Stable Integration of Inducible SPLICS Reporters Enables Spatio-Temporal Analysis of Multiple Organelle Contact Sites upon Modulation of Cholesterol Traffic
Abstract
:1. Introduction
2. Materials and Methods
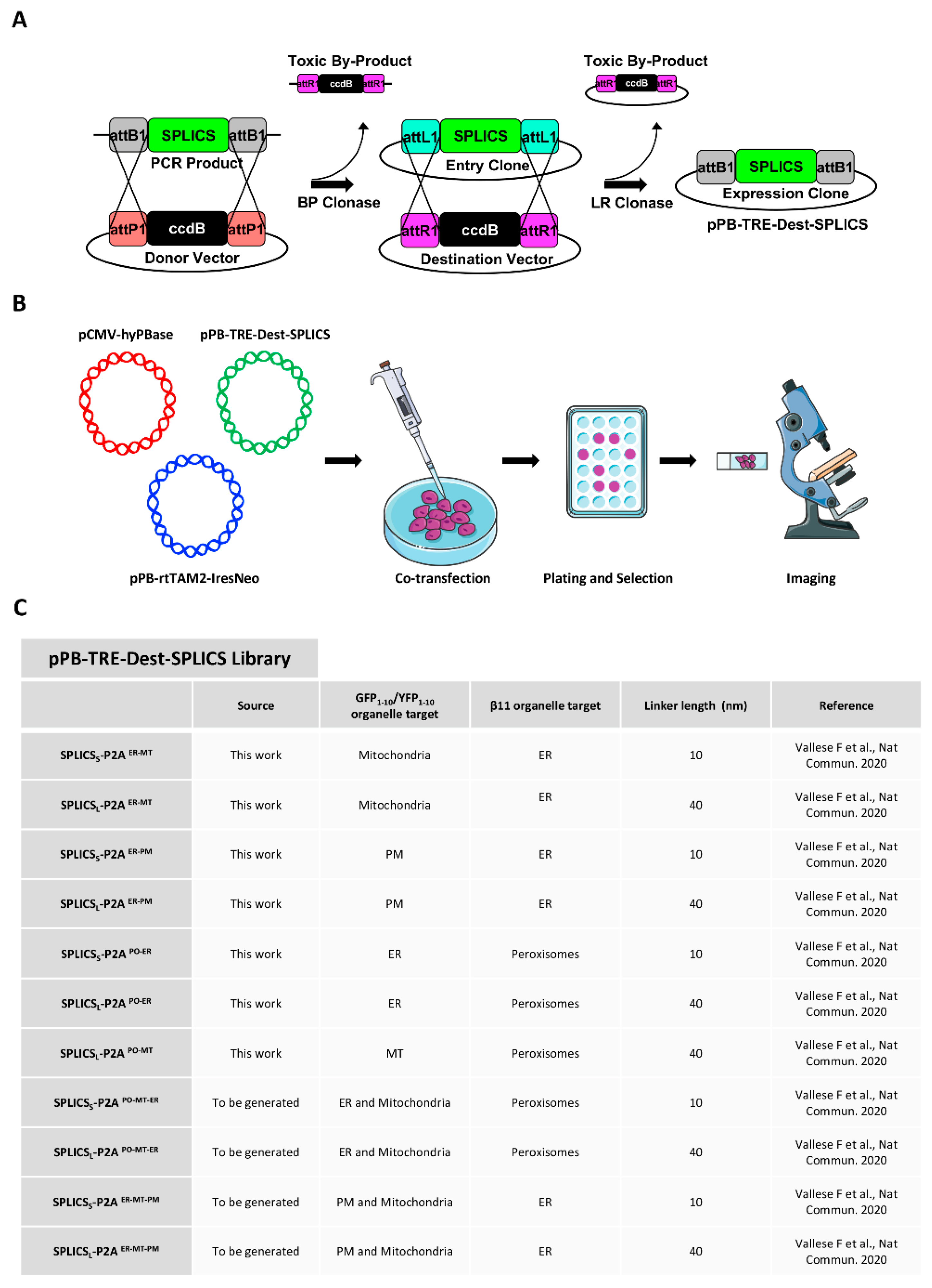
2.1. Cloning Plasmid Construction
2.2. Cell Line
2.3. Stable Transfection
2.4. Immunicytochemistry and Filipin Staining
2.5. Doxycycline Induction
2.6. U18666A Treatment
2.7. Confocal Microscopy and Image Analysis
2.8. Western Blot
2.9. Statistical Analysis
3. Results
3.1. Vector Library Generation and Generation of HeLa Polyclonal Stable Cell Lines
3.2. Western Blotting of Protein Expression in HeLa SPLICSS/LER-MT and SPLICSLPO-MT, SPLICSS/LER-PM and SPLICSS/LPO-ER Stable Cell Lines
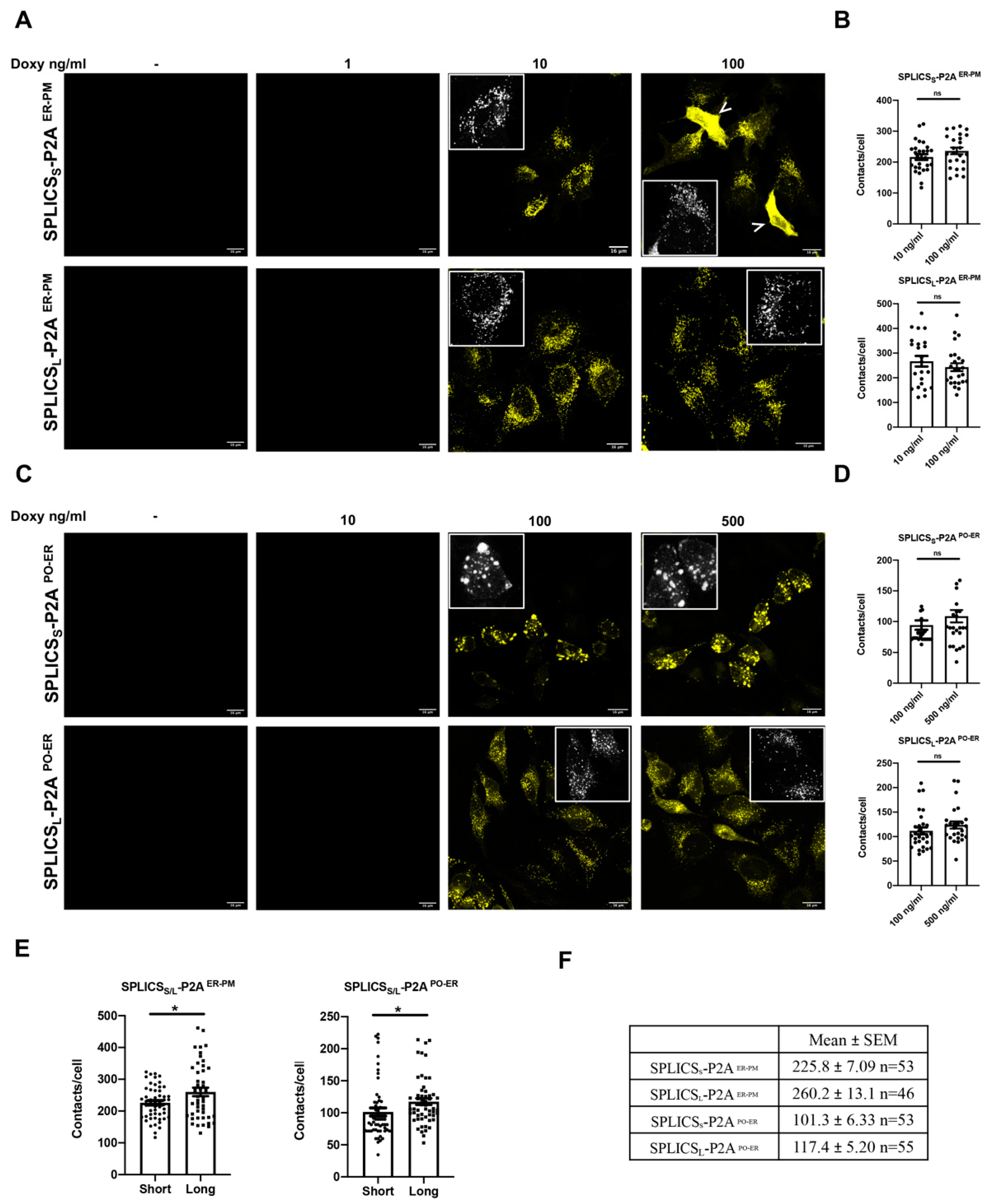
3.3. Short and Long ER-Plasma Membrane and ER-Peroxisomes Interactions in HeLa Polyclonal Stable Cell Lines Express Inducible SPLICS Sensors
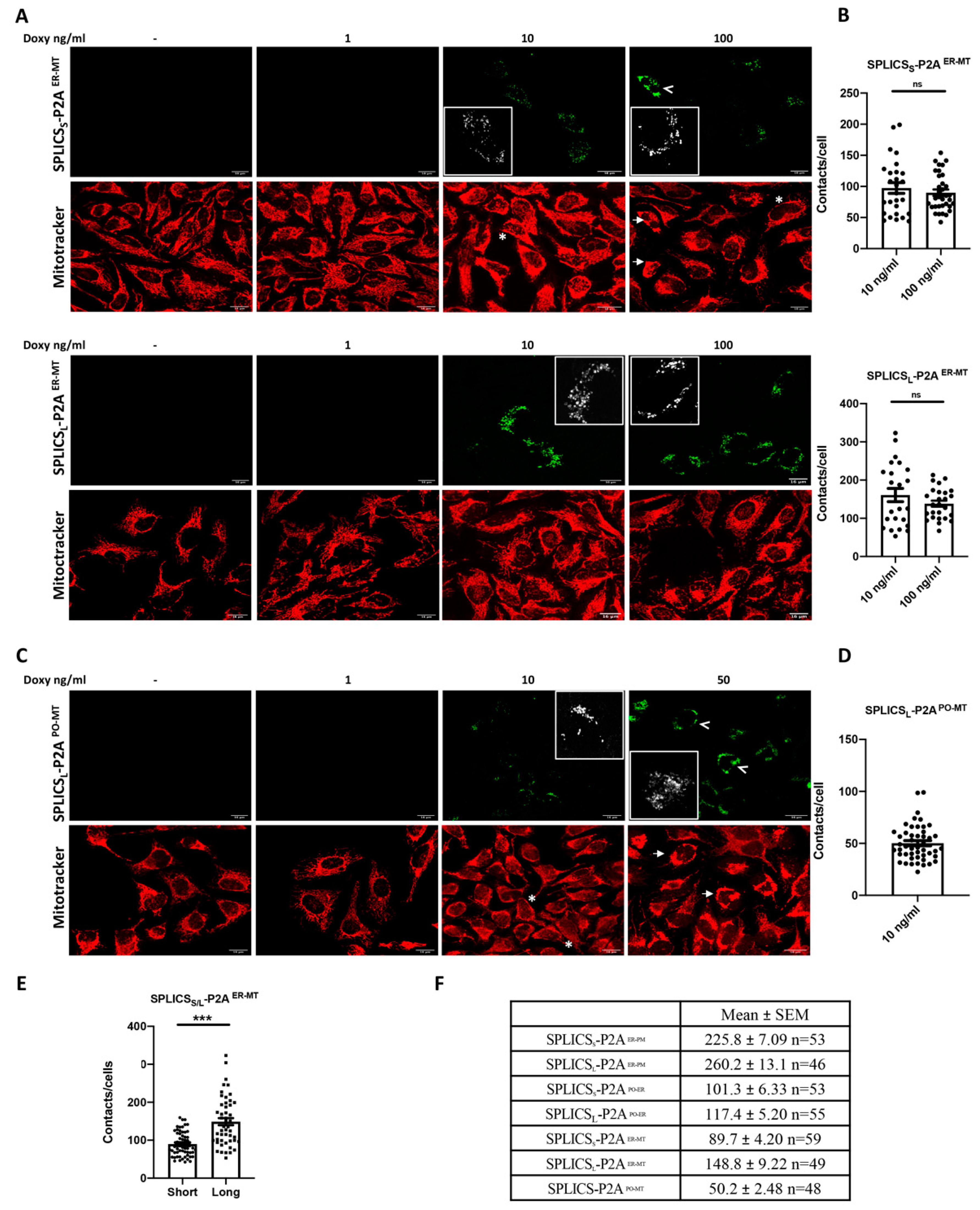
3.4. Short and Long Mitochondria-ER and Mitochondria-Peroxisomes Interactions in HeLa Polyclonal Stable Cell Lines Express Inducible SPLICS Sensors
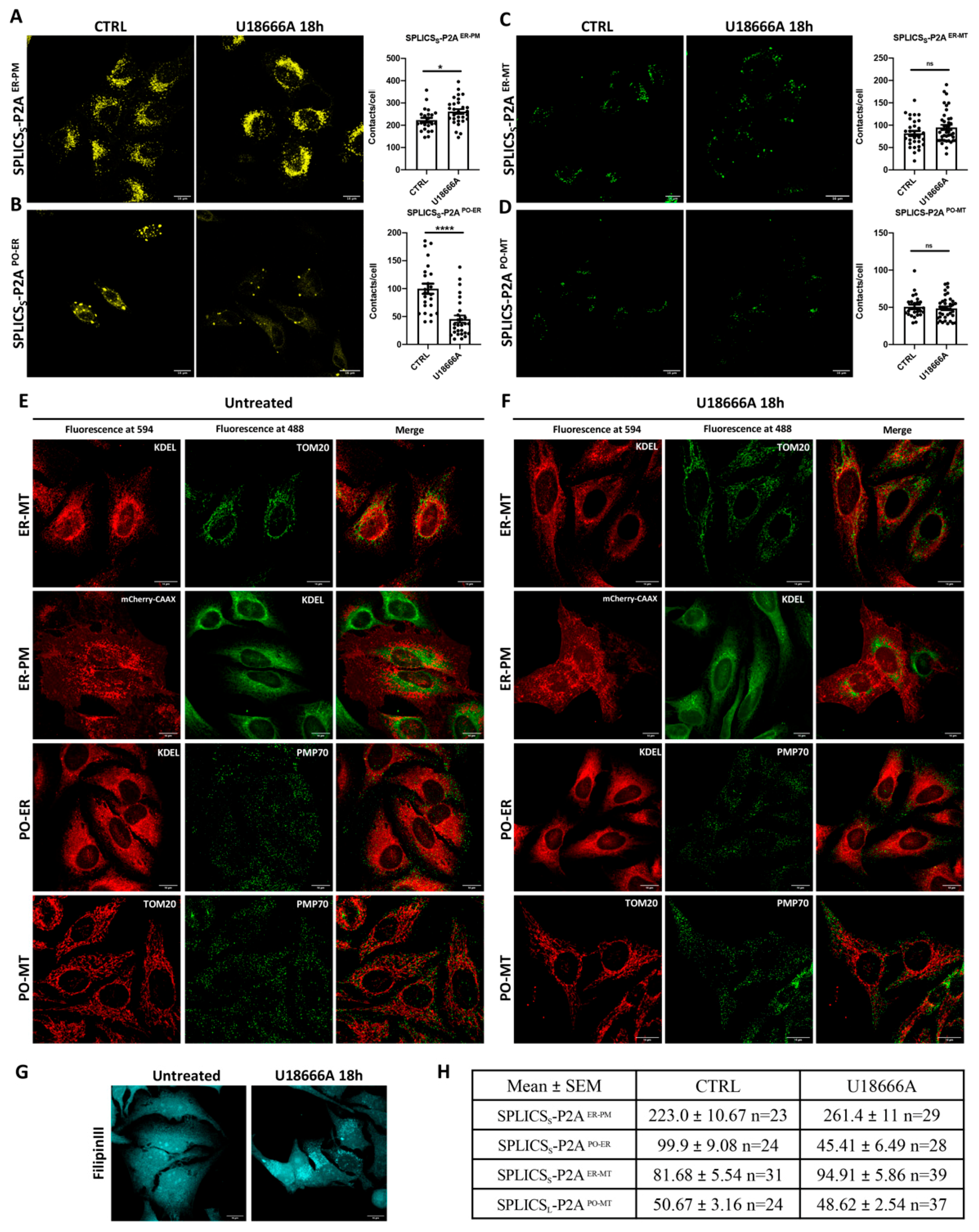
3.5. Effect of U18666A Treatment of Stable HeLa Cell Lines Expressing SPLICSSER-PM and SPLICSSPO-ER, SPLICSSER-MT, and SPLICSLPO-MT
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scorrano, L.; De Matteis, M.A.; Emr, S.; Giordano, F.; Hajnoczky, G.; Kornmann, B.; Lackner, L.L.; Levine, T.P.; Pellegrini, L.; Reinisch, K.; et al. Coming together to define membrane contact sites. Nat. Commun. 2019, 10, 1287. [Google Scholar] [CrossRef] [PubMed]
