KANPHOS: A Database of Kinase-Associated Neural Protein Phosphorylation in the Brain
Abstract
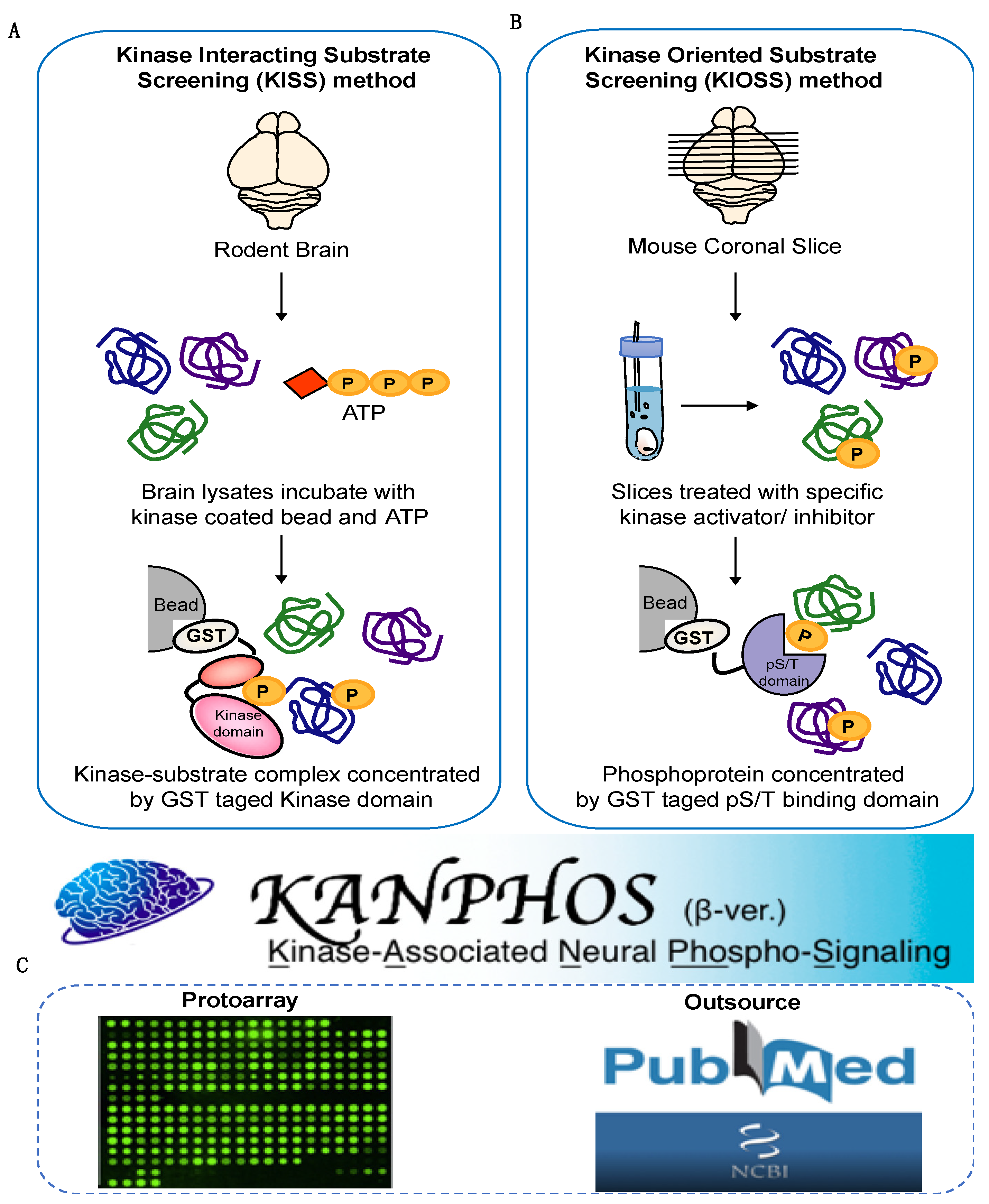
1. Introduction
2. Materials and Methods
2.1. System Configuration and Application Software
2.2. Data Collection, Management and Analysis Tool
2.3. Preparation and Incubation of Coronal Slices
2.4. Mass Spectrometry
2.5. SDS-PAGE and Immunoblotting
3. Results
3.1. Construction and Content of the KANPHOS Database
3.2. Data Accessibility and KANPHOS Workflow
3.3. Case Study: Identification of Adenosine-A2A Receptor Signaling and MAPK-Mediated Signaling Molecules Using KANPHOS
3.4. Pathway Analysis Using KANPHOS Revealed That HCN and Calcium Channel Proteins Are Involved in the MEK-MAPK Pathway Downstream of A2AR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cohen, P. The origins of protein phosphorylation. Nat. Cell. Biol. 2002, 4, E127–E130. [Google Scholar] [CrossRef] [PubMed]
- Pawson, T.; Scott, J.D. Protein phosphorylation in signaling—50 years and counting. Trends Biochem. Sci. 2005, 30, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.O.; Yu, L.; Coba, M.P.; Husi, H.; Campuzano, I.; Blackstock, W.P.; Choudhary, J.S.; Grant, S.G. Proteomic analysis of in vivo phosphorylated synaptic proteins. J. Biol. Chem. 2005, 280, 5972–5982. [Google Scholar] [CrossRef] [PubMed]
- Trinidad, J.C.; Specht, C.G.; Thalhammer, A.; Schoepfer, R.; Burlingame, A.L. Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol. Cell. Proteom. 2006, 5, 914–922. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P. The role of protein phosphorylation in human health and disease. The Sir Hans Krebs Medal Lecture. Eur. J. Biochem. 2001, 268, 5001–5010. [Google Scholar] [CrossRef]
- Manning, G.; Whyte, D.B.; Martinez, R.; Hunter, T.; Sudarsanam, S. The protein kinase complement of the human genome. Science 2002, 298, 1912–1934. [Google Scholar] [CrossRef] [PubMed]
- Nestler, E.J.; Greengard, P. Protein phosphorylation in the brain. Nature 1983, 305, 583–588. [Google Scholar] [CrossRef]
- Ghanizadeh, A.; Sahraeizadeh, A.; Berk, M. A head-to-head comparison of aripiprazole and risperidone for safety and treating autistic disorders, a randomized double blind clinical trial. Child Psychiatry Hum. Dev. 2014, 45, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Posey, D.J.; Stigler, K.A.; Erickson, C.A.; McDougle, C.J. Antipsychotics in the treatment of autism. J. Clin. Investig. 2008, 118, 6–14. [Google Scholar] [CrossRef]
- Woods, S.W. Chlorpromazine equivalent doses for the newer atypical antipsychotics. J. Clin. Psychiatry 2003, 64, 663–667. [Google Scholar] [CrossRef]
