Myxospermy Evolution in Brassicaceae: A Highly Complex and Diverse Trait with Arabidopsis as an Uncommon Model
Abstract
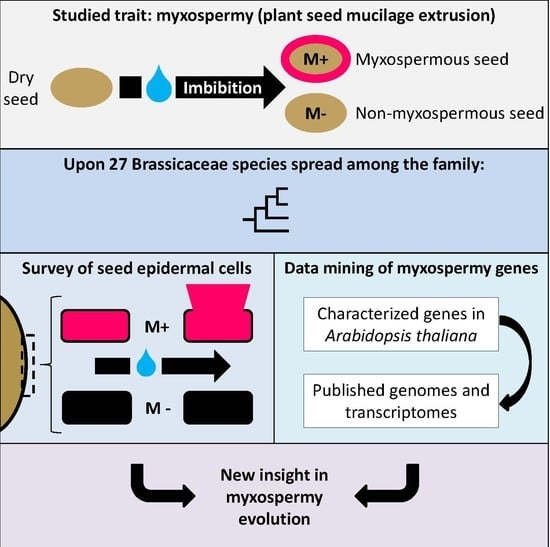
:1. Introduction
2. Materials and Methods
2.1. Plant Material, Seeds and Genomes
2.2. Dry Seed Phenotyping
2.3. Phenotyping of Adherent Mucilage
2.4. Cross Section of Mucilage Secretory Cells, Histochemistry and Immunofluorescence
2.5. Genomic Data Mining of A. thaliana MSC Toolbox Gene Orthologs
2.6. Transcriptomic Data Mining of A. thaliana MSC Toolbox Gene Orthologs from A. thaliana, C. sativa, B. napus and A. arabicum Public Seed Development Transcriptomes
3. Results
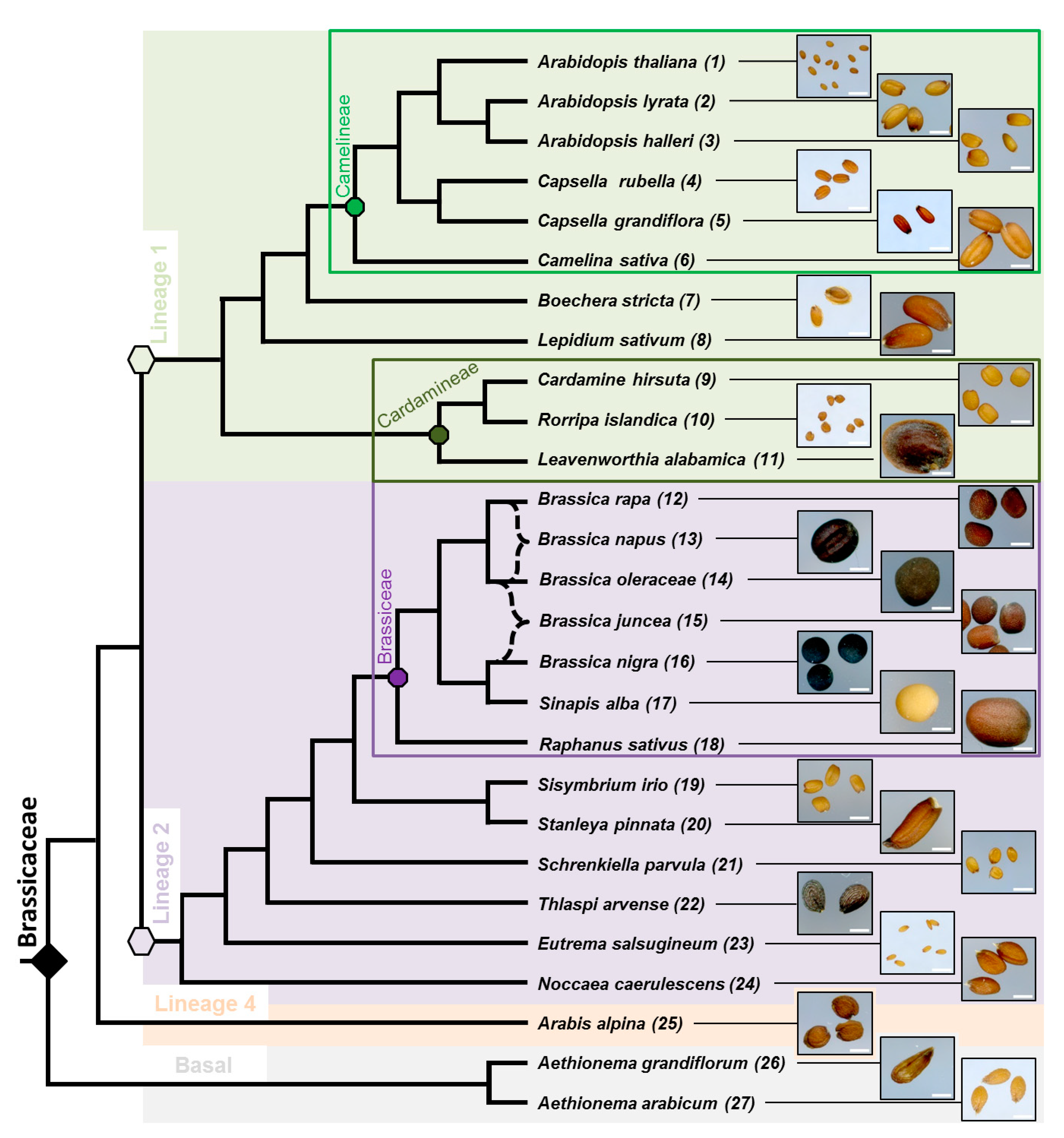
3.1. Phylogenic Relationship between 27 Selected Species and Their Mature Dry Seed Whole Morphology
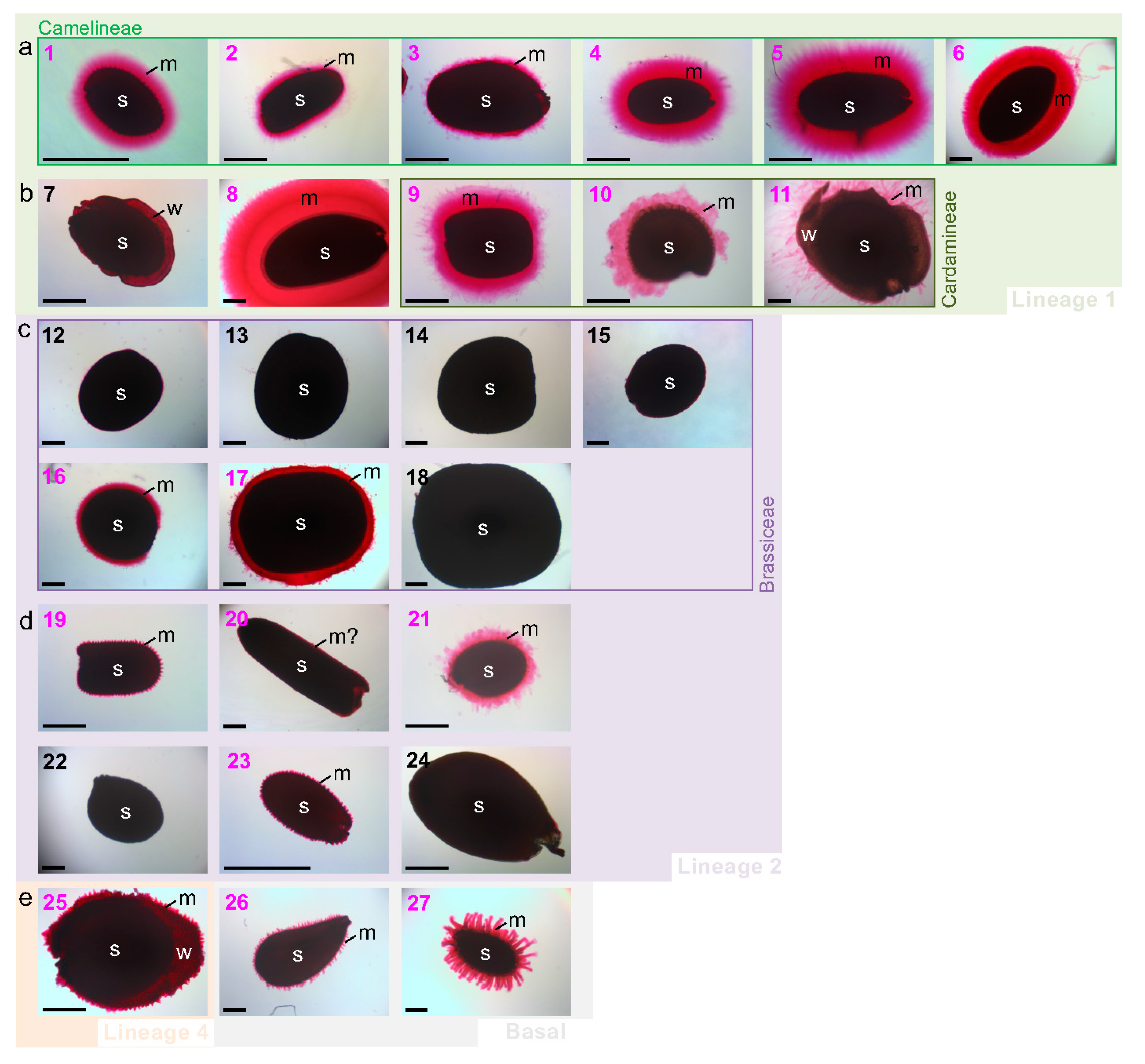
3.2. Myxospermy Occurrence and Morphology at the Whole Seed Level
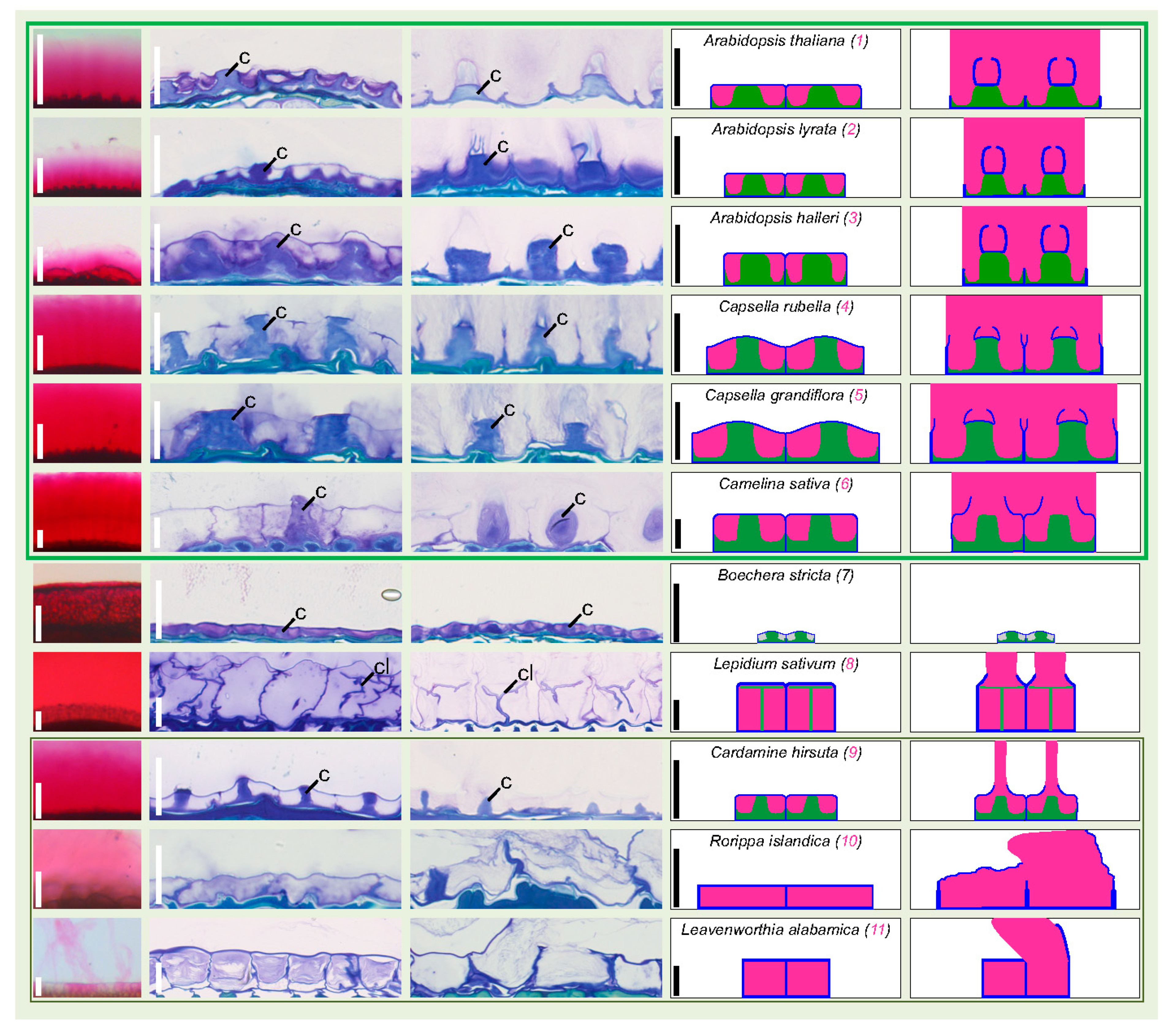
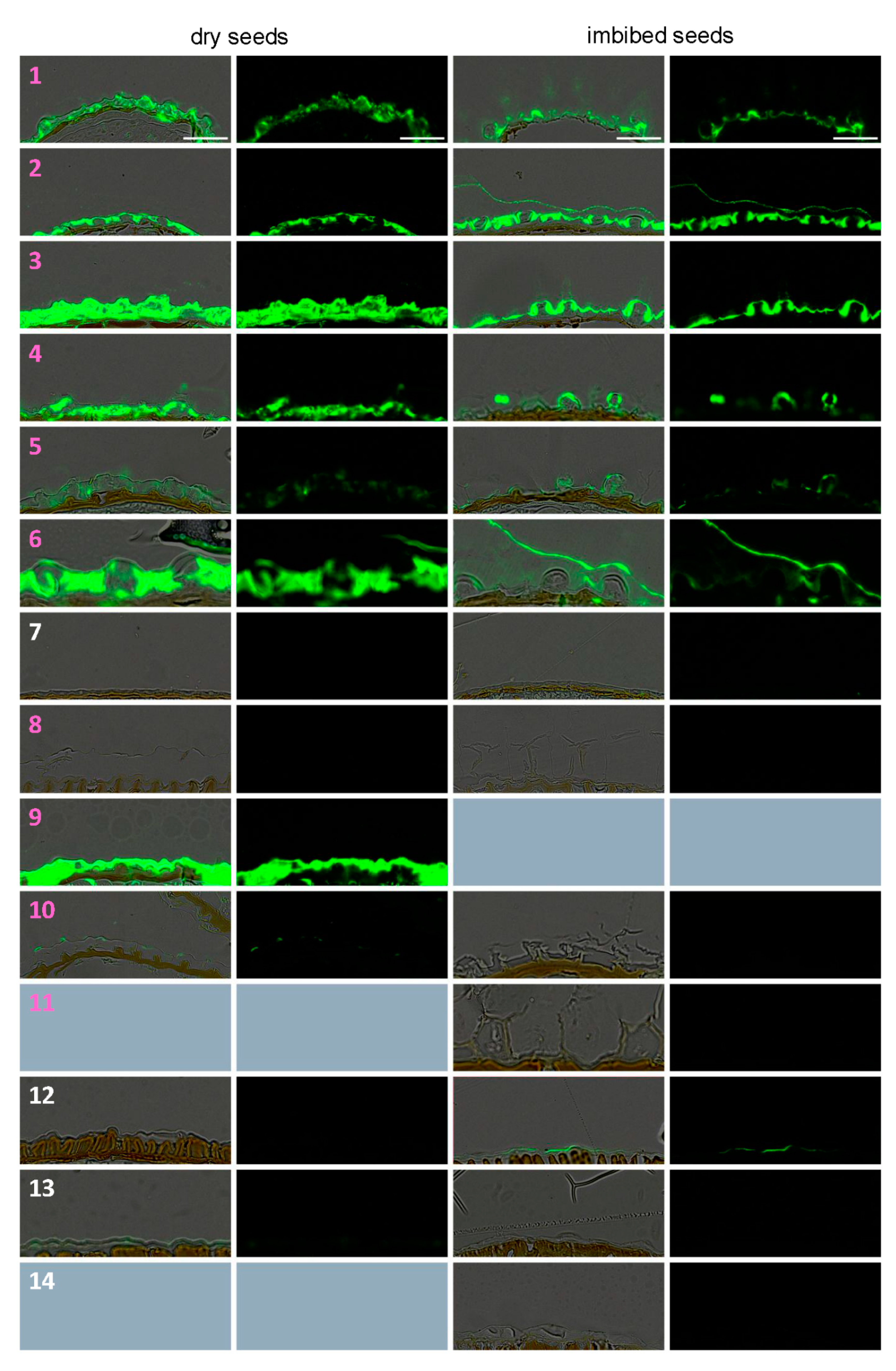
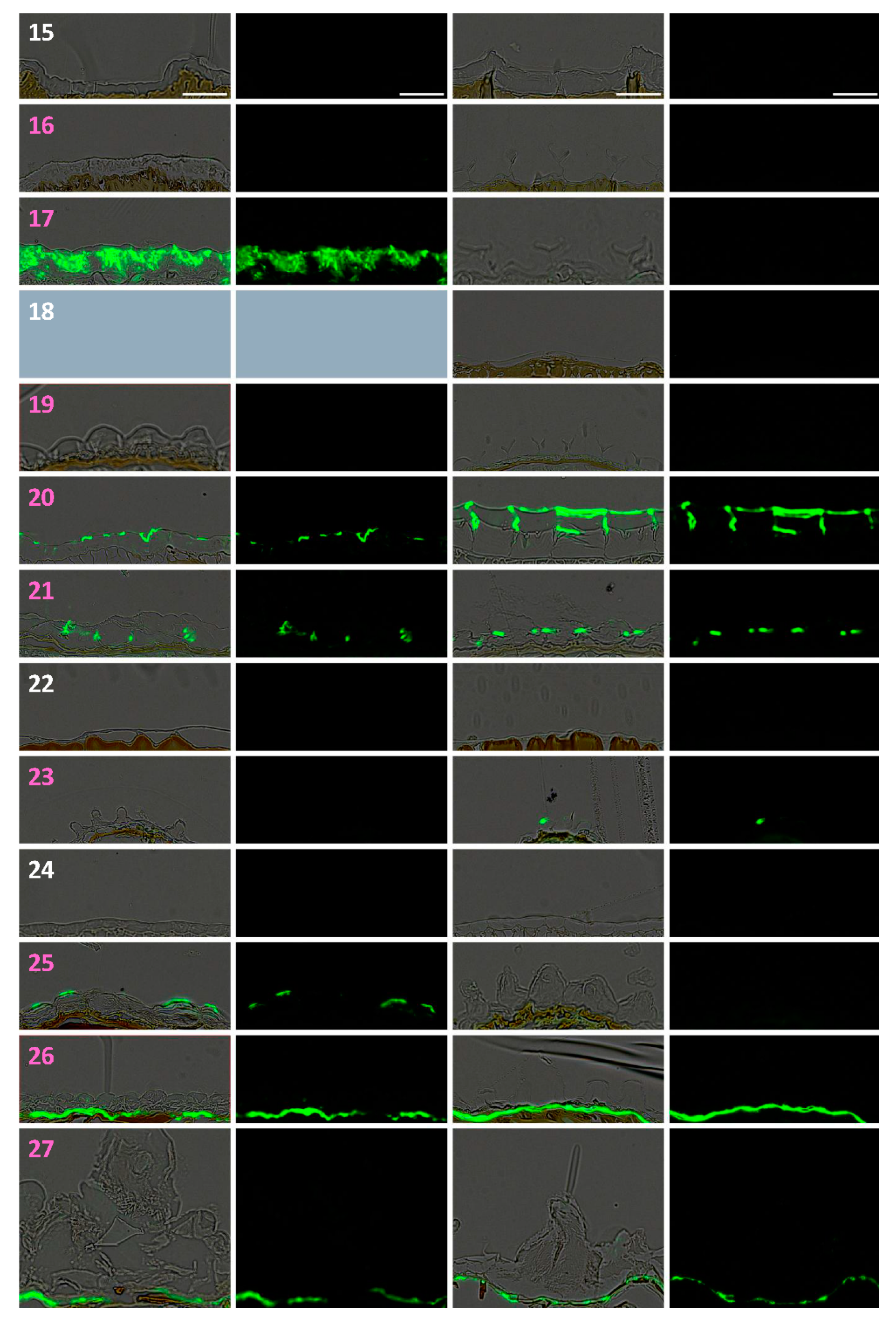
3.3. Dry Seed Epidermis/MSC Morphology and MSC Opening Mode in Imbibed Seeds
3.4. In Situ Immunolabeling of SM Content and MSC Primary Cell Wall Microdomains
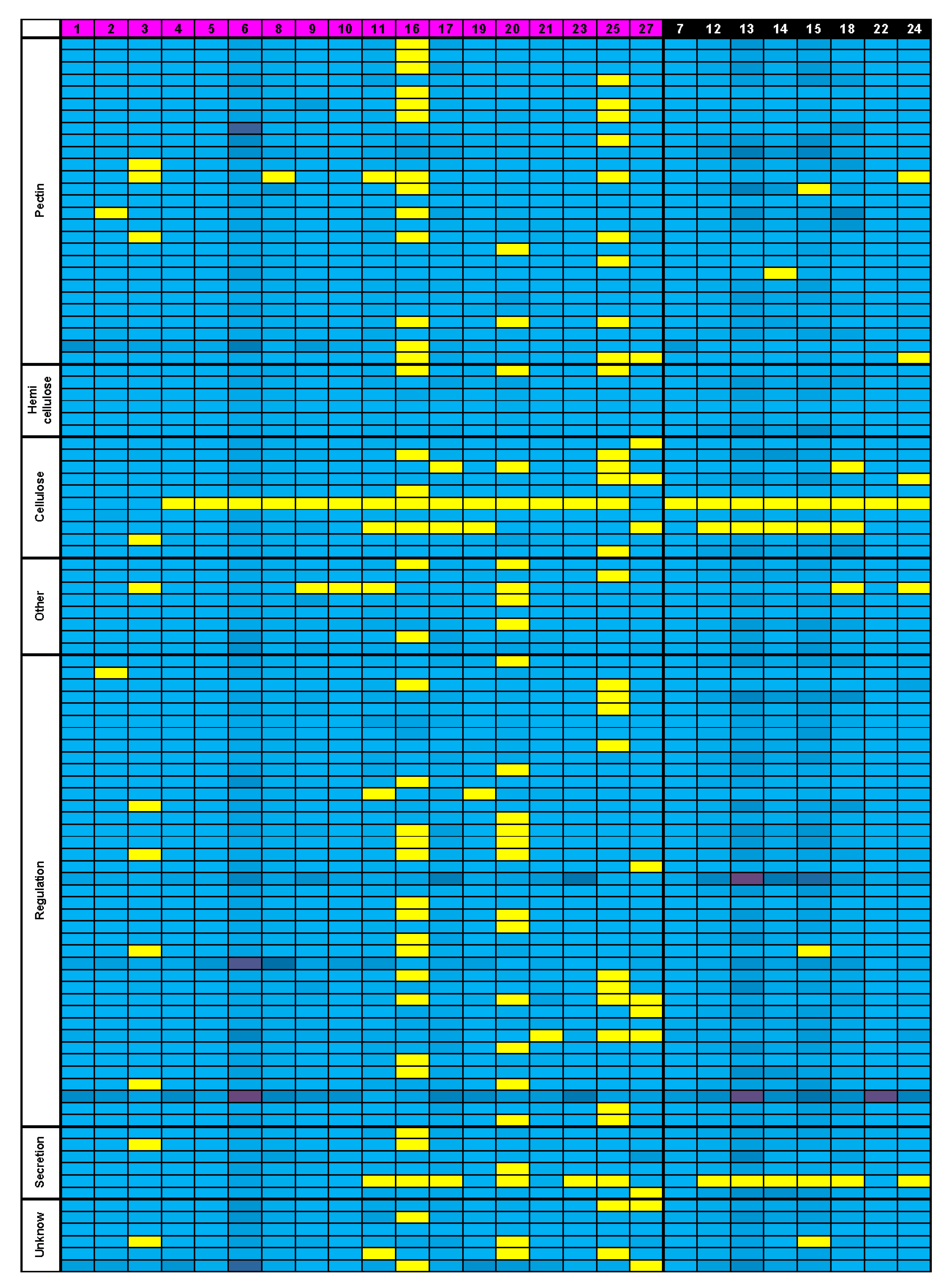
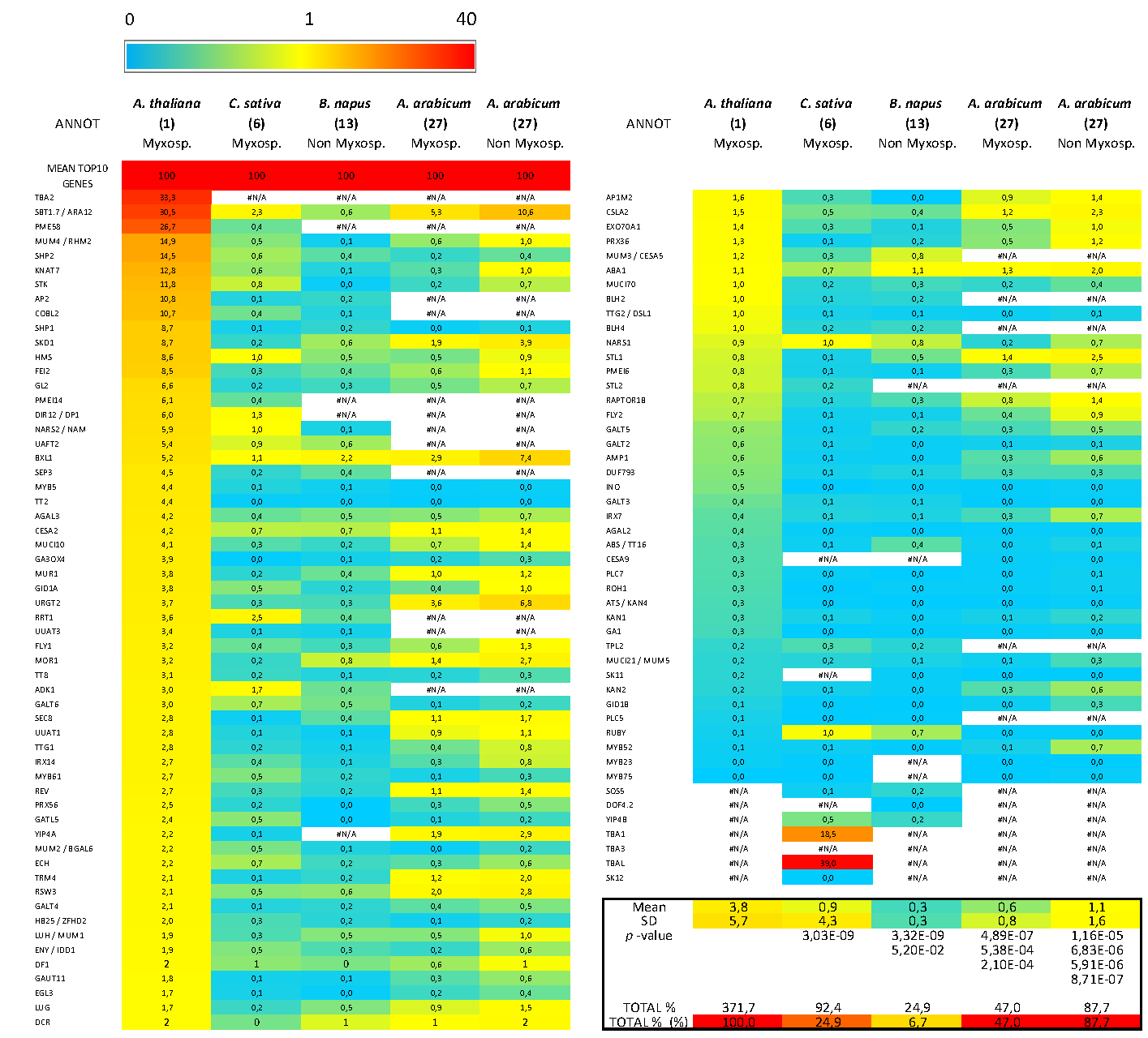
3.5. Bioinformatics Insight on A. thaliana MSC Toolbox Gene Orthologs in Brassicaceae Species
4. Discussion
4.1. Is Myxospermy an Ancestral Feature of the Brassicaceae Family?
4.2. Arabidopsis Species as an Uncommon Model for Mucilage Secretory Cells
4.3. Ecological Perspective
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Doyle, J.A. Phylogenetic analyses and morphological innovations in land plants. Annu. Plant Rev. 2017, 45, 1–50. [Google Scholar]