- Phillips, M.J.; Voeltz, G.K. Structure and function of ER membrane contact sites with other organelles. Nat. Rev. Mol. Cell Biol 2016, 17, 69–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, D.R.; Eid, R.; Miller, K.A.; Boucher, E.; Mandato, C.A.; Greenwood, M.T. Intracellular second messengers mediate stress inducible hormesis and Programmed Cell Death: A review. Biochim. Biophys. Acta. Mol. Cell. Res. 2019, 1866, 773–792. [Google Scholar] [CrossRef] [PubMed]
- Cieri, D.; Vicario, M.; Giacomello, M.; Vallese, F.; Filadi, R.; Wagner, T.; Pozzan, T.; Pizzo, P.; Scorrano, L.; Brini, M.; et al. SPLICS: A split green fluorescent protein-based contact site sensor for narrow and wide heterotypic organelle juxtaposition. Cell Death Differ. 2018, 25, 1131–1145. [Google Scholar] [CrossRef] [Green Version]
- Vallese, F.; Catoni, C.; Cieri, D.; Barazzuol, L.; Ramirez, O.; Calore, V.; Bonora, M.; Giamogante, F.; Pinton, P.; Brini, M.; et al. An expanded palette of improved SPLICS reporters detects multiple organelle contacts in vitro and in vivo. Nat. Commun. 2020, 11, 6069. [Google Scholar] [CrossRef]
- Doghman-Bouguerra, M.; Granatiero, V.; Sbiera, S.; Sbiera, I.; Lacas-Gervais, S.; Brau, F.; Fassnacht, M.; Rizzuto, R.; Lalli, E. FATE1 antagonizes calcium- and drug-induced apoptosis by uncoupling ER and mitochondria. EMBO Rep. 2016, 17, 1264–1280. [Google Scholar] [CrossRef] [Green Version]
- Granatiero, V.; Giorgio, V.; Cali, T.; Patron, M.; Brini, M.; Bernardi, P.; Tiranti, V.; Zeviani, M.; Pallafacchina, G.; De Stefani, D.; et al. Reduced mitochondrial Ca(2+) transients stimulate autophagy in human fibroblasts carrying the 13514A>G mutation of the ND5 subunit of NADH dehydrogenase. Cell Death Differ. 2016, 23, 231–241. [Google Scholar] [CrossRef] [Green Version]
