The RAL Enigma: Distinct Roles of RALA and RALB in Cancer
Abstract
:1. Introduction
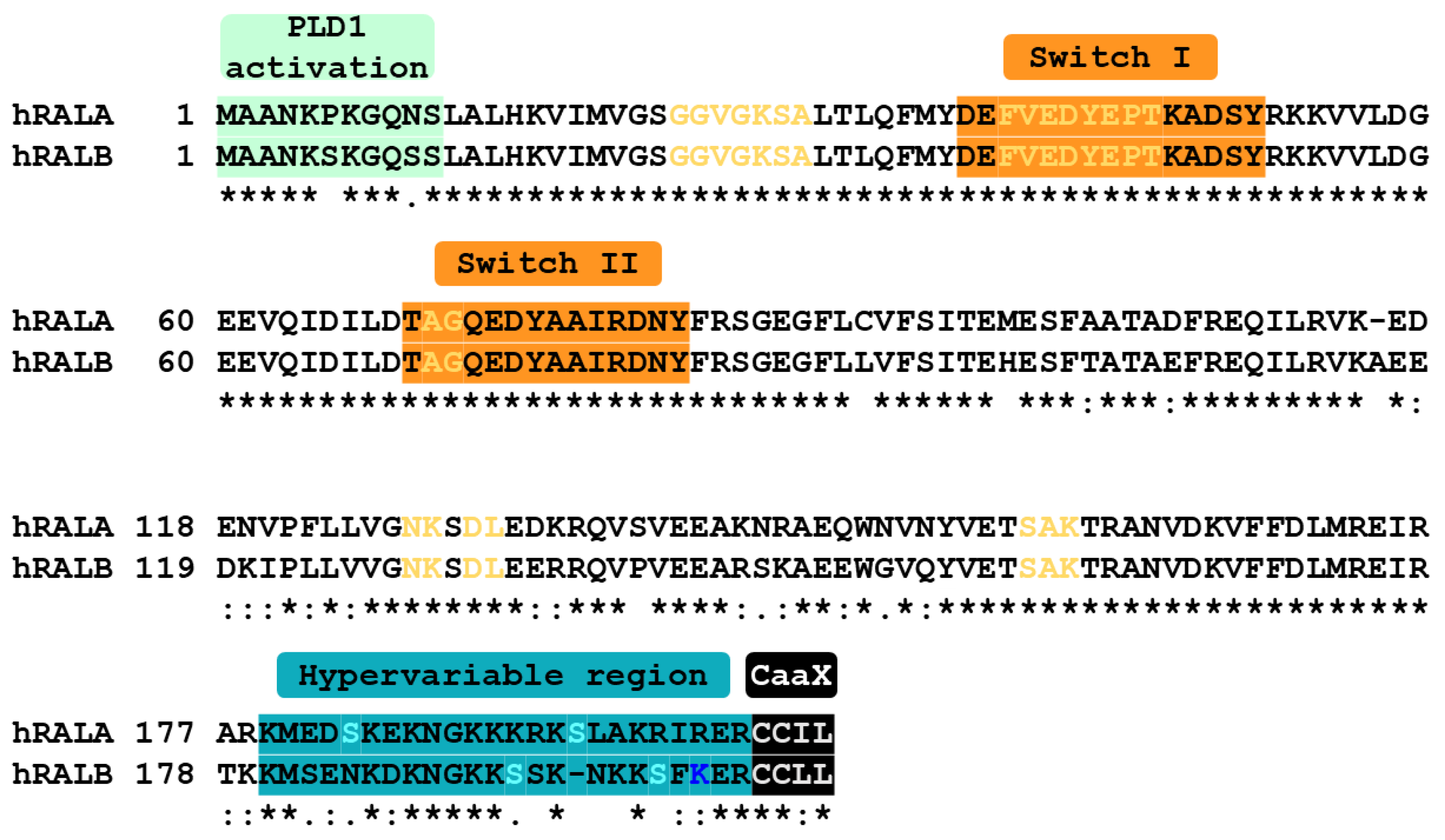
2. Structure and Post-Translational Modifications of RAL-GTPases
2.1. Structure
2.2. Regulation of RAL Activation
2.3. RAL-GTPase Phosphorylation
2.4. RAL-GTPase Ubiquitination
2.5. RAL-GTPase Geranylgeranylation
2.6. Post-Translational Modification Summary
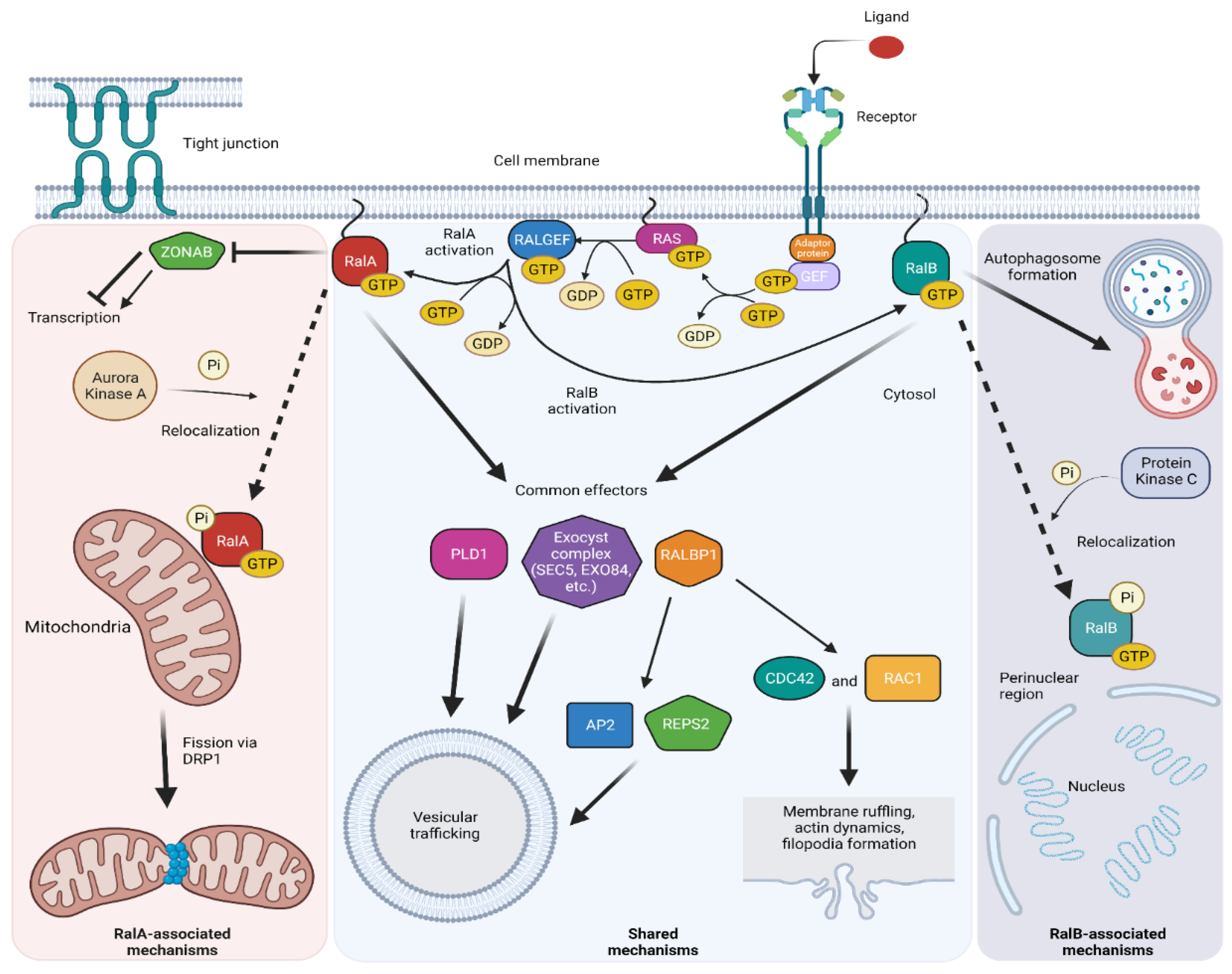
3. RAL-GTPase Effectors
3.1. RALBP1
3.2. EXO84 and SEC5
3.3. PLD1
3.4. Filamin
3.5. Autophagosome
3.6. JNK/Jun Pathway
3.7. ZONAB
4. Divergent Roles of RALA and RALB in Endocytosis, Exocytosis, and Vesicle Trafficking
4.1. Endocytosis
4.2. Exocytosis
4.3. Exosome Secretion
5. Divergent Roles of RALA and RALB in Cancer
5.1. Bladder Cancer
5.2. Blood Cancers
5.3. Breast Cancer
5.4. Colorectal Cancer
5.5. Gastric Cancer
5.6. Hepatic Cancer
5.7. Lung Cancer
5.8. Melanoma
5.9. Nerve Sheath Tumors
5.10. Ovarian Cancer
5.11. Pancreatic Cancer
5.12. Prostate Cancer
5.13. Renal Cancer
5.14. Summary of RAL-GTPases in Cancer and Considerations for Future Research
6. Therapeutic Targeting of RAL-GTPases
6.1. GGTase-I Inhibitors
6.2. Indirect Inhibition
6.3. Small Molecule RAL Inhibitors
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Harvey, J.J. An Unidetified Virus Which Causes the Rapid Production of Tumors in Mice. Nature 1964, 204, 1104–1105. [Google Scholar] [CrossRef] [PubMed]
- Kirsten, W.H.; Mayer, L.A. Morphologic responses to a murine erythroblastosis virus. J. Natl. Cancer Inst. 1967, 39, 311–335. [Google Scholar] [PubMed]
- Karnoub, A.E.; Weinberg, R.A. Ras oncogenes: Split personalities. Nat. Rev. Mol. Cell Biol. 2008, 9, 517–531. [Google Scholar] [CrossRef] [Green Version]
- Qu, L.; Pan, C.; He, S.M.; Lang, B.; Gao, G.D.; Wang, X.L.; Wang, Y. The Ras Superfamily of Small GTPases in Non-neoplastic Cerebral Diseases. Front. Mol. Neurosci. 2019, 12, 121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cherfils, J.; Zeghouf, M. Regulation of small GTPases by GEFs, GAPs, and GDIs. Physiol. Rev. 2013, 93, 269–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reiner, D.J.; Lundquist, E.A. Small GTPases. WormBook 2018, 2018, 1–65. [Google Scholar] [CrossRef]
- Prior, I.A.; Lewis, P.D.; Mattos, C. A comprehensive survey of Ras mutations in cancer. Cancer Res. 2012, 72, 2457–2467. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Pore, N.; Behrooz, A.; Ismail-Beigi, F.; Maity, A. Regulation of glut1 mRNA by hypoxia-inducible factor-1. Interaction between H-ras and hypoxia. J. Biol. Chem. 2001, 276, 9519–9525. [Google Scholar] [CrossRef] [Green Version]
- Gauthier-Rouvière, C.; Fernandez, A.; Lamb, N.J. ras-induced c-fos expression and proliferation in living rat fibroblasts involves C-kinase activation and the serum response element pathway. EMBO J. 1990, 9, 171–180. [Google Scholar] [CrossRef]
- Ouwens, D.M.; de Ruiter, N.D.; van der Zon, G.C.; Carter, A.P.; Schouten, J.; van der Burgt, C.; Kooistra, K.; Bos, J.L.; Maassen, J.A.; van Dam, H. Growth factors can activate ATF2 via a two-step mechanism: Phosphorylation of Thr71 through the Ras-MEK-ERK pathway and of Thr69 through RalGDS-Src-p38. EMBO J. 2002, 21, 3782–3793. [Google Scholar] [CrossRef]
- Westwick, J.K.; Cox, A.D.; Der, C.J.; Cobb, M.H.; Hibi, M.; Karin, M.; Brenner, D.A. Oncogenic Ras activates c-Jun via a separate pathway from the activation of extracellular signal-regulated kinases. Proc. Natl. Acad. Sci. USA 1994, 91, 6030–6034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haga, R.B.; Ridley, A.J. Rho GTPases: Regulation and roles in cancer cell biology. Small GTPases 2016, 7, 207–221. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maldonado, M.D.M.; Dharmawardhane, S. Targeting Rac and Cdc42 GTPases in Cancer. Cancer Res. 2018, 78, 3101–3111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urano, T.; Emkey, R.; Feig, L.A. Ral-GTPases mediate a distinct downstream signaling pathway from Ras that facilitates cellular transformation. EMBO J. 1996, 15, 810–816. [Google Scholar] [CrossRef] [PubMed]