- Baroux, C.; Grossniklaus, U. Seeds—An evolutionary innovation underlying reproductive success in flowering plants. Curr. Top. Dev. Biol. 2019, 131, 605–642. [Google Scholar]
- Coen, O.; Magnani, E. Seed coat thickness in the evolution of Angiosperms. Cell. Mol. Life Sci. 2018, 75, 2509–2518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beeckman, T.; De Rycke, R.; Viane, R.; Inzé, D. Histological study of seed coat development in Arabidopsis thaliana. J. Plant Res. 2000, 113, 139–148. [Google Scholar] [CrossRef]
- Western, T.L. Isolation and characterization of mutants defective in seed coat mucilage secretory cell development in Arabidopsis. Plant Physiol. 2001, 127, 998–1011. [Google Scholar] [PubMed]
- Voiniciuc, C.; Yang, B.; Schmidt, M.H.W.; Günl, M.; Usadel, B. Starting to gel: How Arabidopsis seed coat epidermal cells produce specialized secondary cell walls. Int. J. Mol. Sci. 2015, 16, 3452–3473. [Google Scholar] [CrossRef] [Green Version]
- Francoz, E.; Ranocha, P.; Burlat, V.; Dunand, C. Arabidopsis seed mucilage secretory cells: Regulation and dynamics. Trends Plant Sci. 2015, 20, 515–524. [Google Scholar] [CrossRef]
- Francoz, E.; Ranocha, P.; Le Ru, A.; Martinez, Y.; Fourquaux, I.; Jauneau, A.; Dunand, C.; Burlat, V. Pectin demethylesterification generates platforms that anchor peroxidases to remodel plant cell wall domains. Dev. Cell 2019, 48, 261–276. [Google Scholar] [CrossRef] [Green Version]
- Šola, K.; Dean, G.H.; Haughn, G.W. Arabidopsis seed mucilage: A specialised extracellular matrix that demonstrates the structure–function versatility of cell wall polysaccharides. Annu. Plant Rev. 2019, 2, 1085–1116. [Google Scholar]