- Cieri, D.; Vicario, M.; Vallese, F.; D’Orsi, B.; Berto, P.; Grinzato, A.; Catoni, C.; De Stefani, D.; Rizzuto, R.; Brini, M.; et al. Tau localises within mitochondrial sub-compartments and its caspase cleavage affects ER-mitochondria interactions and cellular Ca(2+) handling. Biochim Biophys Acta. Mol. Basis. Dis. 2018, 1864, 3247–3256. [Google Scholar] [CrossRef]
- Filadi, R.; Leal, N.S.; Schreiner, B.; Rossi, A.; Dentoni, G.; Pinho, C.M.; Wiehager, B.; Cieri, D.; Cali, T.; Pizzo, P.; et al. TOM70 Sustains Cell Bioenergetics by Promoting IP3R3-Mediated ER to Mitochondria Ca(2+) Transfer. Curr. Biol. 2018, 28, 369–382 e366. [Google Scholar] [CrossRef] [Green Version]
- Berenguer-Escuder, C.; Grossmann, D.; Massart, F.; Antony, P.; Burbulla, L.F.; Glaab, E.; Imhoff, S.; Trinh, J.; Seibler, P.; Grunewald, A.; et al. Variants in Miro1 Cause Alterations of ER-Mitochondria Contact Sites in Fibroblasts from Parkinson’s Disease Patients. J. Clin. Med. 2019, 8, 2226. [Google Scholar] [CrossRef] [Green Version]
- Cali, T.; Ottolini, D.; Vicario, M.; Catoni, C.; Vallese, F.; Cieri, D.; Barazzuol, L.; Brini, M. splitGFP Technology Reveals Dose-Dependent ER-Mitochondria Interface Modulation by alpha-Synuclein A53T and A30P Mutants. Cells 2019, 8, 1072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carreras-Sureda, A.; Jana, F.; Urra, H.; Durand, S.; Mortenson, D.E.; Sagredo, A.; Bustos, G.; Hazari, Y.; Ramos-Fernandez, E.; Sassano, M.L.; et al. Non-canonical function of IRE1alpha determines mitochondria-associated endoplasmic reticulum composition to control calcium transfer and bioenergetics. Nat. Cell Biol. 2019, 21, 755–767. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Suaga, P.; Perez-Nievas, B.G.; Glennon, E.B.; Lau, D.H.W.; Paillusson, S.; Morotz, G.M.; Cali, T.; Pizzo, P.; Noble, W.; Miller, C.C.J. The VAPB-PTPIP51 endoplasmic reticulum-mitochondria tethering proteins are present in neuronal synapses and regulate synaptic activity. Acta. Neuropathol. Commun. 2019, 7, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeshaw, W.M.; van der Zwaag, M.; Pinto, F.; Lahaye, L.L.; Faber, A.I.; Gomez-Sanchez, R.; Dolga, A.M.; Poland, C.; Monaco, A.P.; van, I.S.C.; et al. Human VPS13A is associated with multiple organelles and influences mitochondrial morphology and lipid droplet motility. Elife 2019, 8, e43561. [Google Scholar] [CrossRef] [PubMed]
- Dematteis, G.; Vydmantaite, G.; Ruffinatti, F.A.; Chahin, M.; Farruggio, S.; Barberis, E.; Ferrari, E.; Marengo, E.; Distasi, C.; Morkuniene, R.; et al. Proteomic analysis links alterations of bioenergetics, mitochondria-ER interactions and proteostasis in hippocampal astrocytes from 3xTg-AD mice. Cell Death Dis. 2020, 11, 645. [Google Scholar] [CrossRef]
- Gutierrez, T.; Qi, H.; Yap, M.C.; Tahbaz, N.; Milburn, L.A.; Lucchinetti, E.; Lou, P.H.; Zaugg, M.; LaPointe, P.G.; Mercier, P.; et al. The ER chaperone calnexin controls mitochondrial positioning and respiration. Sci. Signal. 2020, 13, eaaax6660. [Google Scholar] [CrossRef]
- Hewitt, V.L.; Miller-Fleming, L.; Andreazza, S.; Mattedi, F.; Prudent, J.; Polleux, F.; Vagnoni, A.; Whitworth, A.J. Decreasing pdzd8-mediated mitochondrial-ER contacts in neurons improves fitness by increasing mitophagy. bioRxiv 2020. [Google Scholar] [CrossRef]