- Chardin, P.; Tavitian, A. The ral gene: A new ras related gene isolated by the use of a synthetic probe. EMBO J. 1986, 5, 2203–2208. [Google Scholar] [CrossRef]
- Chardin, P.; Tavitian, A. Coding sequences of humanralAandralBcDNAs. Nucleic Acids Res. 1989, 17, 4380. [Google Scholar] [CrossRef] [Green Version]
- Cantor, S.B.; Urano, T.; Feig, L.A. Identification and characterization of Ral-binding protein 1, a potential downstream target of Ral GTPases. Mol. Cell Biol. 1995, 15, 4578–4584. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.H.; Lee, S.D.; Han, J.M.; Lee, T.G.; Kim, Y.; Park, J.B.; Lambeth, J.D.; Suh, P.G.; Ryu, S.H. Activation of phospholipase D1 by direct interaction with ADP-ribosylation factor 1 and RalA. FEBS Lett. 1998, 430, 231–235. [Google Scholar] [CrossRef] [Green Version]
- Ohta, Y.; Suzuki, N.; Nakamura, S.; Hartwig, J.H.; Stossel, T.P. The small GTPase RalA targets filamin to induce filopodia. Proc. Natl. Acad. Sci. USA 1999, 96, 2122–2128. [Google Scholar] [CrossRef] [Green Version]
- Fukai, S.; Matern, H.T.; Jagath, J.R.; Scheller, R.H.; Brunger, A.T. Structural basis of the interaction between RalA and Sec5, a subunit of the sec6/8 complex. EMBO J. 2003, 22, 3267–3278. [Google Scholar] [CrossRef] [Green Version]
- Moskalenko, S.; Tong, C.; Rosse, C.; Mirey, G.; Formstecher, E.; Daviet, L.; Camonis, J.; White, M.A. Ral GTPases regulate exocyst assembly through dual subunit interactions. J. Biol. Chem. 2003, 278, 51743–51748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thies, K.A.; Cole, M.W.; Schafer, R.E.; Spehar, J.M.; Richardson, D.S.; Steck, S.A.; Das, M.; Lian, A.W.; Ray, A.; Shakya, R.; et al. The small G-protein RalA promotes progression and metastasis of triple-negative breast cancer. Breast Cancer Res. 2021, 23, 65. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Owens, C.; Chandra, N.; Conaway, M.R.; Brautigan, D.L.; Theodorescu, D. Phosphorylation of RalB Is Important for Bladder Cancer Cell Growth and Metastasis. Cancer Res. 2010, 70, 8760–8769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, J.-Q.; Liu, X.; Frankel, P.; Rotunda, T.; Ramos, M.; Flom, J.; Jiang, H.; Feig, L.A.; Morris, A.J.; Kahn, R.A.; et al. Functional association between Arf and RalA in active phospholipase D complex. Proc. Natl. Acad. Sci. USA 1998, 95, 3632–3637. [Google Scholar] [CrossRef] [Green Version]
- Jiang, H.; Luo, J.-Q.; Urano, T.; Frankel, P.; Lu, Z.; Foster, D.A.; Feig, L.A. Involvement of Ral GTPase in v-Src-induced phospholipase D activation. Nature 1995, 378, 409–412. [Google Scholar] [CrossRef] [PubMed]
- Hiatt, S.M.; Neu, M.B.; Ramaker, R.C.; Hardigan, A.A.; Prokop, J.W.; Hancarova, M.; Prchalova, D.; Havlovicova, M.; Prchal, J.; Stranecky, V.; et al. De novo mutations in the GTP/GDP-binding region of RALA, a RAS-like small GTPase, cause intellectual disability and developmental delay. PLoS Genet. 2018, 14, e1007671. [Google Scholar] [CrossRef] [Green Version]
- Fu, D.; van Dam, E.M.; Brymora, A.; Duggin, I.G.; Robinson, P.J.; Roufogalis, B.D. The small GTPases Rab5 and RalA regulate intracellular traffic of P-glycoprotein. Biochim. Biophys. Acta 2007, 1773, 1062–1072. [Google Scholar] [CrossRef] [Green Version]
- Papini, D.; Langemeyer, L.; Abad, M.A.; Kerr, A.; Samejima, I.; Eyers, P.A.; Jeyaprakash, A.A.; Higgins, J.M.; Barr, F.A.; Earnshaw, W.C. TD-60 links RalA GTPase function to the CPC in mitosis. Nat. Commun. 2015, 6, 7678. [Google Scholar] [CrossRef] [Green Version]
- de Leeuw, H.P.; Fernandez-Borja, M.; Reits, E.A.; Romani de Wit, T.; Wijers-Koster, P.M.; Hordijk, P.L.; Neefjes, J.; van Mourik, J.A.; Voorberg, J. Small GTP-binding protein Ral modulates regulated exocytosis of von Willebrand factor by endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2001, 21, 899–904. [Google Scholar] [CrossRef] [Green Version]
- Vetter, I.R.; Wittinghofer, A. The guanine nucleotide-binding switch in three dimensions. Science 2001, 294, 1299–1304. [Google Scholar] [CrossRef] [Green Version]
- Moskalenko, S.; Henry, D.O.; Rosse, C.; Mirey, G.; Camonis, J.H.; White, M.A. The exocyst is a Ral effector complex. Nat. Cell Biol. 2002, 4, 66–72. [Google Scholar] [CrossRef] [PubMed]
