Cytoskeleton Dependent Mobility Dynamics of FcγRIIA Facilitates Platelet Haptotaxis and Capture of Opsonized Bacteria
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents and Chemicals
2.2. Preparation and Functionalization of Micropatterned Arrays
2.3. Preparation of Heat-Aggregated Human IgG and Functionalization of Micropatterns and Microbeads
2.4. Preparation and Functionalization of Micropatterned Arrays with Live E. coli and Platelet Adhesion Assay
2.5. Platelet Preparation, Adhesion Assays, Ca2+ Mobilization, and Fluorescence Microscopy
2.6. Analysis of Platelet Filopodia Number and Length
2.7. Quantification of Platelet Spread Area and Platelet Morphodynamics
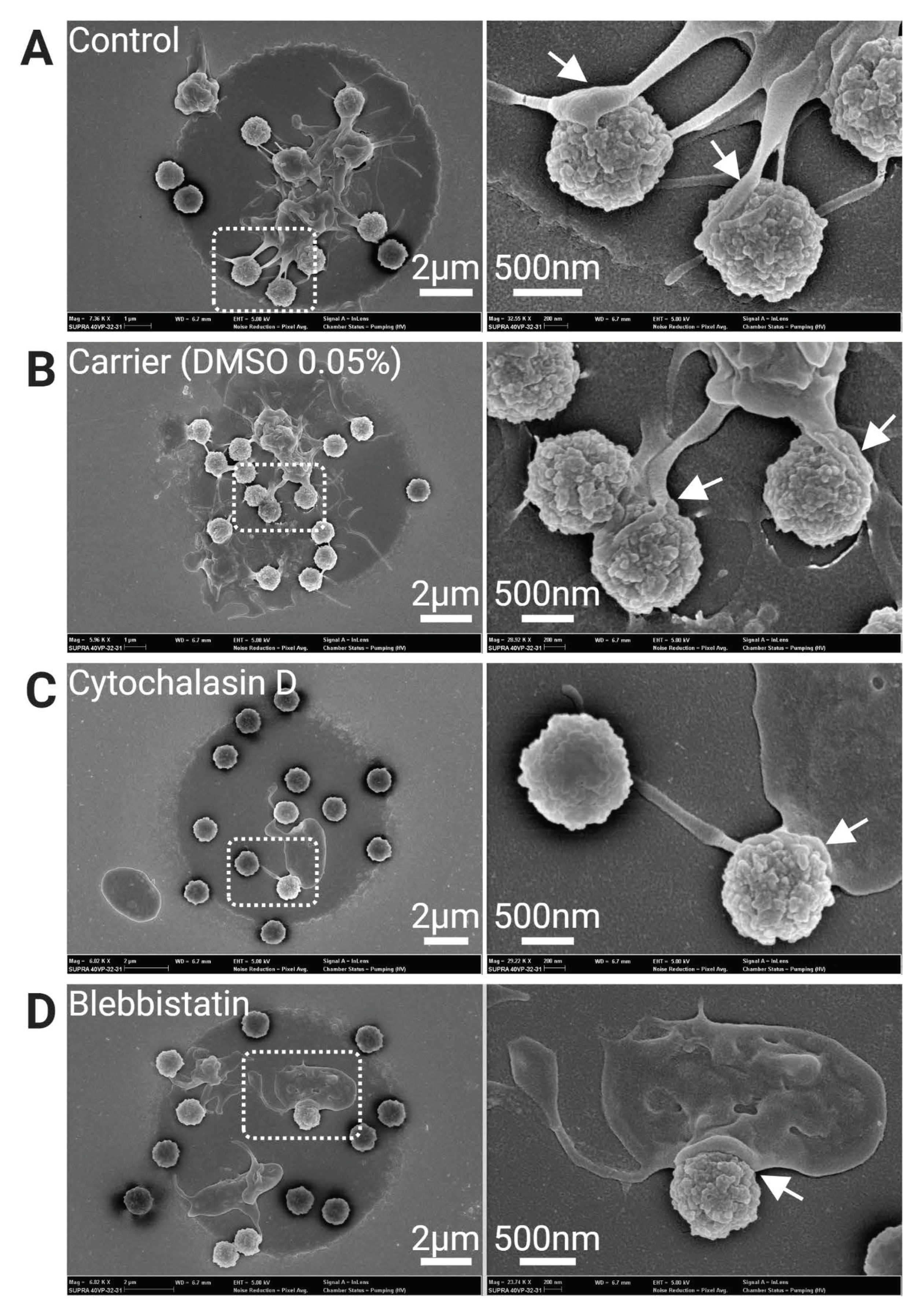
2.8. SEM Analysis
2.9. Preparation of Monovalent QD and Monoclonal anti-Human FcγRIIA Fab Conjugate
2.10. Labelling and Imaging of FcγRIIA on Platelets with Monovalent QD Conjugated to Anti-Human FcγRIIA Fab (QD-Fab)
2.11. Single-Particle Tracking and Analysis of QD-Fab on Platelet Membrane
2.12. Statistical Analysis
3. Results
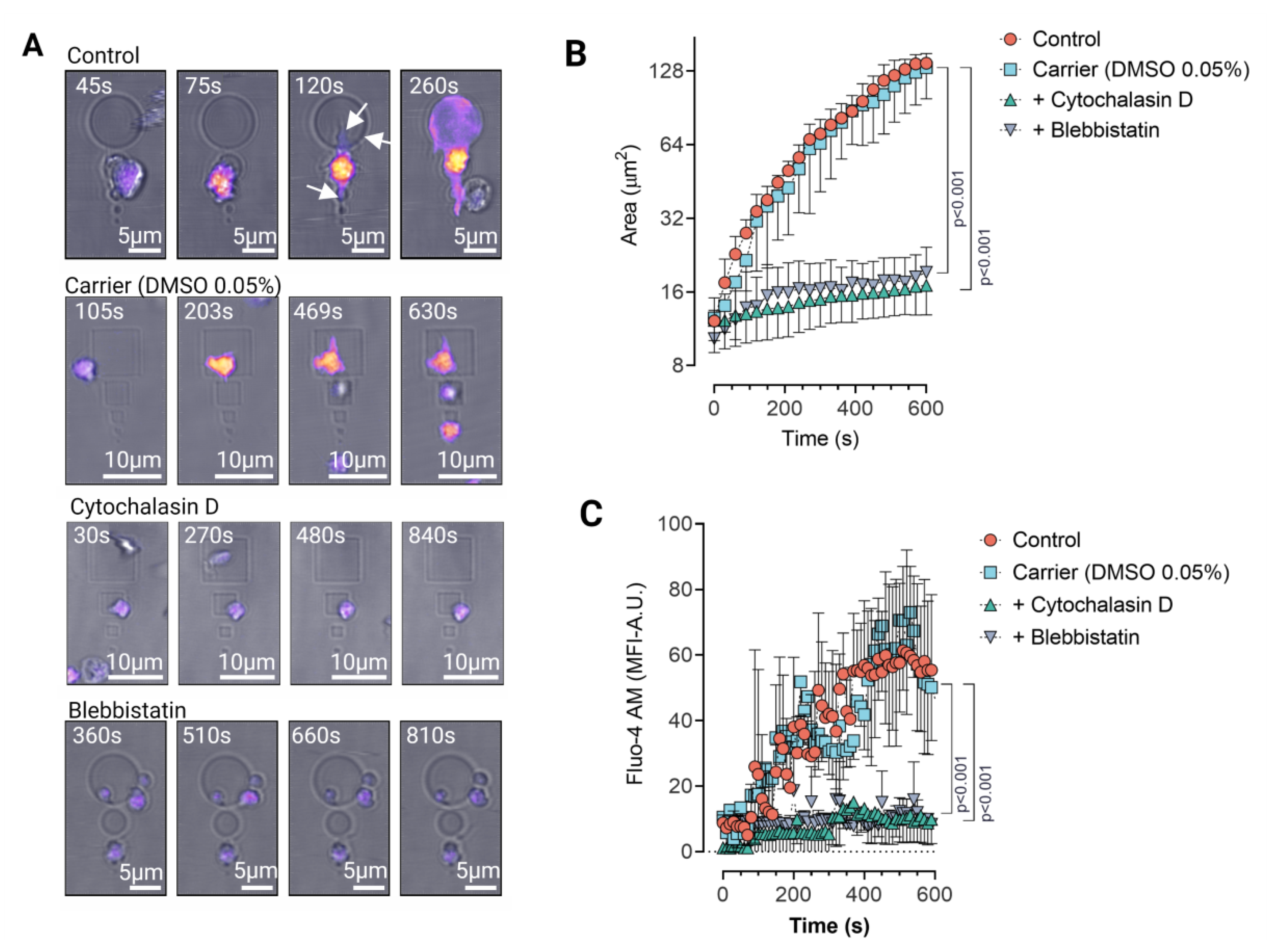
3.1. Platelet Cytoskeletal Integrity Is Indispensable for FcγRIIa Mediated Adhesion and Spreading on IgG Micropatterns
3.2. Platelet Haptotaxis on IgG Planar Micropatterns Is Mediated by Dynamic Membrane Protrusions
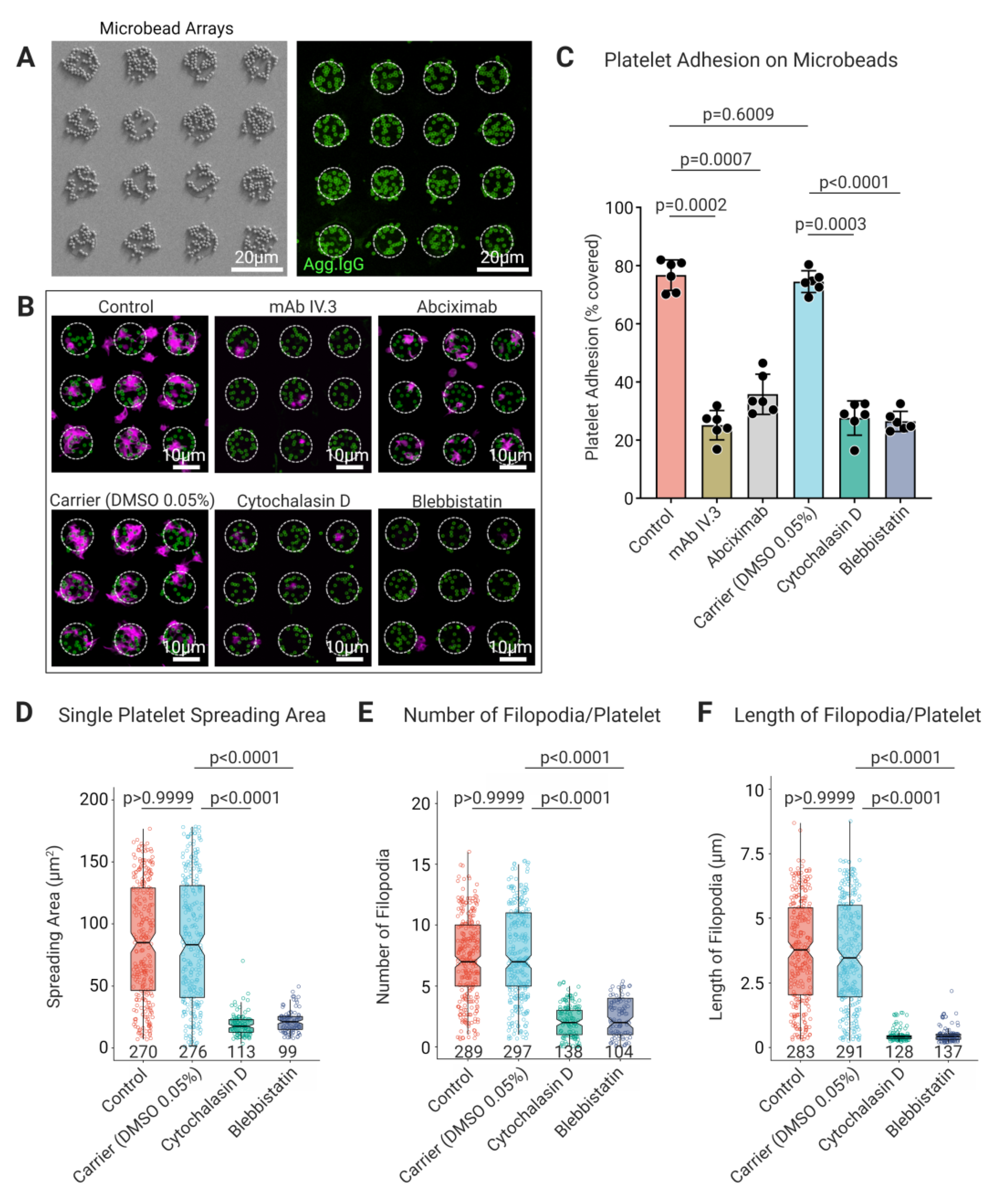
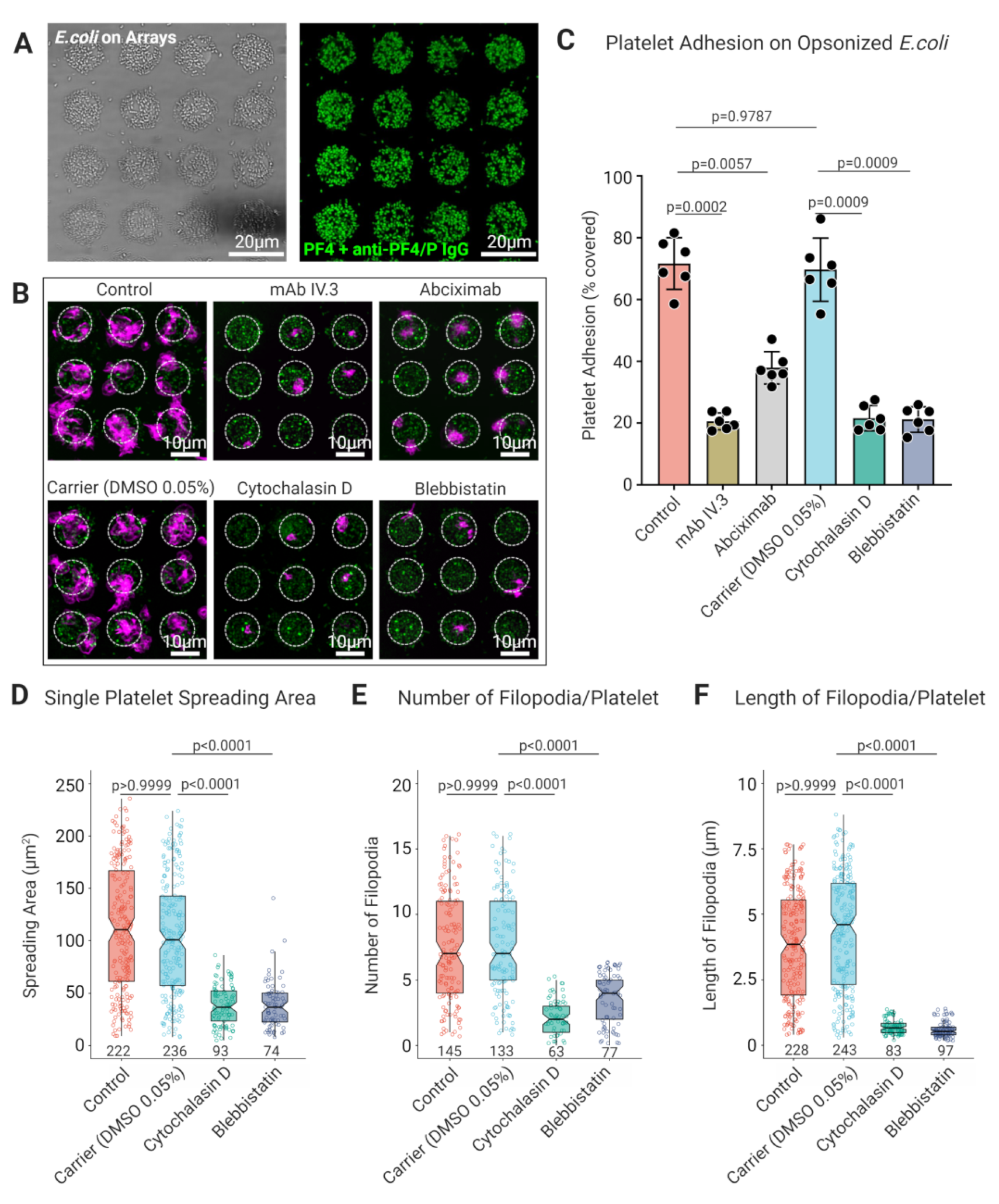
3.3. Platelet FcγRIIA Mediates Adhesion to ‘Bacteriamimetic’ IgG Opsonized Microbeads and Platelet Spreading Is Cytoskeleton Dependent
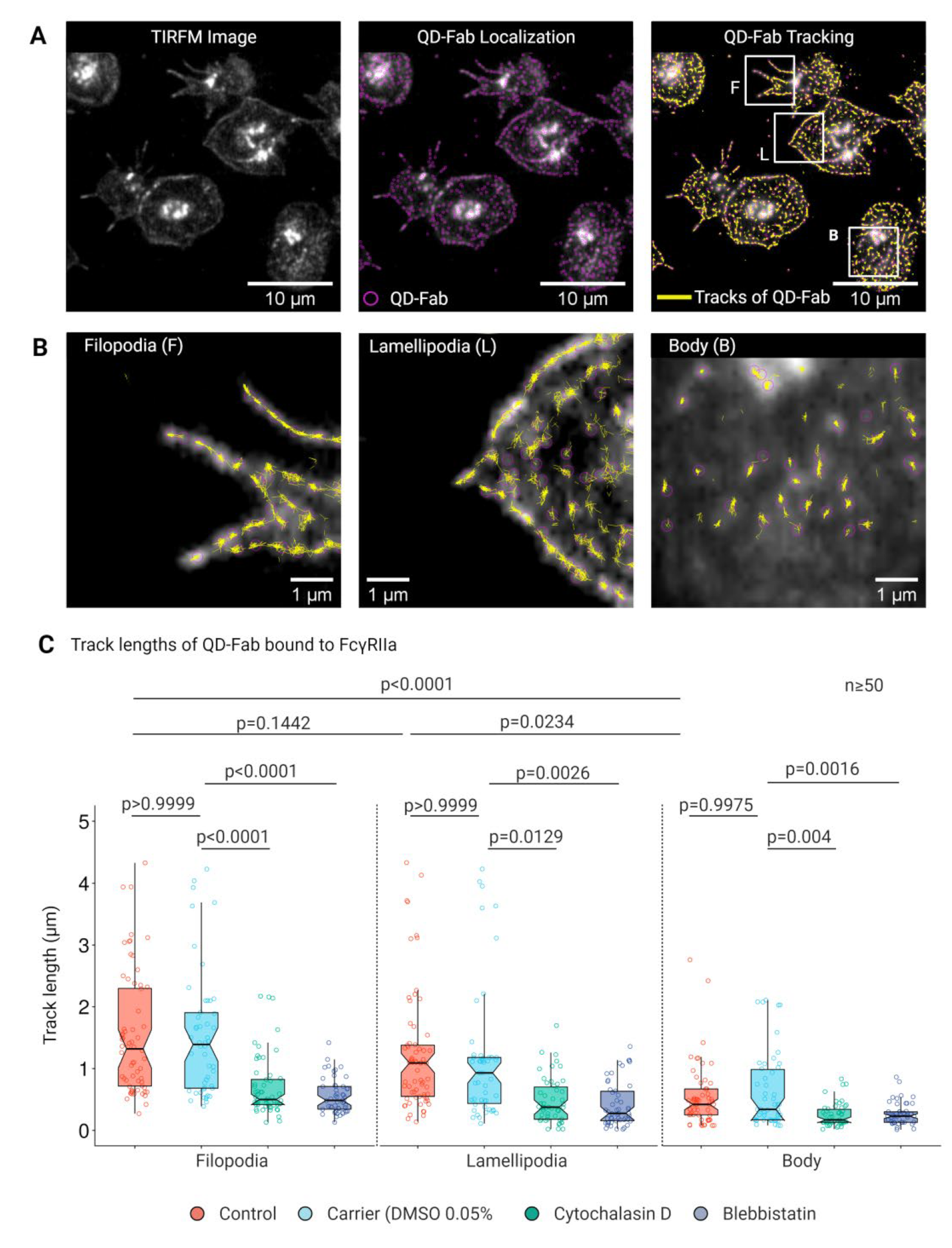
3.4. Lateral Mobility of FcγRIIA Is Dependent on Cytoskeletal Integrity
3.5. Longer Track Lengths and Higher Lateral Mobility of FcγRIIA on Platelet Filopodia and Lamellipodia Facilitate Sensing and Capture IgG Opsonized Bacterial Pathogens by Platelets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Machlus, K.R.; Italiano, J.E., Jr. The incredible journey: From megakaryocyte development to platelet formation. J. Cell Biol. 2013, 201, 785–796. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deppermann, C.; Kubes, P. Start a fire, kill the bug: The role of platelets in inflammation and infection. Innate Immun. 2018, 24, 335–348. [Google Scholar] [CrossRef] [PubMed]
