Structure Modeling
2.6. Molecular Dynamics (MD) Simulations
2.7. Protein Site-Specific Mutagenesis
2.8. Analysis of MD Simulation
3. Results
3.1. Assessment of Missense Mutations by Automated Prediction Tools
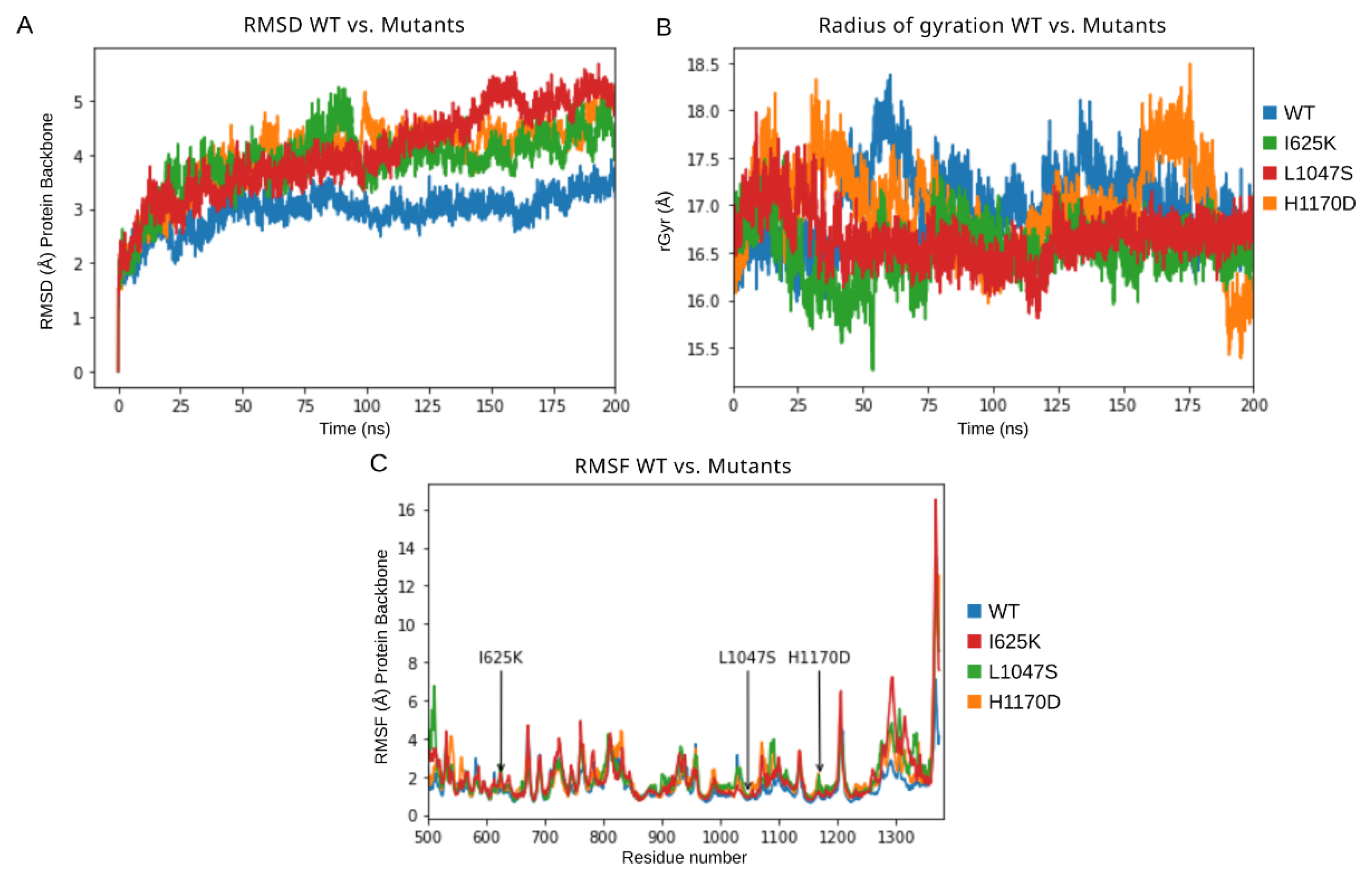
3.2. Molecular Dynamics Simulation of DROSHA Protein
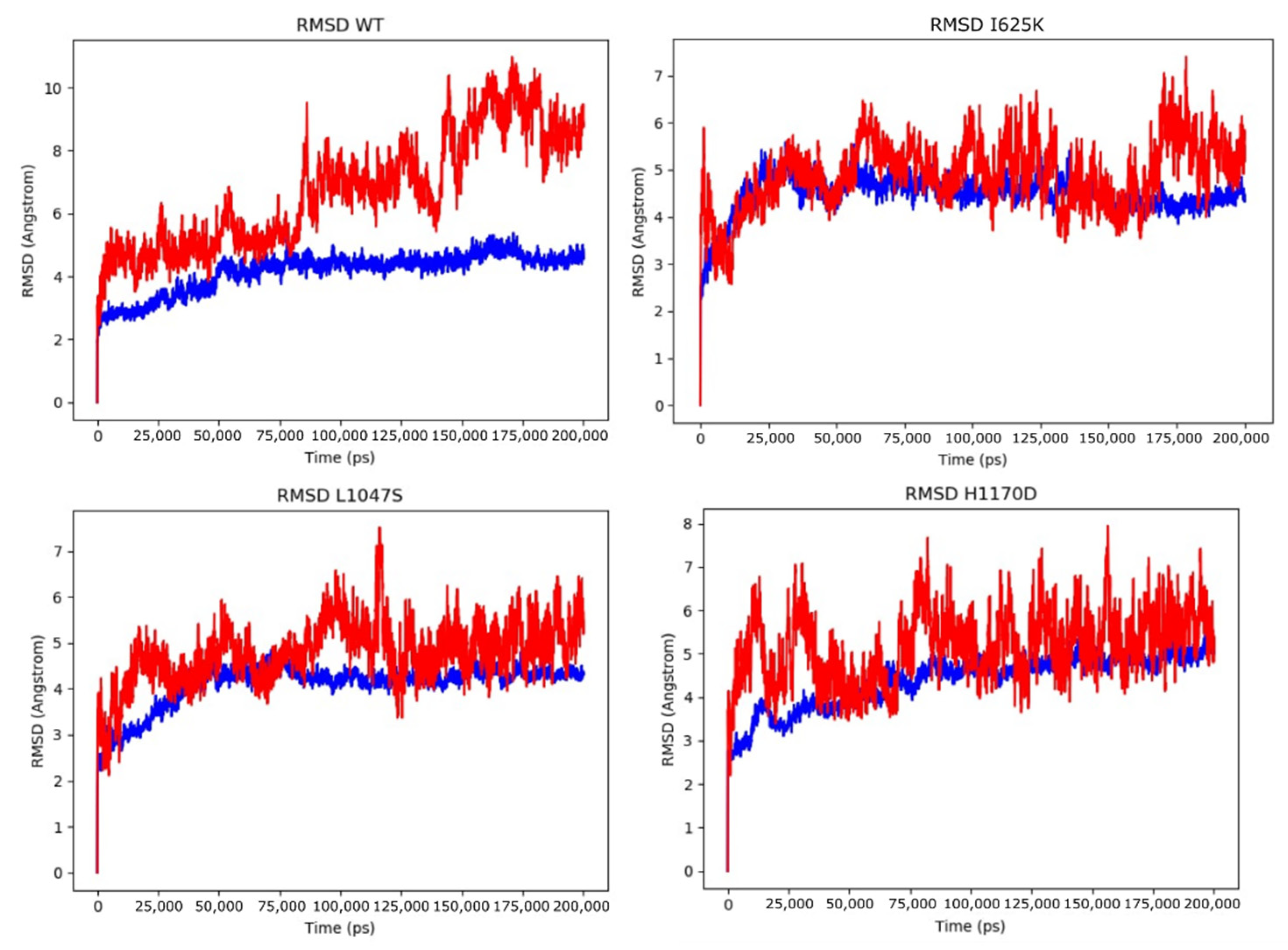
3.3. Molecular Dynamics Simulations of DROSHA-miRNA Complex
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Steensma, D.P.; Bejar, R.; Jaiswal, S.; Lindsley, R.C.; Sekeres, M.; Hasserjian, R.P.; Ebert, B.L. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015, 126, 9–16. [Google Scholar] [CrossRef]
- Meggendorfer, M.; Haferlach, C.; Kern, W.; Haferlach, T. Molecular analysis of myelodysplastic syndrome with isolated deletion of the long arm of chromosome 5 reveals a specific spectrum of molecular mutations with prognostic impact: A study on 123 patients and 27 genes. Haematologica 2017, 102, 1502–1510. [Google Scholar] [CrossRef]
- Santamaria, C.; Muntion, S.; Roson, B.; Blanco, B.; Lopez-Villar, O.; Carrancio, S.; Sanchez-Guijo, F.M.; Diez-Campelo, M.; Alvarez-Fernandez, S.; Sarasquete, M.E.; et al. Impaired expression of Dicer, Drosha, SBDS and some microRNAs in mesenchymal stromal cells from myelodysplastic syndrome patients. Haematologica 2012, 97, 1218–1224. [Google Scholar] [CrossRef] [PubMed]
- Rhyasen, G.W.; Starczynowski, D.T. Deregulation of microRNAs in myelodysplastic syndrome. Leukemia 2011, 26, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Meunier, M.; Guttin, A.; Ancelet, S.; Laurin, D.; Zannoni, J.; Lefebvre, C.; Tondeur, S.; Persoons, V.; Pezet, M.; Pernet-Gallay, K.; et al. Extracellular vesicles from myelodysplastic mesenchymal stromal cells induce DNA damage and mutagenesis of hematopoietic stem cells through miRNA transfer. Leukemia 2020, 34, 2249–2253. [Google Scholar] [CrossRef]
- Raaijmakers, M.H.G.P.; Mukherjee, S.; Guo, S.; Zhang, S.; Kobayashi, T.; Schoonmaker, J.A.; Ebert, B.L.; Al-Shahrour, F.; Hasserjian, R.P.; Scadden, E.O.; et al. Bone progenitor dysfunction induces myelodysplasia and secondary leukemia. Nature 2010, 464, 852–857. [Google Scholar] [CrossRef]
