miR-511 Deficiency Protects Mice from Experimental Colitis by Reducing TLR3 and TLR4 Responses via WD Repeat and FYVE-Domain-Containing Protein 1
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
2.2. DSS-Induced Colitis
2.3. Histology
2.4. Fluorescence Activated Cell Sorting of Colonic Lamina Propria Cells
2.5. BMDM Culture and Stimulations
2.6. mRNA, cDNA and qPCR Analysis
2.7. Cytokine Measurements
2.8. RNA Sequencing of BMDMs
2.9. Westernblot
2.10. Immunohistochemistry
2.11. Statistical Analysis
3. Results
3.1. miR-511 Deficiency Affects Macrophage Responses to Microbial Stimuli
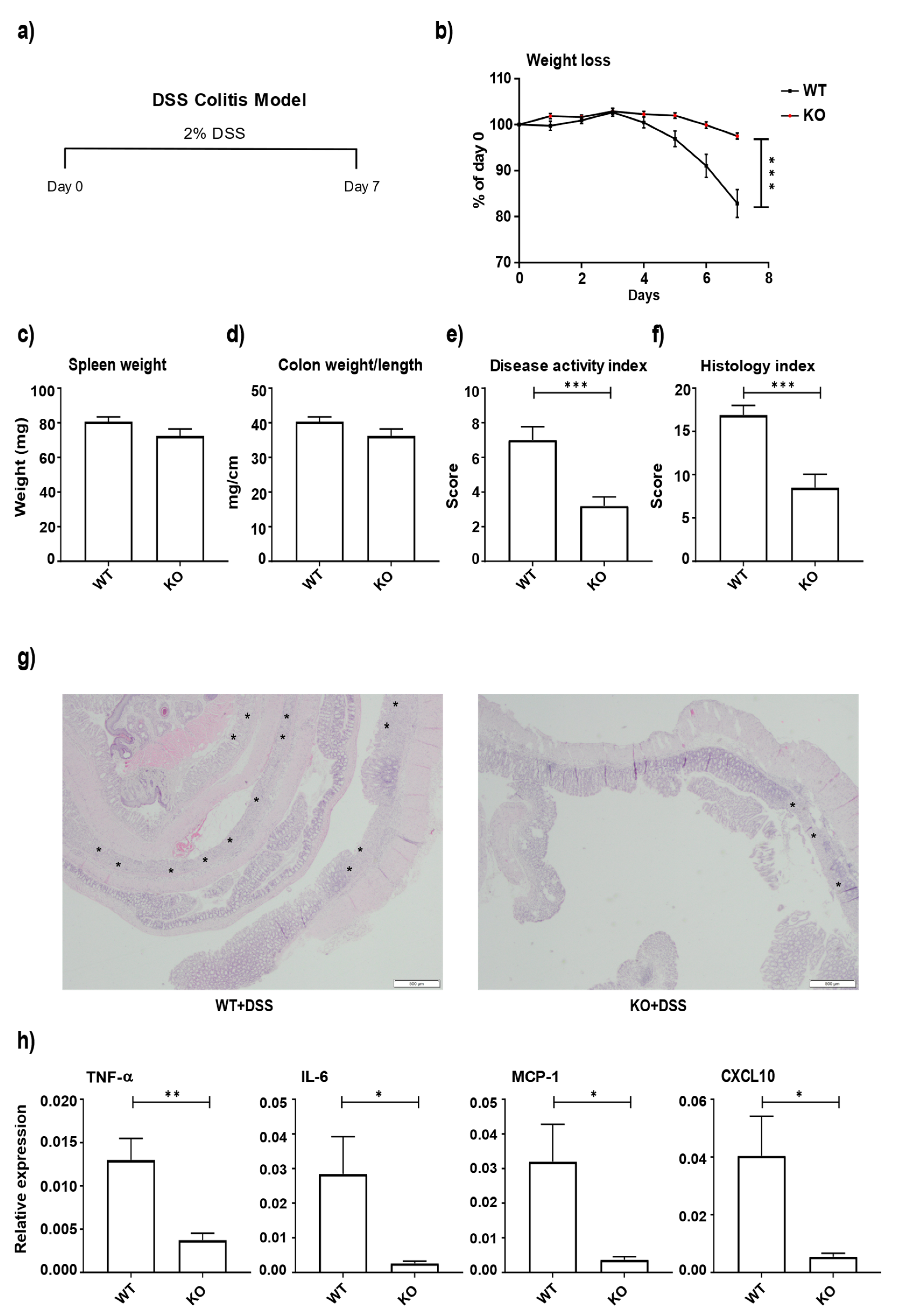
3.2. miR-511-Deficient Mice Are Protected against DSS-Induced Colitis
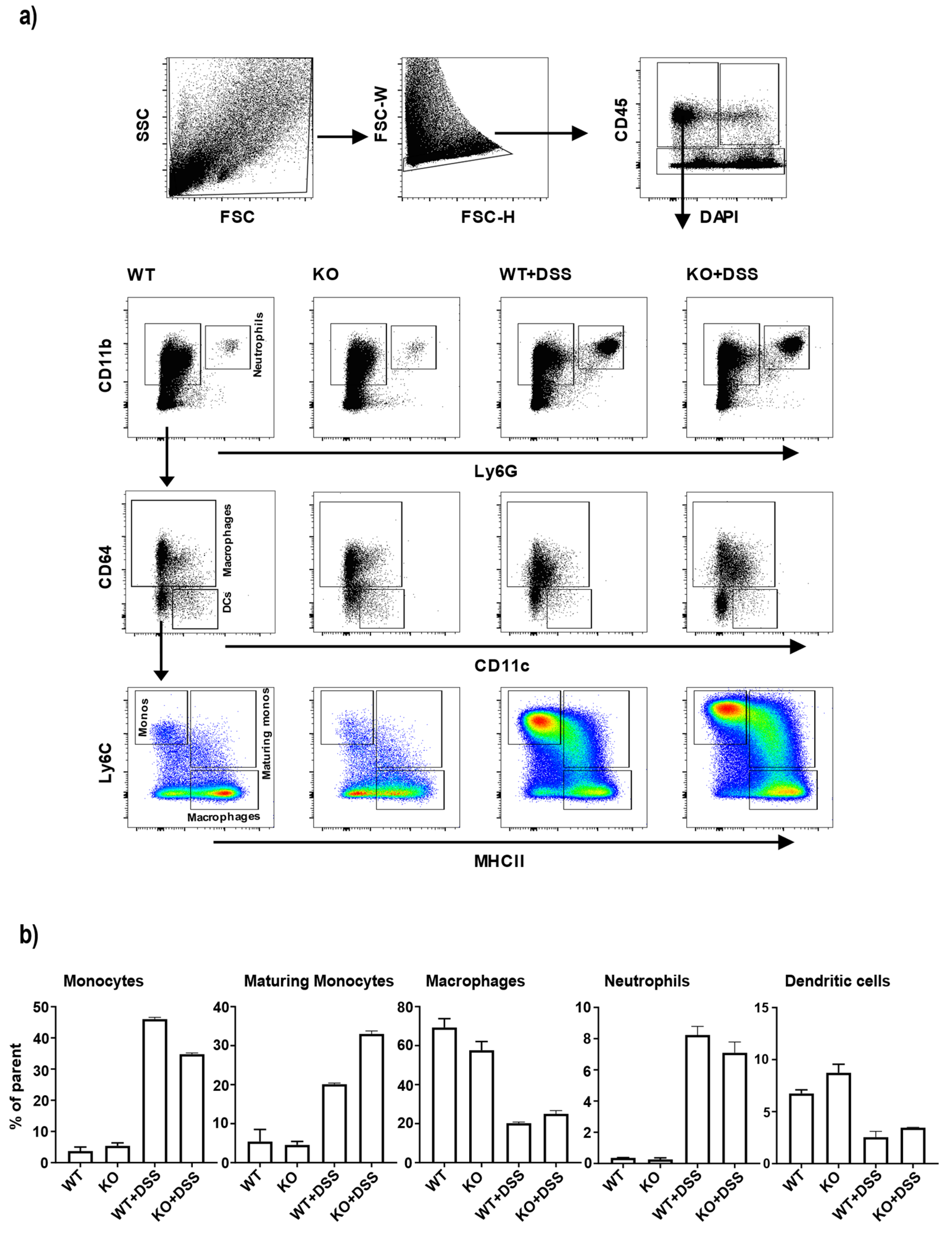
3.3. Changes in Colonic Monocyte and Maturing Monocyte Populations in miR-511-Deficient Mice in Inflamed Condition
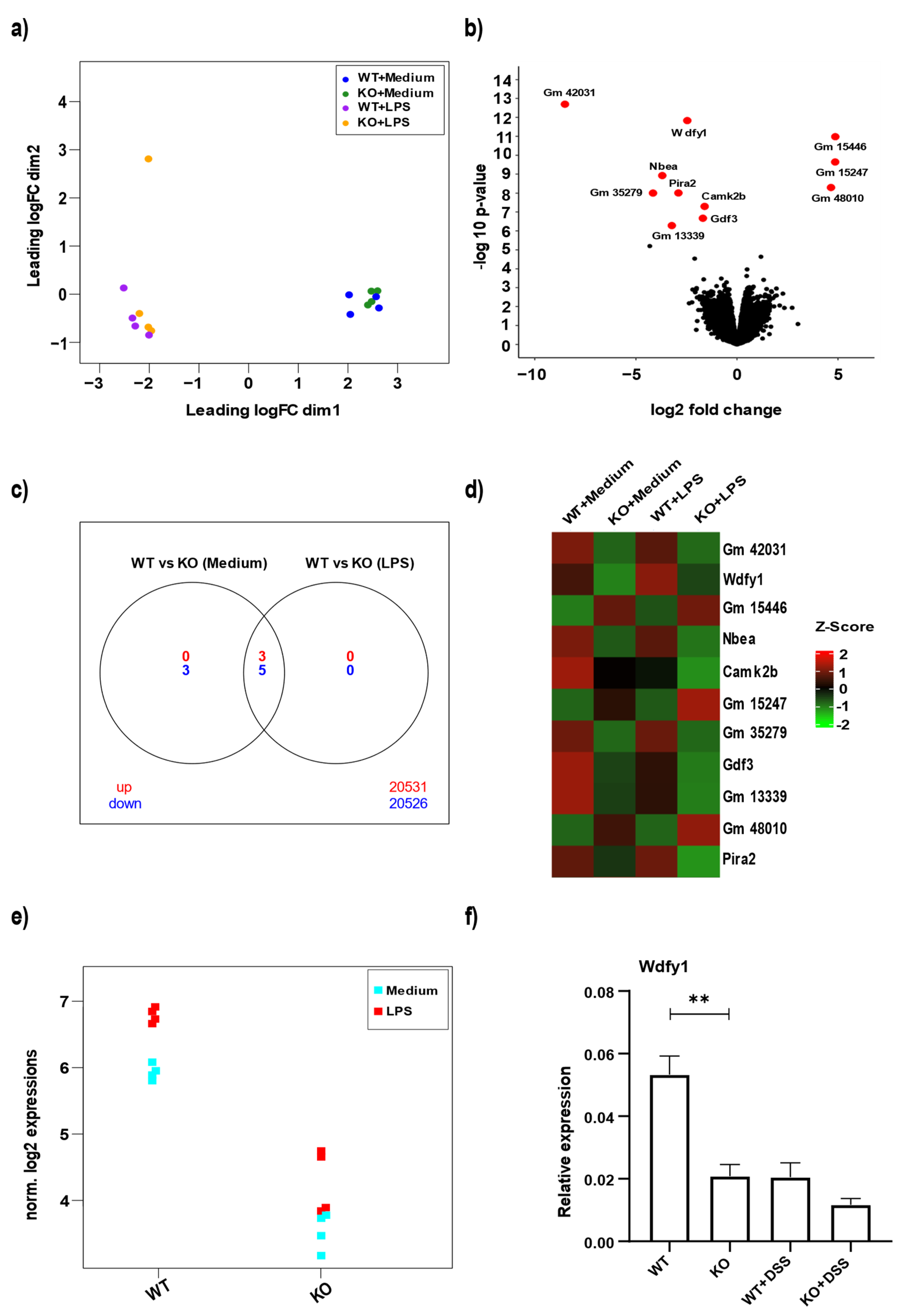
3.4. miR-511 Deficiency Affects Wdfy1
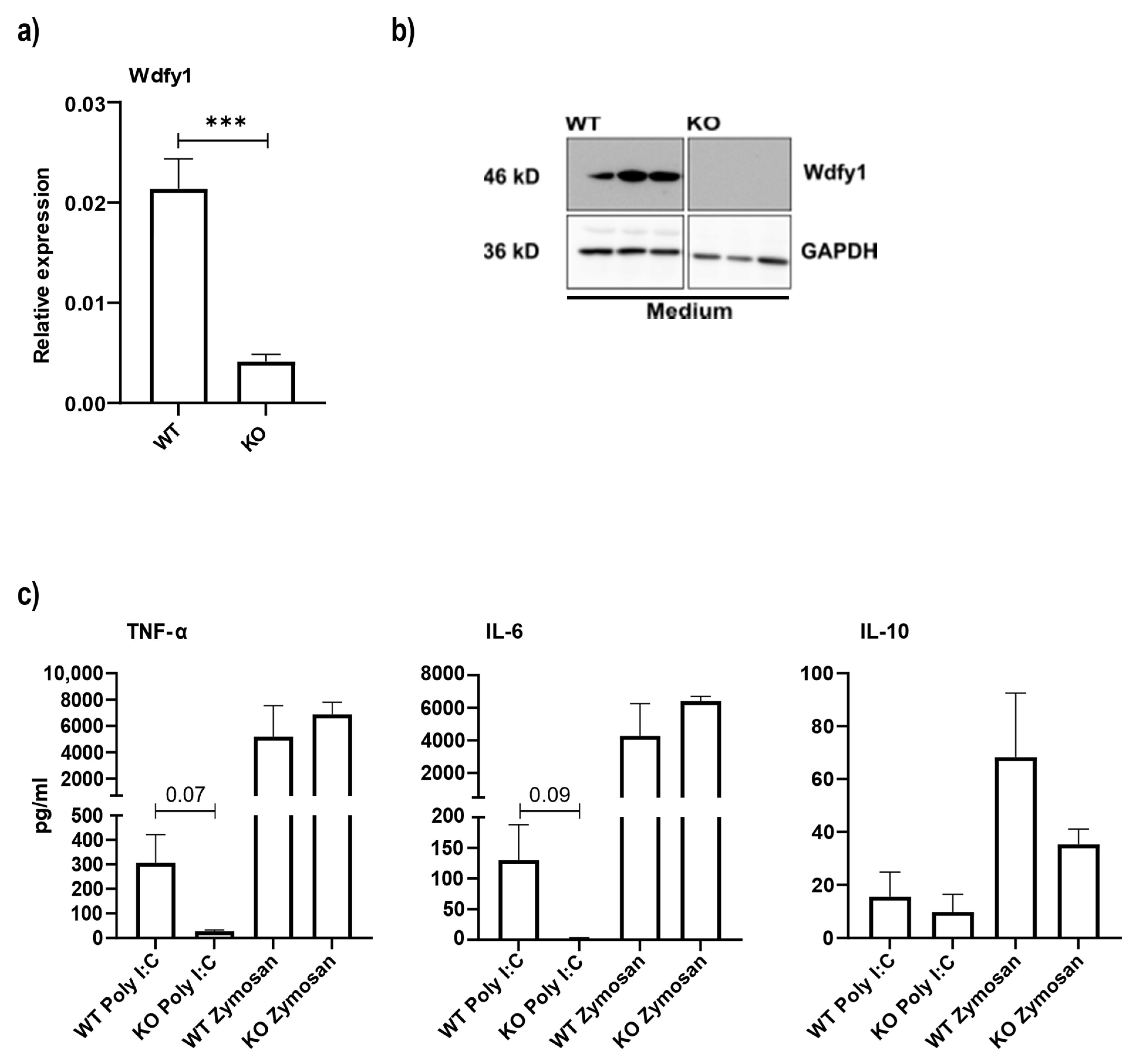
3.5. Macrophages from miR-511-Deficient Mice Have Reduced Wdfy1 Expression, Which Affects Antimicrobial Responses
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bentwich, I.; Avniel, A.; Karov, Y.; Aharonov, R.; Gilad, S.; Barad, O.; Barzilai, A.; Einat, P.; Einav, U.; Meiri, E.; et al. Identification of hundreds of conserved and nonconserved human microRNAs. Nat. Genet. 2005, 37, 766–770. [Google Scholar] [CrossRef]
- Lim, H.J.; Yang, J.L. Regulatory roles and therapeutic potential of microRNA in sarcoma. Crit. Rev. Oncol. Hematol. 2016, 97, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Feinberg, M.W.; Moore, K.J. MicroRNA Regulation of Atherosclerosis. Circ. Res. 2016, 118, 703–720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, E.J.; Shimaoka, M.; Kiyono, H. MicroRNA-mediated dynamic control of mucosal immunity. Int. Immunol. 2017, 29, 157–163. [Google Scholar] [CrossRef]