- van Dam, E.M.; Robinson, P.J. Ral: Mediator of membrane trafficking. Int. J. Biochem. Cell Biol. 2006, 38, 1841–1847. [Google Scholar] [CrossRef] [PubMed]
- Singhal, S.S.; Salgia, R.; Verma, N.; Horne, D.; Awasthi, S. RLIP controls receptor-ligand signaling by regulating clathrin-dependent endocytosis. Biochim. Biophys. Acta Rev. Cancer 2020, 1873, 188337. [Google Scholar] [CrossRef] [PubMed]
- Jin, R.; Junutula, J.R.; Matern, H.T.; Ervin, K.E.; Scheller, R.H.; Brunger, A.T. Exo84 and Sec5 are competitive regulatory Sec6/8 effectors to the RalA GTPase. Embo J. 2005, 24, 2064–2074. [Google Scholar] [CrossRef]
- Jullien-Flores, V.; Mahé, Y.; Mirey, G.; Leprince, C.; Meunier-Bisceuil, B.; Sorkin, A.; Camonis, J.H. RLIP76, an effector of the GTPase Ral, interacts with the AP2 complex: Involvement of the Ral pathway in receptor endocytosis. J. Cell Sci. 2000, 113 Pt 16, 2837–2844. [Google Scholar] [CrossRef]
- Spiegelman, N.A.; Zhang, X.; Jing, H.; Cao, J.; Kotliar, I.B.; Aramsangtienchai, P.; Wang, M.; Tong, Z.; Rosch, K.M.; Lin, H. SIRT2 and Lysine Fatty Acylation Regulate the Activity of RalB and Cell Migration. ACS Chem. Biol. 2019, 14, 2014–2023. [Google Scholar] [CrossRef]
- Nicely, N.I.; Kosak, J.; de Serrano, V.; Mattos, C. Crystal structures of Ral-GppNHp and Ral-GDP reveal two binding sites that are also present in Ras and Rap. Structure 2004, 12, 2025–2036. [Google Scholar] [CrossRef] [Green Version]
- Popovic, M.; Schouten, A.; Rensen-de Leeuw, M.; Rehmann, H. The structure of the Guanine Nucleotide Exchange Factor Rlf in complex with the small G-protein Ral identifies conformational intermediates of the exchange reaction and the basis for the selectivity. J. Struct. Biol. 2016, 193, 106–114. [Google Scholar] [CrossRef]
- Fenwick, R.B.; Prasannan, S.; Campbell, L.J.; Nietlispach, D.; Evetts, K.A.; Camonis, J.; Mott, H.R.; Owen, D. Solution structure and dynamics of the small GTPase RalB in its active conformation: Significance for effector protein binding. Biochemistry 2009, 48, 2192–2206. [Google Scholar] [CrossRef]
- Yan, C.; Theodorescu, D. RAL GTPases: Biology and Potential as Therapeutic Targets in Cancer. Pharmacol. Rev. 2018, 70, 1–11. [Google Scholar] [CrossRef]
- Albright, C.F.; Giddings, B.W.; Liu, J.; Vito, M.; Weinberg, R.A. Characterization of a guanine nucleotide dissociation stimulator for a ras-related GTPase. Embo J. 1993, 12, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Murai, H.; Ikeda, M.; Kishida, S.; Ishida, O.; Okazaki-Kishida, M.; Matsuura, Y.; Kikuchi, A. Characterization of Ral GDP dissociation stimulator-like (RGL) activities to regulate c-fos promoter and the GDP/GTP exchange of Ral. J. Biol. Chem. 1997, 272, 10483–10490. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sood, R.; Makalowska, I.; Carpten, J.D.; Robbins, C.M.; Stephan, D.A.; Connors, T.D.; Morgenbesser, S.D.; Su, K.; Pinkett, H.W.; Graham, C.L.; et al. The human RGL (RalGDS-like) gene: Cloning, expression analysis and genomic organization. Biochim. Biophys. Acta 2000, 1491, 285–288. [Google Scholar] [CrossRef]
- Shao, H.; Andres, D.A. A novel RalGEF-like protein, RGL3, as a candidate effector for rit and Ras. J. Biol. Chem. 2000, 275, 26914–26924. [Google Scholar] [CrossRef]
- Rebhun, J.F.; Chen, H.; Quilliam, L.A. Identification and characterization of a new family of guanine nucleotide exchange factors for the ras-related GTPase Ral. J. Biol. Chem. 2000, 275, 13406–13410. [Google Scholar] [CrossRef] [Green Version]
- Ceriani, M.; Scandiuzzi, C.; Amigoni, L.; Tisi, R.; Berruti, G.; Martegani, E. Functional analysis of RalGPS2, a murine guanine nucleotide exchange factor for RalA GTPase. Exp. Cell Res. 2007, 313, 2293–2307. [Google Scholar] [CrossRef]
- de Bruyn, K.M.; de Rooij, J.; Wolthuis, R.M.; Rehmann, H.; Wesenbeek, J.; Cool, R.H.; Wittinghofer, A.H.; Bos, J.L. RalGEF2, a pleckstrin homology domain containing guanine nucleotide exchange factor for Ral. J. Biol. Chem. 2000, 275, 29761–29766. [Google Scholar] [CrossRef] [Green Version]