- Tsai, A.Y.-L.; Kunieda, T.; Rogalski, J.; Foster, L.J.; Ellis, B.E.; Haughn, G.W. Identification and characterization of Arabidopsis seed coat mucilage proteins. Plant Physiol. 2017, 173, 1059–1074. [Google Scholar] [CrossRef] [Green Version]
- Griffiths, J.S.; Šola, K.; Kushwaha, R.; Lam, P.; Tateno, M.; Young, R.; Voiniciuc, C.; Dean, G.; Mansfield, S.D.; Debolt, S.; et al. Unidirectional movement of cellulose synthase complexes in Arabidopsis seed coat epidermal cells deposit cellulose involved in mucilage extrusion, adherence, and ray formation. Plant Physiol. 2015, 168, 502–520. [Google Scholar] [CrossRef]
- Macquet, A.; Ralet, M.C.; Kronenberger, J.; Marion-Poll, A.; North, H.M. In situ, chemical and macromolecular study of the composition of Arabidopsis thaliana seed coat mucilage. Plant Cell Physiol. 2007, 48, 984–999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ralet, M.C.; Tranquet, O.; Poulain, D.; Moïse, A.; Guillon, F. Monoclonal antibodies to rhamnogalacturonan I backbone. Planta 2010, 231, 1373–1383. [Google Scholar] [CrossRef] [PubMed]
- Saez-Aguayo, S.; Rautengarten, C.; Temple, H.; Sanhueza, D.; Ejsmentewicz, T.; Sandoval-Ibañez, O.; Doñas, D.; Parra-Rojas, J.P.; Ebert, B.; Lehner, A.; et al. UUAT1 is a golgi-localized UDP-uronic acid transporter that modulates the polysaccharide composition of Arabidopsis seed mucilage. Plant Cell 2017, 29, 129–143. [Google Scholar] [CrossRef] [PubMed]
- Phan, J.L.; Burton, R.A. New insights into the composition and structure of seed mucilage. Annu. Plant Rev. 2018, 1, 63–104. [Google Scholar]
- Viudes, S.; Burlat, V.; Dunand, C. Seed mucilage evolution: Diverse molecular mechanisms generate versatile ecological functions for particular environments. Plant Cell Environ. 2020, 43, 2857–2870. [Google Scholar] [CrossRef]