- Luo, J.; Yang, H.; Song, B.L. Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell. Biol. 2020, 21, 225–245. [Google Scholar] [CrossRef]
- Ciscato, F.; Ferrone, L.; Masgras, I.; Laquatra, C.; Rasola, A. Hexokinase 2 in Cancer: A Prima Donna Playing Multiple Characters. Int. J. Mol. Sci 2021, 22, 4716. [Google Scholar] [CrossRef]
- Lopez-Crisosto, C.; Díaz-Vegas, A.; Castro, P.F.; Rothermel, B.A.; Bravo-Sagua, R.; Lavandero, S. Endoplasmic reticulum−mitochondria coupling increases during doxycycline-induced mitochondrial stress in HeLa cells. Cell Death Dis. 2021, 12, 657. [Google Scholar] [CrossRef]
- Naia, L.; Pinho, C.M.; Dentoni, G.; Liu, J.; Leal, N.S.; Ferreira, D.M.S.; Schreiner, B.; Filadi, R.; Fão, L.; Connolly, N.M.C.; et al. Neuronal cell-based high-throughput screen for enhancers of mitochondrial function reveals luteolin as a modulator of mitochondria-endoplasmic reticulum coupling. BMC Biol. 2021, 19, 57. [Google Scholar] [CrossRef] [PubMed]
- Romei, M.G.; Boxer, S.G. Split Green Fluorescent Proteins: Scope, Limitations, and Outlook. Annu. Rev. Biophys. 2019, 48, 19–44. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Zhao, X.; Xu, J.; Shang, W.; Tong, C. A novel fluorescent reporter detects plastic remodeling of mitochondria-ER contact sites. J. Cell. Sci. 2018, 131, jcs208686. [Google Scholar] [CrossRef] [Green Version]
- Tashiro, S.; Kakimoto, Y.; Shinmyo, M.; Fujimoto, S.; Tamura, Y. Improved Split-GFP Systems for Visualizing Organelle Contact Sites in Yeast and Human Cells. Front. Cell. Dev. Biol. 2020, 8, 571388. [Google Scholar] [CrossRef] [PubMed]