- Leucht, S.; Cipriani, A.; Spineli, L.; Mavridis, D.; Orey, D.; Richter, F.; Samara, M.; Barbui, C.; Engel, R.R.; Geddes, J.R.; et al. Comparative efficacy and tolerability of 15 antipsychotic drugs in schizophrenia: A multiple-treatments meta-analysis. Lancet 2013, 382, 951–962. [Google Scholar] [CrossRef]
- Meltzer, H.Y. The role of serotonin in antipsychotic drug action. Neuropsychopharmacology 1999, 21 (Suppl. S2), 106S–115S. [Google Scholar] [CrossRef]
- Millan, M.J. Improving the treatment of schizophrenia: Focus on serotonin (5-HT)(1A) receptors. J. Pharmacol. Exp. Ther. 2000, 295, 853–861. [Google Scholar]
- Mailman, R.B.; Murthy, V. Third generation antipsychotic drugs: Partial agonism or receptor functional selectivity? Curr. Pharm. Des. 2010, 16, 488–501. [Google Scholar] [CrossRef] [PubMed]
- UniProt, C. UniProt: A hub for protein information. Nucleic Acids Res. 2015, 43, D204–D212. [Google Scholar]
- Diella, F.; Gould, C.M.; Chica, C.; Via, A.; Gibson, T.J. Phospho.ELM: A database of phosphorylation sites—Update 2008. Nucleic Acids Res. 2008, 36, D240–D244. [Google Scholar] [CrossRef]
- Hornbeck, P.V.; Kornhauser, J.M.; Tkachev, S.; Zhang, B.; Skrzypek, E.; Murray, B.; Latham, V.; Sullivan, M. PhosphoSitePlus: A comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 2012, 40, D261–D270. [Google Scholar] [CrossRef] [PubMed]
- Gnad, F.; Gunawardena, J.; Mann, M. PHOSIDA 2011: The posttranslational modification database. Nucleic Acids Res. 2011, 39, D253–D260. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xing, X.; Ding, G.; Li, Q.; Wang, C.; Xie, L.; Zeng, R.; Li, Y. SysPTM: A systematic resource for proteomic research on post-translational modifications. Mol. Cell. Proteom. 2009, 8, 1839–1849. [Google Scholar] [CrossRef]
- Moriya, Y.; Kawano, S.; Okuda, S.; Watanabe, Y.; Matsumoto, M.; Takami, T.; Kobayashi, D.; Yamanouchi, Y.; Araki, N.; Yoshizawa, A.C.; et al. The jPOST environment: An integrated proteomics data repository and database. Nucleic Acids Res. 2019, 47, D1218–D1224. [Google Scholar] [CrossRef] [PubMed]
- Peri, S.; Navarro, J.D.; Kristiansen, T.Z.; Amanchy, R.; Surendranath, V.; Muthusamy, B.; Gandhi, T.K.; Chandrika, K.N.; Deshpande, N.; Suresh, S.; et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004, 32, D497–D501. [Google Scholar] [CrossRef] [PubMed]
- Su, M.G.; Lee, T.Y. Incorporating substrate sequence motifs and spatial amino acid composition to identify kinase-specific phosphorylation sites on protein three-dimensional structures. BMC Bioinform. 2013, 14 (Suppl. S16), S2. [Google Scholar] [PubMed]