- Ali, R.A.; Wuescher, L.M.; Dona, K.R.; Worth, R.G. Platelets Mediate Host Defense against Staphylococcus aureus through Direct Bactericidal Activity and by Enhancing Macrophage Activities. J. Immunol. 2017, 198, 344–351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kho, S.; Barber, B.E.; Johar, E.; Andries, B.; Poespoprodjo, J.R.; Kenangalem, E.; Piera, K.A.; Ehmann, A.; Price, R.N.; William, T.; et al. Platelets kill circulating parasites of all major Plasmodium species in human malaria. Blood 2018, 132, 1332–1344. [Google Scholar] [CrossRef]
- Youssefian, T.; Drouin, A.; Masse, J.M.; Guichard, J.; Cramer, E.M. Host defense role of platelets: Engulfment of HIV and Staphylococcus aureus occurs in a specific subcellular compartment and is enhanced by platelet activation. Blood 2002, 99, 4021–4029. [Google Scholar] [CrossRef] [Green Version]
- Beaulieu, L.M.; Clancy, L.; Tanriverdi, K.; Benjamin, E.J.; Kramer, C.D.; Weinberg, E.O.; He, X.; Mekasha, S.; Mick, E.; Ingalls, R.R.; et al. Specific Inflammatory Stimuli Lead to Distinct Platelet Responses in Mice and Humans. PLoS ONE 2015, 10, e0131688. [Google Scholar] [CrossRef]
- Palankar, R.; Binsker, U.; Haracska, B.; Wesche, J.; Greinacher, A.; Hammerschmidt, S. Interaction between the Staphylococcus aureus extracellular adherence protein Eap and its subdomains with platelets. Int. J. Med. Microbiol. 2018, 308, 683–691. [Google Scholar] [CrossRef]
- Binsker, U.; Palankar, R.; Wesche, J.; Kohler, T.P.; Prucha, J.; Burchhardt, G.; Rohde, M.; Schmidt, F.; Broker, B.M.; Mamat, U.; et al. Secreted Immunomodulatory Proteins of Staphylococcus aureus Activate Platelets and Induce Platelet Aggregation. Thromb. Haemost. 2018, 118, 745–757. [Google Scholar] [CrossRef]
- Zhu, W.; Gregory, J.C.; Org, E.; Buffa, J.A.; Gupta, N.; Wang, Z.; Li, L.; Fu, X.; Wu, Y.; Mehrabian, M.; et al. Gut Microbial Metabolite TMAO Enhances Platelet Hyperreactivity and Thrombosis Risk. Cell 2016, 165, 111–124. [Google Scholar] [CrossRef] [Green Version]
- Riaz, A.H.; Tasma, B.E.; Woodman, M.E.; Wooten, R.M.; Worth, R.G. Human platelets efficiently kill IgG-opsonized E. coli. FEMS Immunol. Med. Microbiol. 2012, 65, 78–83. [Google Scholar] [CrossRef] [Green Version]
- Stocker, T.J.; Ishikawa-Ankerhold, H.; Massberg, S.; Schulz, C. Small but mighty: Platelets as central effectors of host defense. Thromb. Haemost. 2017, 117, 651–661. [Google Scholar] [CrossRef] [PubMed]