- Tserel, L.; Runnel, T.; Kisand, K.; Pihlap, M.; Bakhoff, L.; Kolde, R.; Peterson, H.; Vilo, J.; Peterson, P.; Rebane, A. MicroRNA expression profiles of human blood monocyte-derived dendritic cells and macrophages reveal miR-511 as putative positive regulator of Toll-like receptor 4. J. Biol. Chem. 2011, 286, 26487–26495. [Google Scholar] [CrossRef] [Green Version]
- Squadrito, M.L.; Pucci, F.; Magri, L.; Moi, D.; Gilfillan, G.D.; Ranghetti, A.; Casazza, A.; Mazzone, M.; Lyle, R.; Naldini, L.; et al. miR-511-3p Modulates Genetic Programs of Tumor-Associated Macrophages. Cell Rep. 2012, 1, 141–154. [Google Scholar] [CrossRef] [Green Version]
- Gordon, S.; Plüddemann, A.; Mukhopadhyay, S. Plasma membrane receptors of tissue macrophages: Functions and role in pathology. J. Pathol. 2020, 250, 656–666. [Google Scholar] [CrossRef]
- Seyedizade, S.S.; Afshari, K.; Bayat, S.; Rahmani, F.; Momtaz, S.; Rezaei, N.; Abdolghaffari, A.H. Current Status of M1 and M2 Macrophages Pathway as Drug Targets for Inflammatory Bowel Disease. Arch. Immunol. Ther. Exp. 2020, 68, 10. [Google Scholar] [CrossRef]
- Zhao, Y.; Pang, W.; Yang, N.; Hao, L.; Wang, L. MicroRNA-511 inhibits malignant behaviors of breast cancer by directly targeting SOX9 and regulating the PI3K/Akt pathway. Int. J. Oncol. 2018, 53, 2715–2726. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.H.; Pang, M.; Dong, W.; Xin, J.X.; Li, Y.J.; Zhang, Z.C.; Yu, L.; Wang, P.Y.; Li, B.S.; Xie, S.Y. miR-511 induces the apoptosis of radioresistant lung adenocarcinoma cells by triggering BAX. Oncol. Rep. 2014, 31, 1473–1479. [Google Scholar] [CrossRef]