- Yang, X.; Baskin, J.M.; Baskin, C.C.; Huang, Z. More than just a coating: Ecological importance, taxonomic occurrence and phylogenetic relationships of seed coat mucilage. Perspect. Plant Ecol. Evol. Syst. 2012, 14, 434–442. [Google Scholar] [CrossRef]
- North, H.M.; Berger, A.; Saez-Aguayo, S.; Ralet, M.C. Understanding polysaccharide production and properties using seed coat mutants: Future perspectives for the exploitation of natural variants. Ann. Bot. 2014, 114, 1251–1263. [Google Scholar] [CrossRef] [Green Version]
- Phan, J.L.; Tucker, M.R.; Khor, S.F.; Shirley, N.; Lahnstein, J.; Beahan, C.; Bacic, A.; Burton, R.A. Differences in glycosyltransferase family 61 accompany variation in seed coat mucilage composition in Plantago spp. J. Exp. Bot. 2016, 67, 6481–6495. [Google Scholar] [CrossRef] [Green Version]
- Cowley, J.M.; Burton, R.A. The goo-d stuff: Plantago as a myxospermous model with modern utility. New Phytol. 2021, 229, 1917–1923. [Google Scholar] [CrossRef]
- Voiniciuc, C.; Zimmermann, E.; Schmidt, M.H.-W.; Günl, M.; Fu, L.; North, H.M.; Usadel, B. Extensive natural variation in Arabidopsis seed mucilage structure. Front. Plant Sci. 2016, 7, 803. [Google Scholar] [CrossRef] [Green Version]
- Poulain, D.; Botran, L.; North, H.M.; Ralet, M.-C. Composition and physicochemical properties of outer mucilage from seeds of Arabidopsis natural accessions. AoB Plants 2019, 11, plz031. [Google Scholar] [CrossRef]
- Liu, J.; Shim, Y.Y.; Shen, J.; Wang, Y.; Ghosh, S.; Reaney, M.J.T. Variation of composition and functional properties of gum from six Canadian flaxseed (Linum usitatissimum L.) cultivars. Int. J. Food Sci. Technol. 2016, 51, 2313–2326. [Google Scholar] [CrossRef]