- Moiseev, I.S.; Tcvetkov, N.Y.; Barkhatov, I.M.; Barabanshikova, M.V.; Bug, D.S.; Petuhova, N.V.; Tishkov, A.V.; Bakin, E.A.; Izmailova, E.A.; Shakirova, A.I.; et al. High mutation burden in the checkpoint and micro-RNA processing genes in myelodysplastic syndrome. PLoS ONE 2021, 16, e0248430. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Patel, D.J. Drosha and Dicer: Slicers cut from the same cloth. Cell Res. 2016, 26, 511–512. [Google Scholar] [CrossRef][Green Version]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef]
- Morin, R.; Assouline, S.; Alcaide, M.; Mohajeri, A.; Johnston, R.; Chong, L.; Grewal, J.; Yu, S.; Fornika, D.; Bushell, K.; et al. Genetic landscapes of relapsed and refractory diffuse large B-cell lymphomas. Clin. Cancer Res. 2015, 22, 2290–2300. [Google Scholar] [CrossRef] [PubMed]
- Nangalia, J.; Massie, C.; Baxter, E.J.; Nice, F.; Gundem, G.; Wedge, D.; Avezov, E.; Li, J.; Kollmann, K.; Kent, D.; et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N. Engl. J. Med. 2013, 369, 2391–2405. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Gu, Z.-H.; Yan, Z.-X.; Zhao, X.; Xie, Y.-Y.; Zhang, Z.-G.; Pan, C.-M.; Hu, Y.; Cai, C.-P.; Dong, Y.; et al. Exome sequencing identifies somatic mutations of DDX3X in natural killer/T-cell lymphoma. Nat. Genet. 2015, 47, 1061–1066. [Google Scholar] [CrossRef] [PubMed]
- Gunawardana, J.; Chan, F.C.; Telenius, A.; Woolcock, B.; Kridel, R.; Tan, K.L.; Ben-Neriah, S.; Mottok, A.; Lim, R.; Boyle, M.; et al. Recurrent somatic mutations of PTPN1 in primary mediastinal B cell lymphoma and Hodgkin lymphoma. Nat. Genet. 2014, 46, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Kataoka, K.; Nagata, Y.; Kitanaka, A.; Shiraishi, Y.; Shimamura, T.; Yasunaga, J.-I.; Totoki, Y.; Chiba, K.; Sato-Otsubo, A.; Nagae, G.; et al. Integrated molecular analysis of adult T-cell leukemia/lymphoma. Nat. Genet. 2015, 47, 1304–1315. [Google Scholar] [CrossRef] [PubMed]
- Beà, S.; Valdés-Mas, R.; Navarro, A.; Salaverria, I.; Garcia, D.M.; Jares, P.; Giné, E.; Pinyol, M.; Royo, C.; Nadeu, F.; et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc. Natl. Acad. Sci. USA 2013, 110, 18250–18255. [Google Scholar] [CrossRef]