- He, S.; Wang, G.; Ni, J.; Zhuang, J.; Zhuang, S.; Wang, G.; Ye, Y.; Xia, W. MicroRNA-511 inhibits cellular proliferation and invasion in colorectal cancer by directly targeting hepatoma-derived growth factor. Oncol. Res. 2018, 26, 1355–1363. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, C.; Chen, L. MiR-511 mimic transfection inhibits the proliferation, invasion of osteosarcoma cells and reduces metastatic osteosarcoma tumor burden in nude mice via targeting MAPK1. Cancer Biomark. 2019, 26, 343–351. [Google Scholar] [CrossRef] [Green Version]
- Zheng, D.; Sabbagh, J.J.; Blair, L.J.; Darling, A.L.; Wen, X.; Dickey, C.A. MicroRNA-511 Binds to FKBP5 mRNA, Which Encodes a Chaperone Protein, and Regulates Neuronal Differentiation. J. Biol. Chem. 2016, 291, 17897–17906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.; Do, D.C.; Ishmael, F.T.; Squadrito, M.L.; Tang, H.M.; Tang, H.L.; Hsu, M.H.; Qiu, L.; Li, C.; Zhang, Y.; et al. Mannose receptor modulates macrophage polarization and allergic inflammation through miR-511-3p. J. Allergy Clin. Immunol. 2018, 141, 350–364.e358. [Google Scholar] [CrossRef] [Green Version]
- Puimège, L.; Van Hauwermeiren, F.; Steeland, S.; Van Ryckeghem, S.; Vandewalle, J.; Lodens, S.; Dejager, L.; Vandevyver, S.; Staelens, J.; Timmermans, S.; et al. Glucocorticoid-induced microRNA-511 protects against TNF by down-regulating TNFR1. EMBO Mol. Med. 2015, 7, 1004–1017. [Google Scholar] [CrossRef]
- Heinsbroek, S.E.M.; Squadrito, M.L.; Schilderink, R.; Hilbers, F.W.; Verseijden, C.; Hofmann, M.; Helmke, A.; Boon, L.; Wildenberg, M.E.; Roelofs, J.J.T.H.; et al. miR-511-3p, embedded in the macrophage mannose receptor gene, contributes to intestinal inflammation. Mucosal Immunol. 2015, 9, 960–973. [Google Scholar] [CrossRef]
- Curtale, G.; Renzi, T.A.; Drufuca, L.; Rubino, M.; Locati, M. Glucocorticoids downregulate TLR4 signaling activity via its direct targeting by miR-511-5p. Eur. J. Immunol. 2017, 47, 2080–2089. [Google Scholar] [CrossRef] [PubMed]