- Linding, R.; Jensen, L.J.; Ostheimer, G.J.; van Vugt, M.A.; Jorgensen, C.; Miron, I.M.; Diella, F.; Colwill, K.; Taylor, L.; Elder, K.; et al. Systematic discovery of in vivo phosphorylation networks. Cell 2007, 129, 1415–1426. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.D.; Lee, T.Y.; Tzeng, S.W.; Wu, L.C.; Horng, J.T.; Tsou, A.P.; Huang, K.T. Incorporating hidden Markov models for identifying protein kinase-specific phosphorylation sites. J. Comput. Chem. 2005, 26, 1032–1041. [Google Scholar] [CrossRef] [PubMed]
- Amano, M.; Hamaguchi, T.; Shohag, M.H.; Kozawa, K.; Kato, K.; Zhang, X.; Yura, Y.; Matsuura, Y.; Kataoka, C.; Nishioka, T.; et al. Kinase-interacting substrate screening is a novel method to identify kinase substrates. J. Cell. Biol. 2015, 209, 895–912. [Google Scholar] [CrossRef] [PubMed]
- Nagai, T.; Nakamuta, S.; Kuroda, K.; Nakauchi, S.; Nishioka, T.; Takano, T.; Zhang, X.; Tsuboi, D.; Funahashi, Y.; Nakano, T.; et al. Phosphoproteomics of the Dopamine Pathway Enables Discovery of Rap1 Activation as a Reward Signal In Vivo. Neuron 2016, 89, 550–565. [Google Scholar] [CrossRef] [PubMed]
- Nishioka, T.; Nakayama, M.; Amano, M.; Kaibuchi, K. Proteomic screening for Rho-kinase substrates by combining kinase and phosphatase inhibitors with 14-3-3zeta affinity chromatography. Cell Struct. Funct. 2012, 37, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Funahashi, Y.; Watanabe, T.; Kaibuchi, K. Advances in defining signaling networks for the establishment of neuronal polarity. Curr. Opin. Cell Biol. 2020, 63, 76–87. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, K.; Sakai, H.; Okumura, Y.; Usui, S. Customizable neuroinformatics database system: XooNIps and its application to the pupil platform. Comput. Biol. Med. 2007, 37, 1036–1041. [Google Scholar] [CrossRef]
- Nishi, A.; Snyder, G.L.; Greengard, P. Bidirectional regulation of DARPP-32 phosphorylation by dopamine. J. Neurosci. 1997, 17, 8147–8155. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Nagai, T.; Ahammad, R.U.; Kuroda, K.; Nakamuta, S.; Nakano, T.; Yukinawa, N.; Funahashi, Y.; Yamahashi, Y.; Amano, M.; et al. Balance between dopamine and adenosine signals regulates the PKA/Rap1 pathway in striatal medium spiny neurons. Neurochem. Int. 2019, 122, 8–18. [Google Scholar] [CrossRef]
- Amano, M.; Nishioka, T.; Yura, Y.; Kaibuchi, K. Identification of Protein Kinase Substrates by the Kinase-Interacting Substrate Screening (KISS) Approach. Curr. Protoc. Cell Biol. 2016, 72, 11–12. [Google Scholar] [CrossRef] [PubMed]
- Nishioka, T.; Amano, M.; Funahashi, Y.; Tsuboi, D.; Yamahashi, Y.; Kaibuchi, K. In Vivo Identification of Protein Kinase Substrates by Kinase-Oriented Substrate Screening (KIOSS). Curr. Protoc. Chem. Biol. 2019, 11, e60. [Google Scholar] [CrossRef] [PubMed]