- Nishat, S.; Wuescher, L.M.; Worth, R.G. Platelets enhance dendritic cell responses against S. aureus through CD40-CD40L interactions. Infect. Immun. 2018, 86, e00186-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arman, M.; Krauel, K. Human platelet IgG Fc receptor FcgammaRIIA in immunity and thrombosis. J. Thromb. Haemost. 2015, 13, 893–908. [Google Scholar] [CrossRef] [PubMed]
- Qiao, J.; Al-Tamimi, M.; Baker, R.I.; Andrews, R.K.; Gardiner, E.E. The platelet Fc receptor, FcgammaRIIa. Immunol. Rev. 2015, 268, 241–252. [Google Scholar] [CrossRef]
- Palankar, R.; Kohler, T.P.; Krauel, K.; Wesche, J.; Hammerschmidt, S.; Greinacher, A. Platelets kill bacteria by bridging innate and adaptive immunity via platelet factor 4 and FcgammaRIIA. J. Thromb. Haemost. 2018, 16, 1187–1197. [Google Scholar] [CrossRef] [Green Version]
- Perdomo, J.; Leung, H.H.L.; Ahmadi, Z.; Yan, F.; Chong, J.J.H.; Passam, F.H.; Chong, B.H. Neutrophil activation and NETosis are the major drivers of thrombosis in heparin-induced thrombocytopenia. Nat. Commun. 2019, 10, 1322. [Google Scholar] [CrossRef] [Green Version]
- Greinacher, A.; Thiele, T.; Warkentin, T.E.; Weisser, K.; Kyrle, P.A.; Eichinger, S. Thrombotic Thrombocytopenia after ChAdOx1 nCov-19 Vaccination. N. Engl. J. Med. 2021, 384, 2092–2101. [Google Scholar] [CrossRef]
- Greinacher, A.; Selleng, K.; Palankar, R.; Wesche, J.; Handtke, S.; Wolff, M.; Aurich, K.; Lalk, M.; Methling, K.; Volker, U.; et al. Insights in ChAdOx1 nCoV-19 vaccine-induced immune thrombotic thrombocytopenia. Blood 2021, 138, 2256–2268. [Google Scholar] [CrossRef]
- Lowenhaupt, R.W.; Miller, M.A.; Glueck, H.I. Platelet migration and chemotaxis demonstrated in vitro. Thromb. Res. 1973, 3, 477–487. [Google Scholar] [CrossRef]
- Pitchford, S.C.; Momi, S.; Baglioni, S.; Casali, L.; Giannini, S.; Rossi, R.; Page, C.P.; Gresele, P. Allergen induces the migration of platelets to lung tissue in allergic asthma. Am. J. Respir. Crit. Care Med. 2008, 177, 604–612. [Google Scholar] [CrossRef]
- Gaertner, F.; Ahmad, Z.; Rosenberger, G.; Fan, S.; Nicolai, L.; Busch, B.; Yavuz, G.; Luckner, M.; Ishikawa-Ankerhold, H.; Hennel, R.; et al. Migrating Platelets Are Mechano-scavengers that Collect and Bundle Bacteria. Cell 2017, 171, 1368–1382.e23. [Google Scholar] [CrossRef] [PubMed]