- Randhawa, P.K.; Singh, K.; Singh, N.; Jaggi, A.S. A Review on Chemical-Induced Inflammatory Bowel Disease Models in Rodents. Korean J. Physiol. Pharmacol. 2014, 18, 279–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wirtz, S.; Neurath, M.F. Mouse models of inflammatory bowel disease. Adv. Drug Deliv. Rev. 2007, 59, 1073–1083. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.H.; Zhang, Y.; Jiang, L.Q.; Wang, S.; Lei, C.Q.; Sun, M.S.; Shu, H.B.; Liu, Y. WDFY1 mediates TLR3/4 signaling by recruiting TRIF. EMBO Rep. 2015, 16, 447–455. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Hu, Y.H.; Liu, Y. Wdfy1 deficiency impairs Tlr3-mediated immune responses in vivo. Cell. Mol. Immunol. 2020, 17, 1014–1016. [Google Scholar] [CrossRef]
- Yeo, I.J.; Park, M.H.; Son, D.J.; Kim, J.Y.; Nam, K.T.; Hyun, B.K.; Kim, S.Y.; Jung, M.H.; Song, M.J.; Chun, H.O.; et al. PRDX6 Inhibits Neurogenesis through Downregulation of WDFY1-Mediated TLR4 Signal. Mol. Neurobiol. 2019, 56, 3132–3144. [Google Scholar] [CrossRef] [Green Version]
- Mielcarska, M.B.; Bossowska-Nowicka, M.; Toka, F.N. Functional failure of TLR3 and its signaling components contribute to herpes simplex encephalitis. J. Neuroimmunol. 2018, 316, 65–73. [Google Scholar] [CrossRef] [Green Version]
- Golshiri-Isfahani, A.; Amizadeh, M.; Arababadi, M.K. The roles of toll like receptor 3, 7 and 8 in allergic rhinitis pathogenesis. Allergol. Immunopathol. 2018, 46, 503–507. [Google Scholar] [CrossRef]
- Cario, E. Toll-like receptors in inflammatory bowel diseases: A decade later. Inflamm. Bowel Dis. 2010, 16, 1583–1597. [Google Scholar] [CrossRef] [PubMed]
- Meena, N.K.; Verma, R.; Verma, N.; Ahuja, V.; Paul, J. TLR4 D299G polymorphism modulates cytokine expression in ulcerative colitis. J. Clin. Gastroenterol. 2013, 47, 773–780. [Google Scholar] [CrossRef]