- van Meer, G.; Voelker, D.R.; Feigenson, G.W. Membrane lipids: Where they are and how they behave. Nat. Rev. Mol. Cell. Biol. 2008, 9, 112–124. [Google Scholar] [CrossRef] [PubMed]
- Lange, Y.; Steck, T.L. The role of intracellular cholesterol transport in cholesterol homeostasis. Trends Cell Biol. 1996, 6, 205–208. [Google Scholar] [CrossRef]
- Lange, Y.; Steck, T.L. Active cholesterol 20 years on. Traffic 2020, 21, 662–674. [Google Scholar] [CrossRef]
- Meng, Y.; Heybrock, S.; Neculai, D.; Saftig, P. Cholesterol Handling in Lysosomes and Beyond. Trends Cell Biol. 2020, 30, 452–466. [Google Scholar] [CrossRef]
- Trinh, M.N.; Brown, M.S.; Goldstein, J.L.; Han, J.; Vale, G.; McDonald, J.G.; Seemann, J.; Mendell, J.T.; Lu, F. Last step in the path of LDL cholesterol from lysosome to plasma membrane to ER is governed by phosphatidylserine. Proc. Natl. Acad. Sci. USA 2020, 117, 18521–18529. [Google Scholar] [CrossRef]
- Xiao, J.; Luo, J.; Hu, A.; Xiao, T.; Li, M.; Kong, Z.; Jiang, L.; Zhou, Z.; Liao, Y.; Xie, C.; et al. Cholesterol transport through the peroxisome-ER membrane contacts tethered by PI(4,5)P2 and extended synaptotagmins. Sci. China. Life Sci. 2019, 62, 1117–1135. [Google Scholar] [CrossRef]
- Chu, B.B.; Liao, Y.C.; Qi, W.; Xie, C.; Du, X.; Wang, J.; Yang, H.; Miao, H.H.; Li, B.L.; Song, B.L. Cholesterol transport through lysosome-peroxisome membrane contacts. Cell 2015, 161, 291–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoglinger, D.; Burgoyne, T.; Sanchez-Heras, E.; Hartwig, P.; Colaco, A.; Newton, J.; Futter, C.E.; Spiegel, S.; Platt, F.M.; Eden, E.R. NPC1 regulates ER contacts with endocytic organelles to mediate cholesterol egress. Nat. Commun. 