- Chapiro, E.; Pramil, E.; Diop, M.; Roos-Weil, D.; Dillard, C.; Gabillaud, C.; Maloum, K.; Settegrana, C.; Baseggio, L.; Lesesve, J.-F.; et al. Genetic characterization of B-cell prolymphocytic leukemia: A prognostic model involving MYC and TP53. Blood 2019, 134, 1821–1831. [Google Scholar] [CrossRef]
- Laurent, C.; Nicolae, A.; Laurent, C.; Le Bras, F.; Haioun, C.; Fataccioli, V.; Amara, N.; Adélaïde, J.; Guille, A.; Schiano, J.-M.; et al. Gene alterations in epigenetic modifiers and JAK-STAT signaling are frequent in breast implant-associated ALCL. Blood 2019, 135, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Sim, N.-L.; Kumar, P.; Hu, J.; Henikoff, S.; Schneider, G.; Ng, P.C. SIFT web server: Predicting effects of amino acid substitutions on proteins. Nucleic Acids Res. 2012, 40, W452–W457. [Google Scholar] [CrossRef]
- Adzhubei, I.; Jordan, D.; Sunyaev, S.R. Predicting functional effect of human missense mutations using PolyPhen-2. Curr. Protoc. Hum. Genet. 2013, 76, 7.20.1–7.20.41. [Google Scholar] [CrossRef]
- Li, B.; Krishnan, V.G.; Mort, M.E.; Xin, F.; Kamati, K.K.; Cooper, D.N.; Mooney, S.D.; Radivojac, P. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics 2009, 25, 2744–2750. [Google Scholar] [CrossRef]
- Choi, Y.; Chan, A.P. PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 2015, 31, 2745–2747. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Zhou, M.; Cui, Y. nsSNPAnalyzer: Identifying disease-associated nonsynonymous single nucleotide polymorphisms. Nucleic Acids Res. 2005, 33, W480–W482. [Google Scholar] [CrossRef]
- Stone, E.A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 2005, 15, 978–986. [Google Scholar] [CrossRef] [PubMed]
- Capriotti, E.; Calabrese, R.; Casadio, R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics 2006, 22, 2729–2734. [Google Scholar] [CrossRef]
- Bromberg, Y.; Yachdav, G.; Rost, B. SNAP predicts effect of mutations on protein function. Bioinformatics 2008, 24, 2397–2398. [Google Scholar] [CrossRef] [PubMed]
- Tang, H.; Thomas, P. PANTHER-PSEP: Predicting disease-causing genetic variants using position-specific evolutionary preservation. Bioinformatics 2016, 32, 2230–2232. [Google Scholar] [CrossRef]