- Cooper, H.S.; Murthy, S.N.; Shah, R.S.; Sedergran, D.J. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab. Investig. A J. Tech. Methods Pathol. 1993, 69, 238–249. [Google Scholar]
- Ten Hove, T.; Drillenburg, P.; Wijnholds, J.; te Velde, A.A.; van Deventer, S.J.H. Differential Susceptibility of Multidrug Resistance Protein-1 Deficient Mice to DSS and TNBS-Induced Colitis. Dig. Dis. Sci. 2002, 47, 2056–2063. [Google Scholar] [CrossRef]
- Heinsbroek, S.E.; Oei, A.; Roelofs, J.J.; Dhawan, S.; te Velde, A.; Gordon, S.; de Jonge, W.J. Genetic deletion of dectin-1 does not affect the course of murine experimental colitis. BMC Gastroenterol. 2012, 12, 33. [Google Scholar] [CrossRef] [PubMed]
- Heinsbroek, S.E.M.; Williams, D.L.; Welting, O.; Meijer, S.L.; Gordon, S.; de Jonge, W.J. Orally delivered β-glucans aggravate dextran sulfate sodium (DSS)-induced intestinal inflammation. Nutr. Res. 2015, 35, 1106–1112. [Google Scholar] [CrossRef] [Green Version]
- Rahman, S.; Davids, M.; van Hamersveld, P.H.P.; Welting, O.; Rahaoui, H.; Schuren, F.; Meijer, S.L.; van den Wijngaard, R.M.; Hakvoort, T.B.M.; de Jonge, W.J.; et al. Dietary Curdlan Enhances Bifidobacteria and Reduces Intestinal Inflammation in Mice. Nutrients 2021, 13, 1305. [Google Scholar] [CrossRef]
- Ruijter, J.M.; Ramakers, C.; Hoogaars, W.M.; Karlen, Y.; Bakker, O.; van den Hoff, M.J.; Moorman, A.F. Amplification efficiency: Linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009, 37, e45. [Google Scholar] [CrossRef] [Green Version]