2019, 10, 4276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.H.; Lee, S.R.; Li, L.H.; Park, H.J.; Park, J.H.; Lee, K.Y.; Kim, M.K.; Shin, B.A.; Choi, S.Y. High cleavage efficiency of a 2A peptide derived from porcine teschovirus-1 in human cell lines, zebrafish and mice. PLoS ONE 2011, 6, e18556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cali, T.; Brini, M. Quantification of organelle contact sites by split-GFP-based contact site sensors (SPLICS) in living cells. Nat. Protoc. 2021, 16, 5287–5308. [Google Scholar] [CrossRef]
- Ikonen, E.; Zhou, X. Cholesterol transport between cellular membranes: A balancing act between interconnected lipid fluxes. Dev. Cell 2021, 56, 1430–1436. [Google Scholar] [CrossRef]
- Prinz, W.A.; Toulmay, A.; Balla, T. The functional universe of membrane contact sites. Nat. Rev. Mol. Cell. Biol. 2020, 21, 7–24. [Google Scholar] [CrossRef]
- Ariotti, N.; Rae, J.; Giles, N.; Martel, N.; Sierecki, E.; Gambin, Y.; Hall, T.E.; Parton, R.G. Ultrastructural localisation of protein interactions using conditionally stable nanobodies. PLoS Biol. 2018, 16, e2005473. [Google Scholar] [CrossRef] [Green Version]
- Sandhu, J.; Li, S.; Fairall, L.; Pfisterer, S.G.; Gurnett, J.E.; Xiao, X.; Weston, T.A.; Vashi, D.; Ferrari, A.; Orozco, J.L.; et al. Aster Proteins Facilitate Nonvesicular Plasma Membrane to ER Cholesterol Transport in Mammalian Cells. Cell 2018, 175, 514–529.e520. [Google Scholar] [CrossRef] [Green Version]
- Brown, M.S.; Goldstein, J.L. A receptor-mediated pathway for cholesterol homeostasis. Science 1986, 232, 34–47. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H. Lysosomal acid lipase and lipid metabolism: New mechanisms, new questions, and new therapies. Curr. Opin. Lipidol. 2018, 29, 218–223. [Google Scholar] [CrossRef]