- Schwarz, J.M.; Cooper, D.N.; Schuelke, M.; Seelow, D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods 2014, 11, 361–362. [Google Scholar] [CrossRef]
- Bendl, J.; Stourac, J.; Salanda, O.; Pavelka, A.; Wieben, E.D.; Zendulka, J.; Brezovsky, J.; Damborsky, J. PredictSNP: Robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput. Biol. 2014, 10, e1003440. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Ascher, D.; Blundell, T.L. mCSM: Predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 2013, 30, 335–342. [Google Scholar] [CrossRef]
- Cheng, J.; Randall, A.; Baldi, P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins Struct. Funct. Bioinform. 2005, 62, 1125–1132. [Google Scholar] [CrossRef]
- Capriotti, E.; Fariselli, P.; Casadio, R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005, 33, W306–W310. [Google Scholar] [CrossRef] [PubMed]
- Worth, C.; Preissner, R.; Blundell, T.L. SDM—A server for predicting effects of mutations on protein stability and malfunction. Nucleic Acids Res. 2011, 39, W215–W222. [Google Scholar] [CrossRef] [PubMed]
- Pires, D.E.; Ascher, D.; Blundell, T.L. DUET: A server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014, 42, W314–W319. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Lu, H.; Zhang, N.; Zhu, Z.; Wang, S.; Li, M. PremPS: Predicting the impact of missense mutations on protein stability. PLoS Comput. Biol. 2020, 16, e1008543. [Google Scholar] [CrossRef] [PubMed]
- Laimer, J.; Hofer, H.; Fritz, M.; Wegenkittl, S.; Lackner, P.; Laimer, J.; Hofer, H.; Fritz, M.; Wegenkittl, S.; Lackner, P. MAESTRO—Multi agent stability prediction upon point mutations. BMC Bioinform. 2015, 16, 116. [Google Scholar] [CrossRef]
- Pires, D.E.V.; Ascher, D.B. mCSM-NA: Predicting the effects of mutations on protein–nucleic acids interactions. Nucleic Acids Res. 2017, 45, W241–W246. [Google Scholar] [CrossRef]
- Tokuriki, N.; Tawfik, D.S. Stability effects of mutations and protein evolvability. Curr. Opin. Struct. Biol. 2009, 19, 596–604. [Google Scholar] [CrossRef] [PubMed]