- Soto-Cerda, B.J.; Cloutier, S.; Quian, R.; Gajardo, H.A.; Olivos, M.; You, F.M. Genome-wide association analysis of mucilage and hull content in flax (Linum usitatissimum l.) seeds. Int. J. Mol. Sci. 2018, 19, 2870. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabrissin, I.; Cueff, G.; Berger, A.; Granier, F.; Sallé, C.; Poulain, D.; Ralet, M.-C.; North, H.M. Natural variation reveals a key role for rhamnogalacturonan I in seed outer mucilage and underlying genes. Plant Physiol. 2019, 181, 1498–1518. [Google Scholar] [CrossRef] [PubMed]
- Vaughan, J.G.; Whitehouse, J.M. Seed structure and the taxonomy of the Cruciferae. Bot. J. Linn. Soc. 1971, 64, 383–409. [Google Scholar] [CrossRef]
- Western, T.L. The sticky tale of seed coat mucilages: Production, genetics, and role in seed germination and dispersal. Seed Sci. Res. 2012, 22, 1–25. [Google Scholar] [CrossRef]
- Hohmann, N.; Wolf, E.M.; Lysak, M.A.; Koch, M.A. A time-calibrated road map of Brassicaceae species radiation and evolutionary history. Plant Cell 2015, 27, 2770–2784. [Google Scholar] [CrossRef] [Green Version]
- Huang, C.H.; Sun, R.; Hu, Y.; Zeng, L.; Zhang, N.; Cai, L.; Zhang, Q.; Koch, M.A.; Al-Shehbaz, I.; Edger, P.P.; et al. Resolution of Brassicaceae phylogeny using nuclear genes uncovers nested radiations and supports convergent morphological evolution. Mol. Biol. Evol. 2016, 33, 394–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nikolov, L.A.; Shushkov, P.; Nevado, B.; Gan, X.; Al-Shehbaz, I.A.; Filatov, D.; Bailey, C.D.; Tsiantis, M. Resolving the backbone of the Brassicaceae phylogeny for investigating trait diversity. New Phytol. 