- Nicolai, L.; Schiefelbein, K.; Lipsky, S.; Leunig, A.; Hoffknecht, M.; Pekayvaz, K.; Raude, B.; Marx, C.; Ehrlich, A.; Pircher, J.; et al. Vascular surveillance by haptotactic blood platelets in inflammation and infection. Nat. Commun. 2020, 11, 5778. [Google Scholar] [CrossRef]
- Louise Meyer, R.; Zhou, X.; Tang, L.; Arpanaei, A.; Kingshott, P.; Besenbacher, F. Immobilisation of living bacteria for AFM imaging under physiological conditions. Ultramicroscopy 2010, 110, 1349–1357. [Google Scholar] [CrossRef] [PubMed]
- Greinacher, A.; Warkentin, T.E. Heparin-Induced Thrombocytopenia. In Practical Transfusion Medicine; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2013; pp. 322–334. [Google Scholar]
- Urbancic, V.; Butler, R.; Richier, B.; Peter, M.; Mason, J.; Livesey, F.J.; Holt, C.E.; Gallop, J.L. Filopodyan: An open-source pipeline for the analysis of filopodia. J. Cell Biol. 2017, 216, 3405–3422. [Google Scholar] [CrossRef] [Green Version]
- Barry, D.J.; Durkin, C.H.; Abella, J.V.; Way, M. Open source software for quantification of cell migration, protrusions, and fluorescence intensities. J. Cell Biol. 2015, 209, 163–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sage, D.; Donati, L.; Soulez, F.; Fortun, D.; Schmit, G.; Seitz, A.; Guiet, R.; Vonesch, C.; Unser, M. DeconvolutionLab2: An open-source software for deconvolution microscopy. Methods 2017, 115, 28–41. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tinevez, J.Y.; Perry, N.; Schindelin, J.; Hoopes, G.M.; Reynolds, G.D.; Laplantine, E.; Bednarek, S.Y.; Shorte, S.L.; Eliceiri, K.W. TrackMate: An open and extensible platform for single-particle tracking. Methods 2017, 115, 80–90. [Google Scholar] [CrossRef] [PubMed]
- Lund, F.W.; Jensen, M.L.; Christensen, T.; Nielsen, G.K.; Heegaard, C.W.; Wustner, D. SpatTrack: An imaging toolbox for analysis of vesicle motility and distribution in living cells. Traffic 2014, 15, 1406–1429. [Google Scholar] [CrossRef] [Green Version]
- Zhi, H.; Dai, J.; Liu, J.; Zhu, J.; Newman, D.K.; Gao, C.; Newman, P.J. Platelet Activation and Thrombus Formation over IgG Immune Complexes Requires Integrin alphaIIbbeta3 and Lyn Kinase. PLoS ONE 2015, 10, e0135738. [Google Scholar] [CrossRef] [Green Version]