- Takano, T.; Wu, M.; Nakamuta, S.; Naoki, H.; Ishizawa, N.; Namba, T.; Watanabe, T.; Xu, C.; Hamaguchi, T.; Yura, Y.; et al. Discovery of long-range inhibitory signaling to ensure single axon formation. Nat. Commun. 2017, 8, 33. [Google Scholar] [CrossRef] [PubMed]
- Koike, R.; Amano, M.; Kaibuchi, K.; Ota, M. Protein kinases phosphorylate long disordered regions in intrinsically disordered proteins. Protein Sci. 2020, 29, 564–571. [Google Scholar] [CrossRef]
- Shohag, M.H.; Nishioka, T.; Ahammad, R.U.; Nakamuta, S.; Yura, Y.; Hamaguchi, T.; Kaibuchi, K.; Amano, M. Phosphoproteomic Analysis Using the WW and FHA Domains as Biological Filters. Cell Struct. Funct. 2015, 40, 95–104. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Mok, J.; Im, H.; Snyder, M. Global identification of protein kinase substrates by protein microarray analysis. Nat. Protoc. 2009, 4, 1820–1827. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Michaud, G.A.; Merkel, J.S.; Zhou, F.; Huang, J.; Mattoon, D.R.; Schweitzer, B. Protein kinase substrate identification on functional protein arrays. BMC Biotechnol. 2008, 8, 22. [Google Scholar] [CrossRef] [PubMed]
- Wardas, J. Neuroprotective role of adenosine in the CNS. Pol. J. Pharmacol. 2002, 54, 313–326. [Google Scholar] [PubMed]
- Kemp, J.M.; Powell, T.P. The structure of the caudate nucleus of the cat: Light and electron microscopy. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1971, 262, 383–401. [Google Scholar] [PubMed]
- Gerfen, C.R.; Engber, T.M.; Mahan, L.C.; Susel, Z.; Chase, T.N.; Monsma, F.J., Jr.; Sibley, D.R. D1 and D2 dopamine receptor-regulated gene expression of striatonigral and striatopallidal neurons. Science 1990, 250, 1429–1432. [Google Scholar] [CrossRef] [PubMed]
- Fink, J.S.; Weaver, D.R.; Rivkees, S.A.; Peterfreund, R.A.; Pollack, A.E.; Adler, E.M.; Reppert, S.M. Molecular cloning of the rat A2 adenosine receptor: Selective co-expression with D2 dopamine receptors in rat striatum. Brain Res. Mol. Brain Res. 1992, 14, 186–195. [Google Scholar] [CrossRef]
- Ferre, S.; O’Connor, W.T.; Svenningsson, P.; Bjorklund, L.; Lindberg, J.; Tinner, B.; Stromberg, I.; Goldstein, M.; Ogren, S.O.; Ungerstedt, U.; et al. Dopamine D1 receptor-mediated facilitation of GABAergic neurotransmission in the rat strioentopenduncular pathway and its modulation by adenosine A1 receptor-mediated mechanisms. Eur. J. Neurosci. 1996, 8, 1545–1553. [Google Scholar] [CrossRef] [PubMed]
- Kull, B.; Svenningsson, P.; Fredholm, B.B. Adenosine A(2A) receptors are colocalized with and activate g(olf) in rat striatum. Mol. Pharmacol. 2000, 58, 771–777. [Google Scholar] [CrossRef] [PubMed]
- McAvoy, T.; Zhou, M.M.; Greengard, P.; Nairn, A.C. Phosphorylation of Rap1GAP, a striatally enriched protein, by protein kinase A controls Rap1 activity and dendritic spine morphology. Proc. Natl. Acad. Sci. USA 2009, 106, 3531–3536. [Google Scholar] [CrossRef] [PubMed]