- Calloni, G.; Zoffoli, S.; Stefani, M.; Dobson, C.M.; Chiti, F. Investigating the effects of mutations on protein aggregation in the cell. J. Biol. Chem. 2005, 280, 10607–10613. [Google Scholar] [CrossRef]
- Randles, L.G.; Lappalainen, I.; Fowler, S.B.; Moore, B.; Hamill, S.J.; Clarke, J. Using model proteins to quantify the effects of pathogenic mutations in Ig-like proteins. J. Biol. Chem. 2006, 281, 24216–24226. [Google Scholar] [CrossRef]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef]
- Frappier, V.; Chartier, M.; Najmanovich, R.J. ENCoM server: Exploring protein conformational space and the effect of mutations on protein function and stability. Nucleic Acids Res. 2015, 43, W395–W400. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.H.M.; Pires, D.E.V.; Ascher, D.B. DynaMut: Predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018, 46, W350–W355. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, C.H.; Pires, D.E.; Ascher, D.B. DynaMut2: Assessing changes in stability and flexibility upon single and multiple point missense mutations. Protein Sci. 2020, 30, 60–69. [Google Scholar] [CrossRef] [PubMed]
- Partin, A.C.; Zhang, K.; Jeong, B.-C.; Herrell, E.; Li, S.; Chiu, W.; Nam, Y. Cryo-EM structures of human Drosha and DGCR8 in complex with primary microRNA. Mol. Cell 2020, 78, 411–422. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.D.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The protein data bank. Nucleic Acids Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Jacobson, M.P.; Pincus, D.L.; Rapp, C.S.; Day, T.J.; Honig, B.; Shaw, D.E.; Friesner, R.A. A hierarchical approach to all-atom protein loop prediction. Proteins: Struct. Funct. Bioinform. 2004, 55, 351–367. [Google Scholar] [CrossRef]
- Li, J.; Abel, R.; Zhu, K.; Cao, Y.; Zhao, S.; Friesner, R.A. The VSGB 2.0 model: A next generation energy model for high resolution protein structure modeling. Proteins: Struct. Funct. Bioinform. 2011, 79, 2794–2812. [Google Scholar] [CrossRef]