- Hamzeh-Cognasse, H.; Damien, P.; Chabert, A.; Pozzetto, B.; Cognasse, F.; Garraud, O. Platelets and infections-complex interactions with bacteria. Front. Immunol. 2015, 6, 82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swinkels, M.; Rijkers, M.; Voorberg, J.; Vidarsson, G.; Leebeek, F.W.G.; Jansen, A.J.G. Emerging Concepts in Immune Thrombocytopenia. Front. Immunol. 2018, 9, 880. [Google Scholar] [CrossRef] [PubMed]
- Arman, M.; Krauel, K.; Tilley, D.O.; Weber, C.; Cox, D.; Greinacher, A.; Kerrigan, S.W.; Watson, S.P. Amplification of bacteria-induced platelet activation is triggered by FcgammaRIIA, integrin alphaIIbbeta3, and platelet factor 4. Blood 2014, 123, 3166–3174. [Google Scholar] [CrossRef] [PubMed]
- Krauel, K.; Weber, C.; Brandt, S.; Zahringer, U.; Mamat, U.; Greinacher, A.; Hammerschmidt, S. Platelet factor 4 binding to lipid A of Gram-negative bacteria exposes PF4/heparin-like epitopes. Blood 2012, 120, 3345–3352. [Google Scholar] [CrossRef] [Green Version]
- Krauel, K.; Potschke, C.; Weber, C.; Kessler, W.; Furll, B.; Ittermann, T.; Maier, S.; Hammerschmidt, S.; Broker, B.M.; Greinacher, A. Platelet factor 4 binds to bacteria, [corrected] inducing antibodies cross-reacting with the major antigen in heparin-induced thrombocytopenia. Blood 2011, 117, 1370–1378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baker, A.T.; Boyd, R.J.; Sarkar, D.; Teijeira-Crespo, A.; Chan, C.K.; Bates, E.; Waraich, K.; Vant, J.; Wilson, E.; Truong, C.D.; et al. ChAdOx1 interacts with CAR and PF4 with implications for thrombosis with thrombocytopenia syndrome. Sci. Adv. 2021, 7, eabl8213. [Google Scholar] [CrossRef]
- Michalik, S.; Siegerist, F.; Palankar, R.; Franzke, K.; Schindler, M.; Reder, A.; Seifert, U.; Cammann, C.; Wesche, J.; Steil, L.; et al. Comparative analysis of ChAdOx1 nCoV-19 and Ad26.COV2.S SARS-CoV-2 vector vaccines. Haematologica 2022, 107, 947. [Google Scholar] [CrossRef]
- Greinacher, A.; Selleng, K.; Mayerle, J.; Palankar, R.; Wesche, J.; Reiche, S.; Aebischer, A.; Warkentin, T.E.; Muenchhoff, M.; Hellmuth, J.C.; et al. Anti-platelet factor 4 antibodies causing VITT do not cross-react with SARS-CoV-2 spike protein. Blood 2021, 138, 1269–1277. [Google Scholar] [CrossRef]