2019, 222, 1638–1651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clausen, M.H.; Willats, W.G.T.; Knox, J.P. Synthetic methyl hexagalacturonate hapten inhibitors of anti-homogalacturonan monoclonal antibodies LM7, JIM5 and JIM7. Carbohydr. Res. 2003, 338, 1797–1800. [Google Scholar] [CrossRef]
- Verhertbruggen, Y.; Marcus, S.E.; Haeger, A.; Ordaz-Ortiz, J.J.; Knox, J.P. An extended set of monoclonal antibodies to pectic homogalacturonan. Carbohydr. Res. 2009, 344, 1858–1862. [Google Scholar] [CrossRef]
- Solovyev, V.; Kosarev, P.; Seledsov, I.; Vorobyev, D. Automatic annotation of eukaryotic genes, pseudogenes and promoters. Genome Biol. 2006, 7, S10. [Google Scholar] [CrossRef] [Green Version]
- Belmonte, M.F.; Kirkbride, R.C.; Stone, S.L.; Pelletier, J.M.; Bui, A.Q.; Yeung, E.C.; Hashimoto, M.; Fei, J.; Harada, C.M.; Munoz, M.D.; et al. Comprehensive developmental profiles of gene activity in regions and subregions of the Arabidopsis seed. Proc. Natl. Acad. Sci. USA 2013, 110, E435–E444. [Google Scholar] [CrossRef] [Green Version]
- Abdullah, H.M.; Akbari, P.; Paulose, B.; Schnell, D.; Qi, W.; Park, Y.; Pareek, A.; Dhankher, O.P. Transcriptome profiling of Camelina sativa to identify genes involved in triacylglycerol biosynthesis and accumulation in the developing seeds. Biotechnol. Biofuels 2016, 9, 136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kagale, S.; Nixon, J.; Khedikar, Y.; Pasha, A.; Provart, N.J.; Clarke, W.E.; Bollina, V.; Robinson, S.J.; Coutu, C.; Hegedus, D.D.; et al. The developmental transcriptome atlas of the biofuel crop Camelina sativa. Plant J. 2016, 88, 879–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, H.; Cui, Y.; Ding, Y.; Mei, J.; Dong, H.; Zhang, W.; Wu, S.; Liang, Y.; Zhang, C.; Li, J.; et