- Harder, E.; Damm, W.; Maple, J.; Wu, C.; Reboul, M.; Xiang, J.Y.; Wang, L.; Lupyan, D.; Dahlgren, M.K.; Knight, J.L.; et al. OPLS3: A force field providing broad coverage of drug-like small molecules and proteins. J. Chem. Theory Comput. 2015, 12, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Roos, K.; Wu, C.; Damm, W.; Reboul, M.; Stevenson, J.M.; Lu, C.; Dahlgren, M.K.; Mondal, S.; Chen, W.; Wang, L.; et al. OPLS3e: Extending force field coverage for drug-like small molecules. J. Chem. Theory Comput. 2019, 15, 1863–1874. [Google Scholar] [CrossRef] [PubMed]
- Sastry, G.M.; Adzhigirey, M.; Day, T.; Annabhimoju, R.; Sherman, W. Protein and ligand preparation: Parameters, protocols, and influence on virtual screening enrichments. J. Comput. Mol. Des. 2013, 27, 221–234. [Google Scholar] [CrossRef] [PubMed]
- Bowers, K.J.; Chow, D.E.; Xu, H.; Dror, R.O.; Eastwood, M.P.; Gregersen, B.A.; Klepeis, J.L.; Kolossvary, I.; Moraes, M.A.; Sacerdoti, F.D.; et al. Scalable algorithms for molecular dynamics simulations on commodity clusters. In Proceedings of the 2006 ACM/IEEE Conference on Supercomputing, Tampa, FL, USA, 11–17 November 2006; p. 43. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Nosé, S. A unified formulation of the constant temperature molecular dynamics methods. J. Chem. Phys. 1984, 81, 511–519. [Google Scholar] [CrossRef]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef] [PubMed]
- Bissantz, C.; Kuhn, B.; Stahl, M. A medicinal chemist’s guide to molecular interactions. J. Med. Chem. 2010, 53, 5061–5084. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.C.; Nguyen, T.A.; Choi, Y.-G.; Jo, M.H.; Hohng, S.; Kim, V.N.; Woo, J.-S. Structure of human Drosha. Cell 2015, 164, 81–90. [Google Scholar] [CrossRef]
- Gurtner, A.; Falcone, E.; Garibaldi, F.; Piaggio, G. Dysregulation of microRNA biogenesis in cancer: The impact of mutant p53 on Drosha complex activity. J. Exp. Clin. Cancer Res. 2016, 35, 45. [Google Scholar] [CrossRef]
- Yan, M.; Huang, H.-Y.; Wang, T.; Wan, Y.; Cui, S.-D.; Liu, Z.-Z.; Fan, Q.-X. Dysregulated expression of Dicer and Drosha in breast cancer. Pathol. Oncol. Res. 2011, 18, 343–348. [Google Scholar] [CrossRef]
- Hata, A.; Kashima, R. Dysregulation of microRNA biogenesis machinery in cancer. Crit. Rev. Biochem. Mol. Biol. 2015, 51, 121–134. [Google Scholar] [CrossRef]