- Greinacher, A. CLINICAL PRACTICE. Heparin-Induced Thrombocytopenia. N. Engl. J. Med. 2015, 373, 252–261. [Google Scholar] [CrossRef]
- Trepat, X.; Chen, Z.; Jacobson, K. Cell migration. Compr. Physiol. 2012, 2, 2369–2392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carter, S.B. Haptotaxis and the mechanism of cell motility. Nature 1967, 213, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Heckman, C.A.; Plummer, H.K., 3rd. Filopodia as sensors. Cell Signal. 2013, 25, 2298–2311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- King, S.J.; Asokan, S.B.; Haynes, E.M.; Zimmerman, S.P.; Rotty, J.D.; Alb, J.G., Jr.; Tagliatela, A.; Blake, D.R.; Lebedeva, I.P.; Marston, D.; et al. Lamellipodia are crucial for haptotactic sensing and response. J. Cell Sci. 2016, 129, 2329–2342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bender, M.; Palankar, R. Platelet Shape Changes during Thrombus Formation: Role of Actin-Based Protrusions. Hamostaseologie 2021, 41, 14–21. [Google Scholar] [CrossRef]
- Sandmann, R.; Koster, S. Topographic Cues Reveal Two Distinct Spreading Mechanisms in Blood Platelets. Sci. Rep. 2016, 6, 22357. [Google Scholar] [CrossRef]
- Schurr, Y.; Sperr, A.; Volz, J.; Beck, S.; Reil, L.; Kusch, C.; Eiring, P.; Bryson, S.; Sauer, M.; Nieswandt, B.; et al. Platelet lamellipodium formation is not required for thrombus formation and stability. Blood 2019, 134, 2318–2329. [Google Scholar] [CrossRef]
- Kita, A.; Sakurai, Y.; Myers, D.R.; Rounsevell, R.; Huang, J.N.; Seok, T.J.; Yu, K.; Wu, M.C.; Fletcher, D.A.; Lam, W.A. Microenvironmental geometry guides platelet adhesion and spreading: A quantitative analysis at the single cell level. PLoS ONE 2011, 6, e26437. [Google Scholar] [CrossRef] [Green Version]
- Myers, D.R.; Qiu, Y.; Fay, M.E.; Tennenbaum, M.; Chester, D.; Cuadrado, J.; Sakurai, Y.; Baek, J.; Tran, R.; Ciciliano, J.C.; et al. Single-platelet nanomechanics measured by high-throughput cytometry. Nat. Mater. 2017, 16, 230–235. [Google Scholar] [CrossRef]
- Medvedev, N.; Palankar, R.; Krauel, K.; Greinacher, A.; Delcea, M. Micropatterned array to assess the interaction of single platelets with platelet factor 4-heparin-IgG complexes. Thromb. Haemost. 2014, 111, 862–872. [Google Scholar] [CrossRef]