al. Time-series analyses of transcriptomes and proteomes reveal molecular networks underlying oil accumulation in canola. Front. Plant Sci. 2017, 7, 2007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arshad, W.; Lenser, T.; Wilhelmsson, P.K.I.; Chandler, J.O.; Steinbrecher, T.; Marone, F.; Pérez, M.; Collinson, M.E.; Stuppy, W.; Rensing, S.A.; et al. A tale of two morphs: Developmental patterns and mechanisms of seed coat differentiation in the dimorphic diaspore model Aethionema arabicum (Brassicaceae). Plant J. 2021, 107, 166–181. [Google Scholar] [CrossRef] [PubMed]
- Kagale, S.; Koh, C.; Nixon, J.; Bollina, V.; Clarke, W.E.; Tuteja, R.; Spillane, C.; Robinson, S.J.; Links, M.G.; Clarke, C.; et al. The emerging biofuel crop Camelina sativa retains a highly undifferentiated hexaploid genome structure. Nat. Commun. 2014, 5, 3706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lenser, T.; Graeber, K.; Cevik, Ö.S.; Adigüzel, N.; Dönmez, A.A.; Grosche, C.; Kettermann, M.; Mayland-Quellhorst, S.; Mérai, Z.; Mohammadin, S.; et al. Developmental control and plasticity of fruit and seed dimorphism in Aethionema arabicum. Plant Physiol. 2016, 172, 1691–1707. [Google Scholar] [CrossRef] [Green Version]
- Rautengarten, C.; Usadel, B.; Neumetzler, L.; Hartmann, J.; Büssis, D.; Altmann, T. A subtilisin-like serine protease essential for mucilage release from Arabidopsis seed coats. Plant J. 2008, 54, 466–480. [Google Scholar] [CrossRef] [PubMed]