- Zerbino, D.R.; Achuthan, P.; Akanni, W.; Amode, M.R.; Barrell, D.; Bhai, J.; Billis, K.; Cummins, C.; Gall, A.; Girón, C.G.; et al. Ensembl 2018. Nucleic Acids Res. 2018, 46, D754–D761. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; Oshlack, A. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 2010, 11, R25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolde, R. Package ’pheatmap’. Available online: https://mran.microsoft.com/snapshot/2018-08-31/web/packages/pheatmap/pheatmap.pdf (accessed on 20 May 2018).
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [Green Version]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [Green Version]
- Wu, D.; Smyth, G.K. Camera: A competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 2012, 40, e133. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013. [Google Scholar]
- Huber, W.; Carey, V.J.; Gentleman, R.; Anders, S.; Carlson, M.; Carvalho, B.S.; Bravo, H.C.; Davis, S.; Gatto, L.; Girke, T.; et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nat. Methods 2015, 12, 115–121. [Google Scholar] [CrossRef]
- Bain, C.C.; Scott, C.L.; Uronen-Hansson, H.; Gudjonsson, S.; Jansson, O.; Grip, O.; Guilliams, M.; Malissen, B.; Agace, W.W.; Mowat, A.M. Resident and pro-inflammatory macrophages in the colon represent alternative context-dependent fates of the same Ly6Chi monocyte precursors. Mucosal Immunol. 2013, 6, 498–510. [Google Scholar] [CrossRef] [PubMed]
- Cerovic, V.; Bain, C.C.; Mowat, A.M.; Milling, S.W. Intestinal macrophages and dendritic cells: What’s the difference? Trends Immunol. 2014, 35, 270–277. [Google Scholar] [CrossRef]
- Ridley, S.H.; Ktistakis, N.; Davidson, K.; Anderson, K.E.; Manifava, M.; Ellson, C.D.; Lipp, P.; Bootman, M.; Coadwell, J.; Nazarian, A.; et al. FENS-1 and DFCP1 are FYVE domain-containing proteins with distinct functions in the endosomal and Golgi compartments. J. Cell Sci. 2001, 114, 3991–4000. [Google Scholar] [CrossRef]