- Snyder, G.L.; Allen, P.B.; Fienberg, A.A.; Valle, C.G.; Huganir, R.L.; Nairn, A.C.; Greengard, P. Regulation of phosphorylation of the GluR1 AMPA receptor in the neostriatum by dopamine and psychostimulants in vivo. J. Neurosci. 2000, 20, 4480–4488. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.H.; Yamahashi, Y.; Kuroda, K.; Faruk, M.O.; Zhang, X.; Yamada, K.; Yamanaka, A.; Nagai, T.; Kaibuchi, K. Accumbal D2R-medium spiny neurons regulate aversive behaviors through PKA-Rap1 pathway. Neurochem. Int. 2021, 143, 104935. [Google Scholar] [CrossRef] [PubMed]
- Poolos, N.P.; Bullis, J.B.; Roth, M.K. Modulation of h-channels in hippocampal pyramidal neurons by p38 mitogen-activated protein kinase. J. Neurosci. 2006, 26, 7995–8003. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Bullis, J.B.; Lau, I.H.; Jones, T.D.; Warner, L.N.; Poolos, N.P. Downregulation of dendritic HCN channel gating in epilepsy is mediated by altered phosphorylation signaling. J. Neurosci. 2010, 30, 6678–6688. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, E.M. Regulation of voltage-dependent calcium channels in rat sensory neurones involves a Ras-mitogen-activated protein kinase pathway. J. Physiol. 2000, 527, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Cole, A.R.; Knebel, A.; Morrice, N.A.; Robertson, L.A.; Irving, A.J.; Connolly, C.N.; Sutherland, C. GSK-3 phosphorylation of the Alzheimer epitope within collapsin response mediator proteins regulates axon elongation in primary neurons. J. Biol. Chem. 2004, 279, 50176–50180. [Google Scholar] [CrossRef]
- Uchida, Y.; Ohshima, T.; Sasaki, Y.; Suzuki, H.; Yanai, S.; Yamashita, N.; Nakamura, F.; Takei, K.; Ihara, Y.; Mikoshiba, K.; et al. Semaphorin3A signalling is mediated via sequential Cdk5 and GSK3beta phosphorylation of CRMP2: Implication of common phosphorylating mechanism underlying axon guidance and Alzheimer’s disease. Genes Cells 2005, 10, 165–179. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, T.; Kawano, Y.; Arimura, N.; Kawabata, S.; Kikuchi, A.; Kaibuchi, K. GSK-3beta regulates phosphorylation of CRMP-2 and neuronal polarity. Cell 2005, 120, 137–149. [Google Scholar] [CrossRef]
- Cole, A.R.; Noble, W.; van Aalten, L.; Plattner, F.; Meimaridou, R.; Hogan, D.; Taylor, M.; LaFrancois, J.; Gunn-Moore, F.; Verkhratsky, A.; et al. Collapsin response mediator protein-2 hyperphosphorylation is an early event in Alzheimer’s disease progression. J. Neurochem. 2007, 103, 1132–1144. [Google Scholar] [CrossRef]