- Kuang, X.; Chi, J.; Wang, L. Deregulated microRNA expression and its pathogenetic implications for myelodysplastic syndromes. Hematology 2016, 21, 593–602. [Google Scholar] [CrossRef] [PubMed][Green Version]
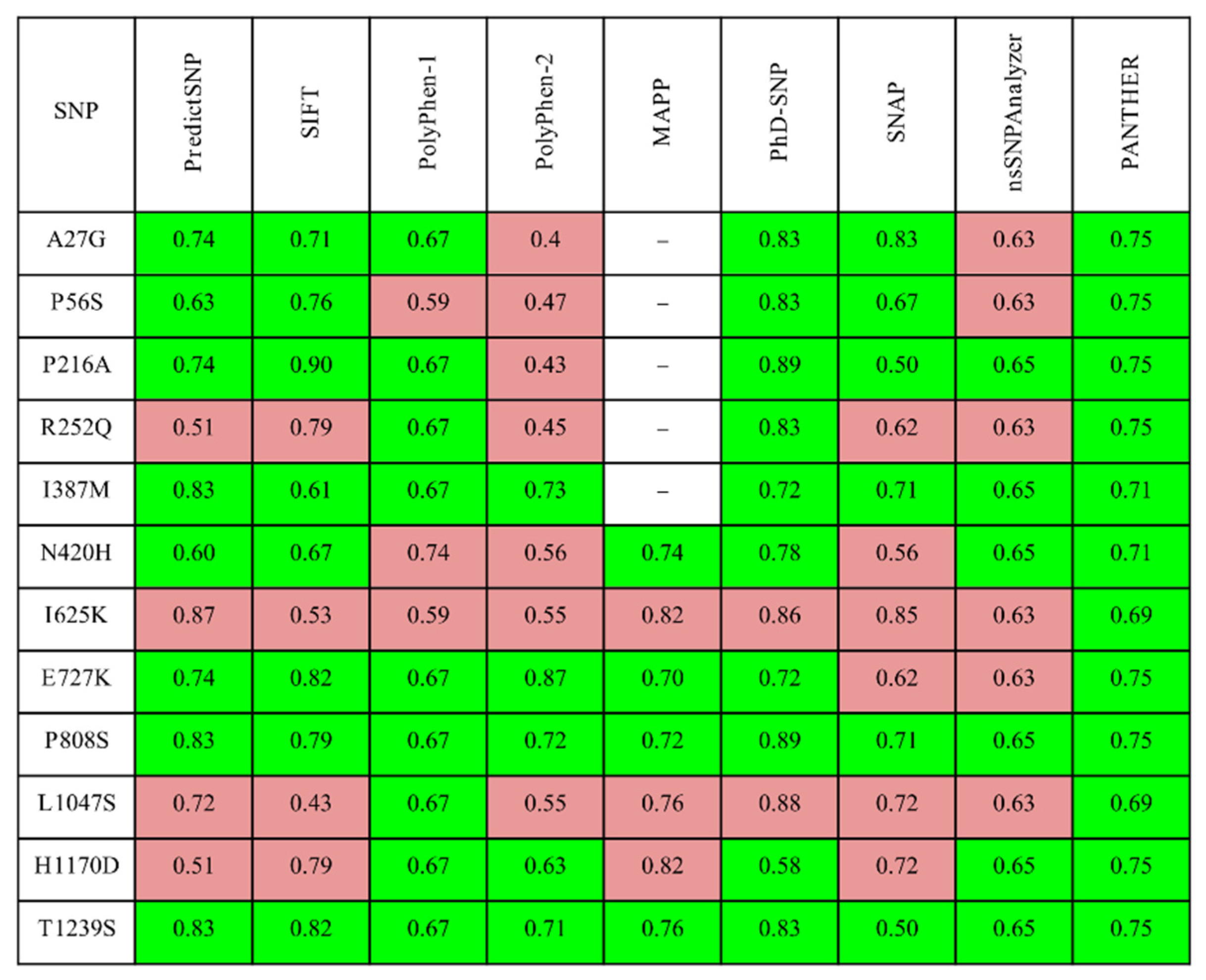

| Variant | mCSM | mCMS- NA | MUpro | i- Mutant | SDM | DUET | PremPS | Maestro | FoldX4 | Dyna Mut | Dyna Mut2 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| I625K | destab (−1.545) | stab (2.543) | destab (−2.234) | destab (−1.850) | destab (−1.080) | destab (−1.437) | destab (1.440) | destab (0.095) | destab (0.334) | stab (0.053) | destab (−1.42) |
| L1047S | destab (−3.220) | stab (3.590) | destab (−2.266) | destab (−1.520) | destab (−4.310) | destab (−3.578) | destab (2.460) | destab (0.614) | destab (4.975) | destab (−1.203) | destab (−3.10) |
| H1170D | destab (−0.837) | destab (−4.107) | destab (−0.539) | destab (−0.70) | destab (−0.660) | destab (−0.812) | destab (1.28) | destab (0.254) | destab (1.325) | destab (−0.216) | destab (−0.45) |
| Model | H-Bonds Intra | rGyr (Å) | RMSD Backbone (Å) | Total Energy (kcal/mol) | |||
|---|---|---|---|---|---|---|---|
| Range | Mean | Range | Mean | Range | Mean | Mean | |
| Drosha wt | (708, 803) | 754.339 | (15.979, 18.376) | 16.995 | (0.000, 3.930) | 2.964 | −33,438 |
| I625K | (720, 821) | 770.246 | (15.257, 17.527) | 16.523 | (0.000, 5.273) | 3.871 | −32,940 |
| L1047S | (715, 817) | 761.297 | (15.81, 17.982) | 16.666 | (0.000, 5.689) | 4.077 | −32,897 |
| H1170 | (729, 833) | 774.806 | (15.385, 18.494) | 16.975 | (0.000, 5.284) | 3.982 | −33,049 |
| Model | H-Bonds RNA–Protein | RMSD Protein (Å) | RMSD RNA (Å) | |||||
|---|---|---|---|---|---|---|---|---|
| Range | Mean | Range | Mean | SD | Range | Mean | SD | |
| Drosha WT | (28, 64) | 47.141 | (0.000, 5.380) | 4.146 | 0.634 | (0.000, 10.969) | 6.697 | 1.803 |
| H1170 | (30, 67) | 45.527 | (0.000, 5.556) | 4.388 | 0.619 | (0.000, 7.947) | 5.151 | 0.807 |
| I625K | (33, 66) | 46.066 | (0.000, 5.588) | 4.475 | 0.417 | (0.000, 7.396) | 4.893 | 0.710 |
| L1047S | (32, 73) | 47.392 | (0.000, 4.852) | 4.099 | 0.468 | (0.000, 7.511) | 4.774 | 0.674 |