- das Chagas, P.F.; de Sousa, G.R.; Veronez, L.C.; Martins-da-Silva, A.; Corrêa, C.A.P.; Cruzeiro, G.A.V.; Nagano, L.F.P.; Queiroz, R.G.P.; Marie, S.K.N.; Brandalise, S.R.; et al. Identification of ITPR1 as a Hub Gene of Group 3 Medulloblastoma and Coregulated Genes with Potential Prognostic Values. J. Mol. Neurosci. 2022, 72, 633–641. [Google Scholar] [CrossRef]
- Sun, M.-S.; Yuan, L.-T.; Kuei, C.-H.; Lin, H.-Y.; Chen, Y.-L.; Chiu, H.-W.; Lin, Y.-F. RGL2 Drives the Metastatic Progression of Colorectal Cancer via Preventing the Protein Degradation of β-Catenin and KRAS. Cancers 2021, 13, 1763. [Google Scholar] [CrossRef]
- Guo, H.; Wang, S.; Xie, A.; Sun, W.; Wei, C.; Xian, S.; Yin, H.; Li, M.; Sun, H.; Li, H.; et al. Ral GEF with the PH Domain and SH3 Binding Motif 1 Regulated by Splicing Factor Junction Plakoglobin and Pyrimidine Metabolism Are Prognostic in Uterine Carcinosarcoma. Dis. Markers 2021, 2021, 1484227. [Google Scholar] [CrossRef]
- Xue, D.; Cheng, P.; Jiang, J.; Ren, Y.; Wu, D.; Chen, W. Systemic Analysis of the Prognosis-Related RNA Alternative Splicing Signals in Melanoma. Med. Sci. Monit. 2020, 26, e921133. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Wu, Y.; Xia, H.; Song, X. RGL2 as an age-dependent factor regulates colon cancer progression. Comput. Struct. Biotechnol. J. 2021, 19, 2190–2201. [Google Scholar] [CrossRef] [PubMed]
- Zago, G.; Veith, I.; Singh, M.K.; Fuhrmann, L.; De Beco, S.; Remorino, A.; Takaoka, S.; Palmeri, M.; Berger, F.; Brandon, N.; et al. RalB directly triggers invasion downstream Ras by mobilizing the Wave complex. eLife 2018, 7, e40474. [Google Scholar] [CrossRef] [PubMed]
- Vigil, D.; Martin, T.D.; Williams, F.; Yeh, J.J.; Campbell, S.L.; Der, C.J. Aberrant overexpression of the Rgl2 Ral small GTPase-specific guanine nucleotide exchange factor promotes pancreatic cancer growth through Ral-dependent and Ral-independent mechanisms. J. Biol. Chem. 2010, 285, 34729–34740. [Google Scholar] [CrossRef] [Green Version]
- González-García, A.; Pritchard, C.A.; Paterson, H.F.; Mavria, G.; Stamp, G.; Marshall, C.J. RalGDS is required for tumor formation in a model of skin carcinogenesis. Cancer Cell 2005, 7, 219–226. [Google Scholar] [CrossRef] [Green Version]
- Santos, A.O.; Parrini, M.C.; Camonis, J. RalGPS2 Is Essential for Survival and Cell Cycle Progression of Lung Cancer Cells Independently of Its Established Substrates Ral GTPases. PLoS ONE 2016, 11, e0154840. [Google Scholar] [CrossRef]
- D’Aloia, A.; Arrigoni, E.; Costa, B.; Berruti, G.; Martegani, E.; Sacco, E.; Ceriani, M. RalGPS2 Interacts with Akt and PDK1 Promoting Tunneling Nanotubes Formation in Bladder Cancer and Kidney Cells Microenvironment. Cancers 2021, 13, 6330. [Google Scholar] [CrossRef]
- D’Aloia, A.; Berruti, G.; Costa, B.; Schiller, C.; Ambrosini, R.; Pastori, V.; Martegani, E.; Ceriani, M. RalGPS2 is involved in tunneling nanotubes formation in 5637 bladder cancer cells. Exp. Cell Res. 2018, 362, 349–361. [Google Scholar] [CrossRef]
- Saito, R.; Shirakawa, R.; Nishiyama, H.; Kobayashi, T.; Kawato, M.; Kanno, T.; Nishizawa, K.; Matsui, Y.; Ohbayashi, T.; Horiguchi, M.; et al. Downregulation of Ral GTPase-activating protein promotes tumor invasion and metastasis of bladder cancer. Oncogene 2013, 32, 894–902. [Google Scholar] [CrossRef] [Green Version]
- Shirakawa, R.; Horiuchi, H. Ral GTPases: Crucial mediators of exocytosis and tumourigenesis. J. Biochem. 2015, 157, 285–299. [Google Scholar] [CrossRef] [Green Version]
- Shirakawa, R.; Fukai, S.; Kawato, M.; Higashi, T.; Kondo, H.; Ikeda, T.; Nakayama, E.; Okawa, K.; Nureki, O.; Kimura, T.; et al. Tuberous sclerosis tumor suppressor complex-like complexes act as GTPase-activating proteins for Ral GTPases. J. Biol. Chem. 2009, 284, 21580–21588. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, L.; Zhao, L.; Sze, K.M.F.; Kam, C.S.; Ming, V.S.I.; Wang, X.; Zhang, V.X.; Ho, D.W.H.; Cheung, T.T.; Chan, L.K.; et al. Dysregulation of RalA signaling through dual regulatory mechanisms exerts its oncogenic functions in hepatocellular carcinoma. Hepatology, 2021; in press. [Google Scholar] [CrossRef] [PubMed]