- Castillon, C.; Gonzalez, L.; Domenichini, F.; Guyon, S.; Da Silva, K.; Durand, C.; Lestaevel, P.; Vaillend, C.; Laroche, S.; Barnier, J.V.; et al. The intellectual disability PAK3 R67C mutation impacts cognitive functions and adult hippocampal neurogenesis. Hum. Mol. Genet. 2020, 29, 1950–1968. [Google Scholar] [CrossRef] [PubMed]
- Pavlowsky, A.; Chelly, J.; Billuart, P. Emerging major synaptic signaling pathways involved in intellectual disability. Mol. Psychiatry 2012, 17, 682–693. [Google Scholar] [CrossRef] [PubMed]
- Weiwer, M.; Xu, Q.; Gale, J.P.; Lewis, M.; Campbell, A.J.; Schroeder, F.A.; Van de Bittner, G.C.; Walk, M.; Amaya, A.; Su, P.; et al. Functionally Biased D2R Antagonists: Targeting the beta-Arrestin Pathway to Improve Antipsychotic Treatment. ACS Chem. Biol. 2018, 13, 1038–1047. [Google Scholar] [CrossRef] [PubMed]
- Nakazawa, K.; Jeevakumar, V.; Nakao, K. Spatial and temporal boundaries of NMDA receptor hypofunction leading to schizophrenia. NPJ Schizophr. 2017, 3, 7. [Google Scholar] [CrossRef] [PubMed]






| Method/Stimulation | Kinase | No. of Phos. Site |
|---|---|---|
| KIOSS (1433z)/Forskolin | PKA, MAPK and other | 278 |
| KIOSS (1433z)/OA&U0126 | MAPK and other | 324 |
| KIOSS (1433z)/D1R Agonist | PKA, MAPK and other | 185 |
| KIOSS (1433z)/A2AR Agonist | PKA, MAPK and other | 62 |
| KISS | CaMKI, CDK5, FYN, LYN, MAPK1, PAK7, PKA, PKN, ROCK2 | 3597 |
| Protoarray | AKT1, CaMK2A, CDK5, LRRK2, LYN, MAPK1, PRKCA, PRKACA, ROCK2 | 1782 |
| Outsource | AurA, AurB, CaMKI, CaMKII, CaMK4, CDK5, GSK3B, LYN, MAPK, MARK1, PKA, AMPK, PRKCA, PRKCE, ROCK, STK11(LKB1) | 3736 |
| Motif | Identified Phosphoprotein and Phosphorylation Site | |||||
|---|---|---|---|---|---|---|
| PKA | Arhgap21 | Arhgap21 | Arhgap21 | Arhgap23 | Brsk1 | Brsk2 |
| S472 | S851 | S875 | S401 | S324 | S383 | |
| Bsn | Bsn | Cacna1b | Ccdc177 | Ccny | Cdk17 | |
| S1987 | S2860 | S2219 | S303 | S301 | S180 | |
| Chrna4 | Fam126b | Gpr75 | Hcn2 | Kcnh7 | Kif21b | |
| S540 | S521 | S317 | S840 | S174 | S1150 | |
| Madd | Mark1 | Nedd4l | Nf1 | Prickle2 | Prrt3 | |
| S1038 | S394 | T353 | S2524 | S752 | S846 | |
| Rap1gap | Rims1 | Sik3 | Syn1 | Taf4b | Ttc32 | |
| S245 | S887 | S454 | S427 | S707 | S24 | |
| MAPK | Ablim1 | Arhgap21 | Bsn | Cacna1e | Ccdc177 | Cdk17 |
| S89 | S42 | Y3020 | S791 | S308 | T11 | |
| Chrna4 | Chrna4 | Hcn3 | Lrfn4 | Madd | Madd | |
| S540 | S543 | S633 | S627 | S1038 | T1045 | |
| Nedd4l | Prrt3 | Rap1gap | Sipa1l1 | Sorbs2 | Spata2 | |
| S329 | S834 | S213 | S1528 | S944 | S247 | |
| Syn1 | ||||||
| S427 | ||||||
| Others | Arhgap21 | Atp6v1h | Cep170 | Gpr75 | Kcnh2 | Khnyn |
| S856 | Y125 | S353 | T313 | S322 | S43 | |
| Madd | Mapk8 | Rsl1d1 | Rsl1d1 | Sh2d5 | Ttc32 | |
| S1089 | S210 | T314 | S316 | S126 | T18 | |
| Identified MAPK Candidate Substrates List | |||||||