| Model | Minimal Total Energy of the Complex (kcal/mol) | Mean Total Energy of the Complex (kcal/mol) | MM-GBSA ΔG Bind (kcal/mol) |
|---|---|---|---|
| Drosha WT | −46,193.114 | −46,049.497 | −380.929 |
| I625K | −45,830.501 | −45,711.641 | −386.547 |
| L1047S | −45,787.443 | −45,687.955 | −439.187 |
| H1170D | −45,941.373 | −45,869.388 | −334.761 |
| Mutation | Histology-Based Diagnosis | Functional Domains | Sequence-Based Tools Predicting Functional Impairment | Structure-Based Tools Predicting Molecular Destabilization | ΔRMSD Backbone (Free Protein, Å) | ΔG (Free Protein, kcal/mol) | ΔG (miRNA Protein Complex, kcal/mol) | ΔG Bind (miRNA, kcal/mol) |
|---|---|---|---|---|---|---|---|---|
| I625K | Adult T cell lymphoma-leukemia | Central domain (Platform) | 8/9 | 9/11 | 0.91 | 498 (destabilization) | 337.86 (destabilization) | −5.62 (increased affinity) |
| L1047S | B cell prolymphocytic leukemia | RNAse IIIa | 7/9 | 10/11 | 1.11 | 541 (destabilization) | 361.54 (destabilization) | −58.26 (increased affinity) |
| H1170D | Breast implant-associated anaplastic large cell lymphoma, invasive | RNAse IIIb | 4/9 | 11/11 | 1.02 | 389 (destabilization) | 180.11 (destabilization) | 46.17 (decreased affinity) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bug, D.S.; Tishkov, A.V.; Moiseev, I.S.; Porozov, Y.B.; Petukhova, N.V. Towards Understanding the Pathogenicity of DROSHA Mutations in Oncohematology. Cells 2021, 10, 2357. https://doi.org/10.3390/cells10092357
Bug DS, Tishkov AV, Moiseev IS, Porozov YB, Petukhova NV. Towards Understanding the Pathogenicity of DROSHA Mutations in Oncohematology. Cells. 2021; 10(9):2357. https://doi.org/10.3390/cells10092357
Chicago/Turabian StyleBug, Dmitrii S., Artem V. Tishkov, Ivan S. Moiseev, Yuri B. Porozov, and Natalia V. Petukhova. 2021. "Towards Understanding the Pathogenicity of DROSHA Mutations in Oncohematology" Cells 10, no. 9: 2357. https://doi.org/10.3390/cells10092357
APA StyleBug, D. S., Tishkov, A. V., Moiseev, I. S., Porozov, Y. B., & Petukhova, N. V. (2021). Towards Understanding the Pathogenicity of DROSHA Mutations in Oncohematology. Cells, 10(9), 2357. https://doi.org/10.3390/cells10092357