- Gantner, B.N.; Simmons, R.M.; Canavera, S.J.; Akira, S.; Underhill, D.M. Collaborative induction of inflammatory responses by dectin-1 and Toll-like receptor 2. J. Exp. Med. 2003, 197, 1107–1117. [Google Scholar] [CrossRef] [Green Version]
- Frasnelli, M.E.; Tarussio, D.; Chobaz-Péclat, V.; Busso, N.; So, A. TLR2 modulates inflammation in zymosan-induced arthritis in mice. Arthritis Res. Ther. 2005, 7, R370–R379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serbina, N.V.; Jia, T.; Hohl, T.M.; Pamer, E.G. Monocyte-mediated defense against microbial pathogens. Annu. Rev. Immunol. 2008, 26, 421–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desalegn, G.; Pabst, O. Inflammation triggers immediate rather than progressive changes in monocyte differentiation in the small intestine. Nat. Commun. 2019, 10, 3229. [Google Scholar] [CrossRef]
- Stanton, M.J.; Dutta, S.; Polavaram, N.S.; Roy, S.; Muders, M.H.; Datta, K. Angiogenic growth factor axis in autophagy regulation. Autophagy 2013, 9, 789–790. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dutta, S.; Roy, S.; Polavaram, N.S.; Baretton, G.B.; Muders, M.H.; Batra, S.; Datta, K. NRP2 transcriptionally regulates its downstream effector WDFY1. Sci. Rep. 2016, 6, 23588. [Google Scholar] [CrossRef] [Green Version]
- Arisi, I.; D‘Onofrio, M.; Brandi, R.; Felsani, A.; Capsoni, S.; Drovandi, G.; Felici, G.; Weitschek, E.; Bertolazzi, P.; Cattaneo, A. Gene expression biomarkers in the brain of a mouse model for Alzheimer‘s disease: Mining of microarray data by logic classification and feature selection. J. Alzheimers Dis. 2011, 24, 721–738. [Google Scholar] [CrossRef] [Green Version]
- O‘Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [Green Version]

| Gene | 5′-Forward Sequence | 5′-Reverse Sequence |
|---|---|---|
| GAPDH | ATGTGTCCGTCGTGGATCTGA | ATGCCTGCTTCACCACCTTCT |