- Uegaki, M.; Kita, Y.; Shirakawa, R.; Teramoto, Y.; Kamiyama, Y.; Saito, R.; Yoshikawa, T.; Sakamoto, H.; Goto, T.; Akamatsu, S.; et al. Downregulation of RalGTPase-activating protein promotes invasion of prostatic epithelial cells and progression from intraepithelial neoplasia to cancer during prostate carcinogenesis. Carcinogenesis 2019, 40, 1535–1544. [Google Scholar] [CrossRef] [PubMed]
- Gao, P.; Liu, S.; Yoshida, R.; Shi, C.Y.; Yoshimachi, S.; Sakata, N.; Goto, K.; Kimura, T.; Shirakawa, R.; Nakayama, H.; et al. Ral GTPase Activation by Downregulation of RalGAP Enhances Oral Squamous Cell Carcinoma Progression. J. Dent. Res. 2019, 98, 1011–1019. [Google Scholar] [CrossRef]
- Martin, T.D.; Chen, X.W.; Kaplan, R.E.; Saltiel, A.R.; Walker, C.L.; Reiner, D.J.; Der, C.J. Ral and Rheb GTPase Activating Proteins Integrate mTOR and GTPase Signaling in Aging, Autophagy, and Tumor Cell Invasion. Mol. Cell 2014, 53, 209–220. [Google Scholar] [CrossRef] [Green Version]
- Iida, T.; Hirayama, D.; Minami, N.; Matsuura, M.; Wagatsuma, K.; Kawakami, K.; Nagaishi, K.; Nojima, M.; Ikeuchi, H.; Hirota, S.; et al. Down-regulation of RalGTPase-Activating Protein Promotes Colitis-Associated Cancer via NLRP3 Inflammasome Activation. Cell. Mol. Gastroenterol. Hepatol. 2020, 9, 277–293. [Google Scholar] [CrossRef]
- Yoshimachi, S.; Shirakawa, R.; Cao, M.; Trinh, D.A.; Gao, P.; Sakata, N.; Miyazaki, K.; Goto, K.; Miura, T.; Ariake, K.; et al. Ral GTPase-activating protein regulates the malignancy of pancreatic ductal adenocarcinoma. Cancer Sci. 2021, 112, 3064–3073. [Google Scholar] [CrossRef]
- Wolthuis, R.M.; Zwartkruis, F.; Moen, T.C.; Bos, J.L. Ras-dependent activation of the small GTPase Ral. Curr. Biol. 1998, 8, 471–474. [Google Scholar] [CrossRef] [Green Version]
- Takaya, A.; Ohba, Y.; Kurokawa, K.; Matsuda, M. RalA Activation at Nascent Lamellipodia of Epidermal Growth Factor-stimulated Cos7 Cells and Migrating Madin-Darby Canine Kidney Cells. Mol. Biol. Cell 2004, 15, 2549–2557. [Google Scholar] [CrossRef] [Green Version]
- Gildea, J.J.; Harding, M.A.; Seraj, M.J.; Gulding, K.M.; Theodorescu, D. The role of Ral A in epidermal growth factor receptor-regulated cell motility. Cancer Res. 2002, 62, 982–985. [Google Scholar]
- Zwartkruis, F.J.; Wolthuis, R.M.; Nabben, N.M.; Franke, B.; Bos, J.L. Extracellular signal-regulated activation of Rap1 fails to interfere in Ras effector signalling. Embo J. 1998, 17, 5905–5912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.L.; Roufogalis, B.D. Ca2+/calmodulin stimulates GTP binding to the ras-related protein ral-A. J. Biol. Chem. 1999, 274, 14525–14528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clough, R.R.; Sidhu, R.S.; Bhullar, R.P. Calmodulin binds RalA and RalB and is required for the thrombin-induced activation of Ral in human platelets. J. Biol. Chem. 2002, 277, 28972–28980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.B. Regulation of GTP-binding state in RalA through Ca2+ and calmodulin. Exp. Mol. Med. 2001, 33, 54–58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, K.L.; Khan, M.T.; Roufogalis, B.D. Identification and characterization of a calmodulin-binding domain in Ral-A, a Ras-related GTP-binding protein purified from human erythrocyte membrane. J. Biol. Chem. 1997, 272, 16002–16009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feldmann, G.; Mishra, A.; Hong, S.M.; Bisht, S.; Strock, C.J.; Ball, D.W.; Goggins, M.; Maitra, A.; Nelkin, B.D. Inhibiting the cyclin-dependent kinase CDK5 blocks pancreatic cancer formation and progression through the suppression of Ras-Ral signaling. Cancer Res. 2010, 70, 4460–4469. [Google Scholar] [CrossRef] [Green Version]
- Lim, K.-H.; Brady, D.C.; Kashatus, D.F.; Ancrile, B.B.; Der, C.J.; Cox, A.D.; Counter, C.M. Aurora-A Phosphorylates, Activates, and Relocalizes the Small GTPase RalA. Mol. Cell. Biol. 2010, 30, 508–523. [Google Scholar] [CrossRef] [Green Version]
- Sablina, A.A.; Chen, W.; Arroyo, J.D.; Corral, L.; Hector, M.; Bulmer, S.E.; Decaprio, J.A.; Hahn, W.C. The Tumor Suppressor PP2A Aβ Regulates the RalA GTPase. Cell 2007, 129, 969–982. [Google Scholar] [CrossRef] [Green Version]