- Hradilová, I.; Trněný, O.; Válková, M.; Cechová, M.; Janská, A.; Prokešová, L.; Aamir, K.; Krezdorn, N.; Rotter, B.; Winter, P.; et al. A combined comparative transcriptomic, metabolomic, and anatomical analyses of two key domestication traits: Pod dehiscence and seed dormancy in Pea (Pisum sp.). Front. Plant Sci. 2017, 8, 542. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.; Tan, D.; Baskin, J.M.; Baskin, C.C. Fruit and seed heteromorphism in the cold desert annual ephemeral Diptychocarpus strictus (Brassicaceae) and possible adaptive significance. Ann. Bot. 2010, 105, 999–1014. [Google Scholar] [CrossRef] [PubMed]
- Macquet, A.; Ralet, M.-C.; Loudet, O.; Kronenberger, J.; Mouille, G.; Marion-Poll, A.; North, H.M. A naturally occurring mutation in an Arabidopsis accession affects a β-d-Galactosidase that increases the hydrophilic potential of rhamnogalacturonan I in seed mucilage. Plant Cell 2007, 19, 3990–4006. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Viudes, S.; Dunand, C.; Burlat, V. Myxospermy Evolution in Brassicaceae: A Highly Complex and Diverse Trait with Arabidopsis as an Uncommon Model. Cells 2021, 10, 2470. https://doi.org/10.3390/cells10092470
Viudes S, Dunand C, Burlat V. Myxospermy Evolution in Brassicaceae: A Highly Complex and Diverse Trait with Arabidopsis as an Uncommon Model. Cells. 2021; 10(9):2470. https://doi.org/10.3390/cells10092470
Chicago/Turabian StyleViudes, Sébastien, Christophe Dunand, and Vincent Burlat. 2021. "Myxospermy Evolution in Brassicaceae: A Highly Complex and Diverse Trait with Arabidopsis as an Uncommon Model" Cells 10, no. 9: 2470. https://doi.org/10.3390/cells10092470
APA StyleViudes, S., Dunand, C., & Burlat, V. (2021). Myxospermy Evolution in Brassicaceae: A Highly Complex and Diverse Trait with Arabidopsis as an Uncommon Model. Cells, 10(9), 2470. https://doi.org/10.3390/cells10092470