- Thelen, A.M.; Zoncu, R. Emerging Roles for the Lysosome in Lipid Metabolism. Trends Cell. Biol. 2017, 27, 833–850. [Google Scholar] [CrossRef] [PubMed]
- Ridgway, N.D.; Zhao, K. Cholesterol transfer at endosomal-organelle membrane contact sites. Curr. Opin. Lipidol. 2018, 29, 212–217. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giamogante, F.; Barazzuol, L.; Poggio, E.; Tromboni, M.; Brini, M.; Calì, T. Stable Integration of Inducible SPLICS Reporters Enables Spatio-Temporal Analysis of Multiple Organelle Contact Sites upon Modulation of Cholesterol Traffic. Cells 2022, 11, 1643. https://doi.org/10.3390/cells11101643
Giamogante F, Barazzuol L, Poggio E, Tromboni M, Brini M, Calì T. Stable Integration of Inducible SPLICS Reporters Enables Spatio-Temporal Analysis of Multiple Organelle Contact Sites upon Modulation of Cholesterol Traffic. Cells. 2022; 11(10):1643. https://doi.org/10.3390/cells11101643
Chicago/Turabian StyleGiamogante, Flavia, Lucia Barazzuol, Elena Poggio, Marta Tromboni, Marisa Brini, and Tito Calì. 2022. "Stable Integration of Inducible SPLICS Reporters Enables Spatio-Temporal Analysis of Multiple Organelle Contact Sites upon Modulation of Cholesterol Traffic" Cells 11, no. 10: 1643. https://doi.org/10.3390/cells11101643
APA StyleGiamogante, F., Barazzuol, L., Poggio, E., Tromboni, M., Brini, M., & Calì, T. (2022). Stable Integration of Inducible SPLICS Reporters Enables Spatio-Temporal Analysis of Multiple Organelle Contact Sites upon Modulation of Cholesterol Traffic. Cells, 11(10), 1643. https://doi.org/10.3390/cells11101643