- Kashatus, D.F.; Lim, K.-H.; Brady, D.C.; Pershing, N.L.K.; Cox, A.D.; Counter, C.M. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat. Cell Biol. 2011, 13, 1108–1115. [Google Scholar] [CrossRef] [Green Version]
- Martin, T.D.; Mitin, N.; Cox, A.D.; Yeh, J.J.; Der, C.J. Phosphorylation by Protein Kinase Cα Regulates RalB Small GTPase Protein Activation, Subcellular Localization, and Effector Utilization. J. Biol. Chem. 2012, 287, 14827–14836. [Google Scholar] [CrossRef] [Green Version]
- Grumati, P.; Dikic, I. Ubiquitin signaling and autophagy. J. Biol. Chem. 2018, 293, 5404–5413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neyraud, V.; Aushev, V.N.; Hatzoglou, A.; Meunier, B.; Cascone, I.; Camonis, J. RalA and RalB Proteins Are Ubiquitinated GTPases, and Ubiquitinated RalA Increases Lipid Raft Exposure at the Plasma Membrane. J. Biol. Chem. 2012, 287, 29397–29405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simicek, M.; Lievens, S.; Laga, M.; Guzenko, D.; Aushev, V.N.; Kalev, P.; Baietti, M.F.; Strelkov, S.V.; Gevaert, K.; Tavernier, J.; et al. The deubiquitylase USP33 discriminates between RALB functions in autophagy and innate immune response. Nat. Cell Biol. 2013, 15, 1220–1230. [Google Scholar] [CrossRef] [PubMed]
- Falsetti, S.C.; Wang, D.-A.; Peng, H.; Carrico, D.; Cox, A.D.; Der, C.J.; Hamilton, A.D.; Sebti, S.D.M. Geranylgeranyltransferase I Inhibitors Target RalB To Inhibit Anchorage-Dependent Growth and Induce Apoptosis and RalA To Inhibit Anchorage-Independent Growth. Mol. Cell. Biol. 2007, 27, 8003–8014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, N.; Shen, N.; Wang, X.; Jiang, S.; Xue, B.; Li, C. Protein prenylation and human diseases: A balance of protein farnesylation and geranylgeranylation. Sci. China Life Sci. 2015, 58, 328–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jullien-Flores, V.; Dorseuil, O.; Romero, F.; Letourneur, F.; Saragosti, S.; Berger, R.; Tavitian, A.; Gacon, G.; Camonis, J.H. Bridging Ral GTPase to Rho pathways. RLIP76, a Ral effector with CDC42/Rac GTPase-activating protein activity. J. Biol. Chem. 1995, 270, 22473–22477. [Google Scholar] [CrossRef] [Green Version]
- Awasthi, S.; Cheng, J.Z.; Singhal, S.S.; Pandya, U.; Sharma, R.; Singh, S.V.; Zimniak, P.; Awasthi, Y.C. Functional reassembly of ATP-dependent xenobiotic transport by the N- and C-terminal domains of RLIP76 and identification of ATP binding sequences. Biochemistry 2001, 40, 4159–4168. [Google Scholar] [CrossRef]
- Fenwick, R.B.; Campbell, L.J.; Rajasekar, K.; Prasannan, S.; Nietlispach, D.; Camonis, J.; Owen, D.; Mott, H.R. The RalB-RLIP76 Complex Reveals a Novel Mode of Ral-Effector Interaction. Structure 2010, 18, 985–995. [Google Scholar] [CrossRef] [Green Version]
- Matsubara, K.; Hinoi, T.; Koyama, S.; Kikuchi, A. The post-translational modifications of Ral and Rac1 are important for the action of Ral-binding protein 1, a putative effector protein of Ral. FEBS Lett. 1997, 410, 169–174. [Google Scholar] [CrossRef] [Green Version]
- Park, S.H.; Weinberg, R.A. A putative effector of Ral has homology to Rho/Rac GTPase activating proteins. Oncogene 1995, 11, 2349–2355. [Google Scholar]
- Neel, N.F.; Rossman, K.L.; Martin, T.D.; Hayes, T.K.; Yeh, J.J.; Der, C.J. The RalB Small GTPase Mediates Formation of Invadopodia through a GTPase-Activating Protein-Independent Function of the RalBP1/RLIP76 Effector. Mol. Cell. Biol. 2012, 32, 1374–1386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chien, Y.; Kim, S.; Bumeister, R.; Loo, Y.M.; Kwon, S.W.; Johnson, C.L.; Balakireva, M.G.; Romeo, Y.; Kopelovich, L.; Gale, M.; et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 2006, 127, 157–170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hazelett, C.C.; Yeaman, C. Sec5 and Exo84 Mediate Distinct Aspects of RalA-Dependent Cell Polarization. PLoS ONE 2012, 7, e39602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camonis, J.H.; White, M.A. Ral GTPases: Corrupting the exocyst in cancer cells. Trends Cell Biol. 2005, 15, 327–332. [Google Scholar] [CrossRef]