| Cyclophilin | ATGGTCAACCCCACCGTGT | TTCTGCTGTCTTTGGAACTTTGTC |
| EEF2 | TGTCAGTCATCGCCCATGTG | CATCCTTGCGAGTGTCAGTGA |
| RPLP0 | CCAGCGAGGCCACACTGCTG | ACACTGGCCACGTTGCGGAC |
| TNF-α | AAAGCATGATCCGCGACGT | TGCAAGCAGGAATGAGAA |
| IL-6 | GAGTTGTGCAATGGCAATTCTG | TGGTAGCATCCATCATTTCTTTGT |
| IL-10 | TGTCAAATTCATTCATGGCCT | ATCGATTTCTCCCCTGTGAA |
| CXCL10 | CCAAGTGCTGCCGTCATTTTC | TCCCTATGGCCCTCATTCTCA |
| MCP-1 | AGGCTGGAGAGCTACAAGAGGAT | TCTCATTTGGTTCCGATCCAGG |
| TLR4 | TGTCATCAGGGACTTTGCTG | TGTTCTTCTCCTGCCTGACA |
| CD206 | TGTGGTGAGCTGAAAGGTGA | CAGGTGTGGGCTCAGGTAGT |
| Wdfy1 | AGAGTGCAGTCACTGTGCTACC | CTGCTCACACTTCTGACAGGAG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, S.; Vandewalle, J.; van Hamersveld, P.H.P.; Verseijden, C.; Welting, O.; Jongejan, A.; Casanova, P.; Meijer, S.L.; Libert, C.; Hakvoort, T.B.M.; et al. miR-511 Deficiency Protects Mice from Experimental Colitis by Reducing TLR3 and TLR4 Responses via WD Repeat and FYVE-Domain-Containing Protein 1. Cells 2022, 11, 58. https://doi.org/10.3390/cells11010058
Rahman S, Vandewalle J, van Hamersveld PHP, Verseijden C, Welting O, Jongejan A, Casanova P, Meijer SL, Libert C, Hakvoort TBM, et al. miR-511 Deficiency Protects Mice from Experimental Colitis by Reducing TLR3 and TLR4 Responses via WD Repeat and FYVE-Domain-Containing Protein 1. Cells. 2022; 11(1):58. https://doi.org/10.3390/cells11010058
Chicago/Turabian StyleRahman, Shafaque, Jolien Vandewalle, Patricia H. P. van Hamersveld, Caroline Verseijden, Olaf Welting, Aldo Jongejan, Pierina Casanova, Sybren L. Meijer, Claude Libert, Theodorus B. M. Hakvoort, and et al. 2022. "miR-511 Deficiency Protects Mice from Experimental Colitis by Reducing TLR3 and TLR4 Responses via WD Repeat and FYVE-Domain-Containing Protein 1" Cells 11, no. 1: 58. https://doi.org/10.3390/cells11010058