- Boylan, B.; Gao, C.; Rathore, V.; Gill, J.C.; Newman, D.K.; Newman, P.J. Identification of FcgammaRIIa as the ITAM-bearing receptor mediating alphaIIbbeta3 outside-in integrin signaling in human platelets. Blood 2008, 112, 2780–2786. [Google Scholar] [CrossRef] [Green Version]
- Ostrowski, P.P.; Grinstein, S.; Freeman, S.A. Diffusion Barriers, Mechanical Forces, and the Biophysics of Phagocytosis. Dev. Cell 2016, 38, 135–146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, T.; Callan-Jones, A.; Fedorov, E.; Ravasio, A.; Brugués, A.; Ong, H.T.; Toyama, Y.; Low, B.C.; Trepat, X.; Shemesh, T.; et al. Large-scale curvature sensing by directional actin flow drives cellular migration mode switching. Nat. Phys. 2019, 15, 393–402. [Google Scholar] [CrossRef] [PubMed]
- Sakurai, Y.; Fitch-Tewfik, J.L.; Qiu, Y.; Ahn, B.; Myers, D.R.; Tran, R.; Fay, M.E.; Ding, L.; Spearman, P.W.; Michelson, A.D.; et al. Platelet geometry sensing spatially regulates alpha-granule secretion to enable matrix self-deposition. Blood 2015, 126, 531–538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kairdolf, B.A.; Smith, A.M.; Stokes, T.H.; Wang, M.D.; Young, A.N.; Nie, S. Semiconductor quantum dots for bioimaging and biodiagnostic applications. Annu. Rev. Anal. Chem. 2013, 6, 143–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lidke, D.S.; Nagy, P.; Heintzmann, R.; Arndt-Jovin, D.J.; Post, J.N.; Grecco, H.E.; Jares-Erijman, E.A.; Jovin, T.M. Quantum dot ligands provide new insights into erbB/HER receptor-mediated signal transduction. Nat. Biotechnol. 2004, 22, 198–203. [Google Scholar] [CrossRef]
- Paknikar, A.K.; Eltzner, B.; Koster, S. Direct characterization of cytoskeletal reorganization during blood platelet spreading. Prog. Biophys. Mol. Biol. 2018, 144, 166–176. [Google Scholar] [CrossRef]
- Raz-Ben Aroush, D.; Ofer, N.; Abu-Shah, E.; Allard, J.; Krichevsky, O.; Mogilner, A.; Keren, K. Actin Turnover in Lamellipodial Fragments. Curr. Biol. 2017, 27, 2963–2973.e14. [Google Scholar] [CrossRef] [Green Version]
- Finkenstaedt-Quinn, S.A.; Ge, S.; Haynes, C.L. Cytoskeleton dynamics in drug-treated platelets. Anal. Bioanal. Chem. 2015, 407, 2803–2809. [Google Scholar] [CrossRef]