- Li, G.; Han, L.; Chou, T.C.; Fujita, Y.; Arunachalam, L.; Xu, A.; Wong, A.; Chiew, S.K.; Wan, Q.; Wang, L.; et al. RalA and RalB function as the critical GTP sensors for GTP-dependent exocytosis. J. Neurosci. 2007, 27, 190–202. [Google Scholar] [CrossRef] [Green Version]
- Spiczka, K.S.; Yeaman, C. Ral-regulated interaction between Sec5 and paxillin targets Exocyst to focal complexes during cell migration. J. Cell Sci. 2008, 121, 2880–2891. [Google Scholar] [CrossRef] [Green Version]
- Oxford, G.; Owens, C.R.; Titus, B.J.; Foreman, T.L.; Herlevsen, M.C.; Smith, S.C.; Theodorescu, D. RalA and RalB: Antagonistic Relatives in Cancer Cell Migration. Cancer Res. 2005, 65, 7111–7120. [Google Scholar] [CrossRef] [Green Version]
- Rossé, C.; Hatzoglou, A.; Parrini, M.C.; White, M.A.; Chavrier, P.; Camonis, J. RalB mobilizes the exocyst to drive cell migration. Mol. Cell Biol. 2006, 26, 727–734. [Google Scholar] [CrossRef] [Green Version]
- De Gorter, D.J.J.; Reijmers, R.M.; Beuling, E.A.; Naber, H.P.H.; Kuil, A.; Kersten, M.J.; Pals, S.T.; Spaargaren, M. The small GTPase Ral mediates SDF-1–induced migration of B cells and multiple myeloma cells. Blood 2008, 111, 3364–3372. [Google Scholar] [CrossRef] [Green Version]
- Exton, J.H. Phospholipase D: Enzymology, mechanisms of regulation, and function. Physiol. Rev. 1997, 77, 303–320. [Google Scholar] [CrossRef]
- Balch, W.E.; Kahn, R.A.; Schwaninger, R. ADP-ribosylation factor is required for vesicular trafficking between the endoplasmic reticulum and the cis-Golgi compartment. J. Biol. Chem. 1992, 267, 13053–13061. [Google Scholar] [CrossRef]
- Rybko, V.A.; Knizhnik, A.V.; Komelkov, A.V.; Aushev, V.N.; Trukhanova, L.S.; Tchevkina, E.M. Different metastasis promotive potency of small G-proteins RalA and RalB in in vivo hamster tumor model. Cancer Cell Int. 2011, 11, 22. [Google Scholar] [CrossRef] [Green Version]
- Ghoroghi, S.; Mary, B.; Larnicol, A.; Asokan, N.; Klein, A.; Osmani, N.; Busnelli, I.; Delalande, F.; Paul, N.; Halary, S.; et al. Ral GTPases promote breast cancer metastasis by controlling biogenesis and organ targeting of exosomes. eLife 2021, 10, e61539. [Google Scholar] [CrossRef] [PubMed]
- Mizushima, N.; Ohsumi, Y.; Yoshimori, T. Autophagosome formation in mammalian cells. Cell Struct. Funct. 2002, 27, 421–429. [Google Scholar] [CrossRef] [Green Version]
- Bodemann, B.O.; Orvedahl, A.; Cheng, T.; Ram, R.R.; Ou, Y.H.; Formstecher, E.; Maiti, M.; Hazelett, C.C.; Wauson, E.M.; Balakireva, M.; et al. RalB and the exocyst mediate the cellular starvation response by direct activation of autophagosome assembly. Cell 2011, 144, 253–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sawamoto, K.; Winge, P.; Koyama, S.; Hirota, Y.; Yamada, C.; Miyao, S.; Yoshikawa, S.; Jin, M.H.; Kikuchi, A.; Okano, H. The Drosophila Ral GTPase regulates developmental cell shape changes through the Jun NH(2)-terminal kinase pathway. J. Cell Biol. 1999, 146, 361–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balakireva, M.; Rossé, C.; Langevin, J.; Chien, Y.C.; Gho, M.; Gonzy-Treboul, G.; Voegeling-Lemaire, S.; Aresta, S.; Lepesant, J.A.; Bellaiche, Y.; et al. The Ral/exocyst effector complex counters c-Jun N-terminal kinase-dependent apoptosis in Drosophila melanogaster. Mol. Cell Biol. 2006, 26, 8953–8963. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Ruiter, N.D.; Wolthuis, R.M.; van Dam, H.; Burgering, B.M.; Bos, J.L. Ras-dependent regulation of c-Jun phosphorylation is mediated by the Ral guanine nucleotide exchange factor-Ral pathway. Mol. Cell Biol. 2000, 20, 8480–8488. [Google Scholar] [CrossRef] [Green Version]
- Balda, M.S.; Matter, K. The tight junction protein ZO-1 and an interacting transcription factor regulate ErbB-2 expression. EMBO J. 2000, 19, 2024–2033. [Google Scholar] [CrossRef]
- Sourisseau, T.; Georgiadis, A.; Tsapara, A.; Ali, R.R.; Pestell, R.; Matter, K.; Balda, M.S. Regulation of PCNA and cyclin D1 expression and epithelial morphogenesis by the ZO-1-regulated transcription factor ZONAB/DbpA. Mol. Cell Biol. 2006, 26, 2387–2398. [Google Scholar] [CrossRef] [Green Version]