|---|---|---|---|---|---|---|---|
| 2010300C02Rik | Atg9a | Cdk18 | Epb4.1l3 | Lppr3 | Osbpl6 | Rasgrp2 | Ssh2 |
| Aak1 | Bai1 | Cdkl5 | Epb49 | Lrfn4 | Pak7 | Rem2 | Stim1 |
| Abi1 | Baiap2 | Cep170 | Erc2 | Lrrc7 | Panx2 | Rims1 | Syn1 |
| Abi2 | Bcr | Chrna4 | Etl4 | Madd | Pclo | Rims2 | Syn2 |
| Agap2 | Begain | Clasp2 | Fam171a1 | Map3k5 | Pfkfb2 | Rph3a | Syn3 |
| Agfg1 | Brsk1 | Crtc2 | Fam171a2 | Map4 | Pip5k1c | Rrad | Synpo |
| Akap6 | Brsk2 | Ctnnd2 | Fam171b | Mark1 | Pitpnm3 | Sh2d5 | Syt7 |
| Als2 | Bsn | Cyth3 | Frmd4a | Mark3 | Plekha5 | Shank3 | Tanc2 |
| Ampd2 | C17orf59 | Dab2ip | Gab2 | Mast1 | Plekho2 | Shisa7 | Tpd52 |
| Ank2 | C2cd4c | Dennd1a | Git1 | Mast3 | Pola1 | Sipa1l1 | Tpd52l1 |
| Ankrd34a | Cacna1b | Dennd4c | Gm1568 | Mbp | Ppfia2 | Sipa1l2 | Tsc2 |
| Ankrd34b | Cacna1e | Dgkq | Gm15800 | Mink1 | Ppfia3 | Slc4a4 | Uhrf1bp1l |
| Anks1b | Camk2g | Dlgap2 | Gpr158 | Mllt4 | Ppfia4 | Sorbs1 | Ulk1 |
| Arhgap21 | Camkk1 | Dlgap3 | Hcn2 | Mprip | Prrt3 | Sorbs2 | Ulk2 |
| Arhgap23 | Camkk2 | Dos | Hcn3 | Nav1 | Psd | Spata2 | Usp31 |
| Arhgap32 | Camsap2 | Dpysl2 | Iqsec1 | Nav3 | Psd3 | Specc1 | Usp8 |
| Arhgap39 | Caskin1 | Dpysl5 | Kcnb1 | Ndel1 | Rab11fip2 | Speg | Wdr47 |
| Arhgef2 | Ccdc22 | Dstn | Kiaa0284 | Nelf | Ralgapa1 | Srcin1 | |
| Arhgef6 | Ccny | Dtna | Kiaa0528 | Nhsl2 | Rap1gap | Srgap3 | |
| Atat1 | Ccnyl1 | Eif3d | Kiaa1211 | Nyap2 | Rapgef2 | Ssfa2 | |
| Total 157 | |||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahammad, R.U.; Nishioka, T.; Yoshimoto, J.; Kannon, T.; Amano, M.; Funahashi, Y.; Tsuboi, D.; Faruk, M.O.; Yamahashi, Y.; Yamada, K.; et al. KANPHOS: A Database of Kinase-Associated Neural Protein Phosphorylation in the Brain. Cells 2022, 11, 47. https://doi.org/10.3390/cells11010047
Ahammad RU, Nishioka T, Yoshimoto J, Kannon T, Amano M, Funahashi Y, Tsuboi D, Faruk MO, Yamahashi Y, Yamada K, et al. KANPHOS: A Database of Kinase-Associated Neural Protein Phosphorylation in the Brain. Cells. 2022; 11(1):47. https://doi.org/10.3390/cells11010047
Chicago/Turabian StyleAhammad, Rijwan Uddin, Tomoki Nishioka, Junichiro Yoshimoto, Takayuki Kannon, Mutsuki Amano, Yasuhiro Funahashi, Daisuke Tsuboi, Md. Omar Faruk, Yukie Yamahashi, Kiyofumi Yamada, and et al. 2022. "KANPHOS: A Database of Kinase-Associated Neural Protein Phosphorylation in the Brain" Cells 11, no. 1: 47. https://doi.org/10.3390/cells11010047
APA StyleAhammad, R. U., Nishioka, T., Yoshimoto, J., Kannon, T., Amano, M., Funahashi, Y., Tsuboi, D., Faruk, M. O., Yamahashi, Y., Yamada, K., Nagai, T., & Kaibuchi, K. (2022). KANPHOS: A Database of Kinase-Associated Neural Protein Phosphorylation in the Brain. Cells, 11(1), 47. https://doi.org/10.3390/cells11010047