- Jaumouille, V.; Farkash, Y.; Jaqaman, K.; Das, R.; Lowell, C.A.; Grinstein, S. Actin cytoskeleton reorganization by Syk regulates Fcgamma receptor responsiveness by increasing its lateral mobility and clustering. Dev. Cell 2014, 29, 534–546. [Google Scholar] [CrossRef] [Green Version]
- Freeman, S.A.; Vega, A.; Riedl, M.; Collins, R.F.; Ostrowski, P.P.; Woods, E.C.; Bertozzi, C.R.; Tammi, M.I.; Lidke, D.S.; Johnson, P.; et al. Transmembrane Pickets Connect Cyto- and Pericellular Skeletons Forming Barriers to Receptor Engagement. Cell 2018, 172, 305–317.e10. [Google Scholar] [CrossRef] [Green Version]
- Freeman, S.A.; Goyette, J.; Furuya, W.; Woods, E.C.; Bertozzi, C.R.; Bergmeier, W.; Hinz, B.; van der Merwe, P.A.; Das, R.; Grinstein, S. Integrins Form an Expanding Diffusional Barrier that Coordinates Phagocytosis. Cell 2016, 164, 128–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.; Kurilova, S.; Scott, B.L.; Bosworth, E.; Iverson, B.E.; Bailey, E.M.; Hoppe, A.D. TIRF imaging of Fc gamma receptor microclusters dynamics and signaling on macrophages during frustrated phagocytosis. BMC Immunol. 2016, 17, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locke, D.; Chen, H.; Liu, Y.; Liu, C.; Kahn, M.L. Lipid rafts orchestrate signaling by the platelet receptor glycoprotein VI. J. Biol. Chem. 2002, 277, 18801–18809. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palankar, R.; Sachs, L.; Wesche, J.; Greinacher, A. Cytoskeleton Dependent Mobility Dynamics of FcγRIIA Facilitates Platelet Haptotaxis and Capture of Opsonized Bacteria. Cells 2022, 11, 1615. https://doi.org/10.3390/cells11101615
Palankar R, Sachs L, Wesche J, Greinacher A. Cytoskeleton Dependent Mobility Dynamics of FcγRIIA Facilitates Platelet Haptotaxis and Capture of Opsonized Bacteria. Cells. 2022; 11(10):1615. https://doi.org/10.3390/cells11101615
Chicago/Turabian StylePalankar, Raghavendra, Laura Sachs, Jan Wesche, and Andreas Greinacher. 2022. "Cytoskeleton Dependent Mobility Dynamics of FcγRIIA Facilitates Platelet Haptotaxis and Capture of Opsonized Bacteria" Cells 11, no. 10: 1615. https://doi.org/10.3390/cells11101615
APA StylePalankar, R., Sachs, L., Wesche, J., & Greinacher, A. (2022). Cytoskeleton Dependent Mobility Dynamics of FcγRIIA Facilitates Platelet Haptotaxis and Capture of Opsonized Bacteria. Cells, 11(10), 1615. https://doi.org/10.3390/cells11101615