- Frankel, P.; Aronheim, A.; Kavanagh, E.; Balda, M.S.; Matter, K.; Bunney, T.D.; Marshall, C.J. RalA interacts with ZONAB in a cell density-dependent manner and regulates its transcriptional activity. EMBO J. 2005, 24, 54–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, I.; Steeg, P.S. Endocytosis: A pivotal pathway for regulating metastasis. Br. J. Cancer 2021, 124, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Lanzetti, L.; Di Fiore, P.P. Behind the Scenes: Endo/Exocytosis in the Acquisition of Metastatic Traits. Cancer Res. 2017, 77, 1813–1817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosesson, Y.; Mills, G.B.; Yarden, Y. Derailed endocytosis: An emerging feature of cancer. Nat. Rev. Cancer 2008, 8, 835–850. [Google Scholar] [CrossRef]
- Vieira, A.V.; Lamaze, C.; Schmid, S.L. Control of EGF receptor signaling by clathrin-mediated endocytosis. Science 1996, 274, 2086–2089. [Google Scholar] [CrossRef] [Green Version]
- De Donatis, A.; Comito, G.; Buricchi, F.; Vinci, M.C.; Parenti, A.; Caselli, A.; Camici, G.; Manao, G.; Ramponi, G.; Cirri, P. Proliferation versus migration in platelet-derived growth factor signaling: The key role of endocytosis. J. Biol. Chem. 2008, 283, 19948–19956. [Google Scholar] [CrossRef] [Green Version]
- Nakashima, S.; Morinaka, K.; Koyama, S.; Ikeda, M.; Kishida, M.; Okawa, K.; Iwamatsu, A.; Kishida, S.; Kikuchi, A. Small G protein Ral and its downstream molecules regulate endocytosis of EGF and insulin receptors. EMBO J. 1999, 18, 3629–3642. [Google Scholar] [CrossRef]
- Jiang, Y.; Sverdlov, M.S.; Toth, P.T.; Huang, L.S.; Du, G.; Liu, Y.; Natarajan, V.; Minshall, R.D. Phosphatidic Acid Produced by RalA-activated PLD2 Stimulates Caveolae-mediated Endocytosis and Trafficking in Endothelial Cells. J. Biol. Chem. 2016, 291, 20729–20738. [Google Scholar] [CrossRef] [Green Version]
- Nászai, M.; Bellec, K.; Yu, Y.; Román-Fernández, A.; Sandilands, E.; Johansson, J.; Campbell, A.D.; Norman, J.C.; Sansom, O.J.; Bryant, D.M.; et al. RAL GTPases mediate EGFR-driven intestinal stem cell proliferation and tumourigenesis. Elife 2021, 10, e63807. [Google Scholar] [CrossRef]
- Han, K.; Kim, M.H.; Seeburg, D.; Seo, J.; Verpelli, C.; Han, S.; Chung, H.S.; Ko, J.; Lee, H.W.; Kim, K.; et al. Regulated RalBP1 binding to RalA and PSD-95 controls AMPA receptor endocytosis and LTD. PLoS Biol. 2009, 7, e1000187. [Google Scholar] [CrossRef] [Green Version]
- Johansson, J.; Naszai, M.; Hodder, M.C.; Pickering, K.A.; Miller, B.W.; Ridgway, R.A.; Yu, Y.; Peschard, P.; Brachmann, S.; Campbell, A.D.; et al. RAL GTPases Drive Intestinal Stem Cell Function and Regeneration through Internalization of WNT Signalosomes. Cell Stem Cell 2019, 24, 592–607.e597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, M.; Vasioukhin, V. Cell polarity and cancer--cell and tissue polarity as a non-canonical tumor suppressor. J. Cell Sci. 2008, 121, 1141–1150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vermeer, P.D.; Einwalter, L.A.; Moninger, T.O.; Rokhlina, T.; Kern, J.A.; Zabner, J.; Welsh, M.J. Segregation of receptor and ligand regulates activation of epithelial growth factor receptor. Nature 2003, 422, 322–326. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yue, P.; Artym, V.V.; Mueller, S.C.; Guo, W. The role of the exocyst in matrix metalloproteinase secretion and actin dynamics during tumor cell invadopodia formation. Mol. Biol. Cell 2009, 20, 3763–3771. [Google Scholar] [CrossRef] [Green Version]
- Sakurai-Yageta, M.; Recchi, C.; Le Dez, G.; Sibarita, J.B.; Daviet, L.; Camonis, J.; D’Souza-Schorey, C.; Chavrier, P. The interaction of IQGAP1 with the exocyst complex is required for tumor cell invasion downstream of Cdc42 and RhoA. J. Cell Biol. 2008, 181, 985–998. [Google Scholar] [CrossRef] [Green Version]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Lopez, J.A.; Kwan, E.P.; Xie, L.; He, Y.; James, D.E.; Gaisano, H.Y. The RalA GTPase is a central regulator of insulin exocytosis from pancreatic islet beta cells. J. Biol. Chem. 2008, 283, 17939–17945. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.W.; Leto, D.; Chiang, S.H.; Wang, Q.; Saltiel, A.R. Activation of RalA is required for insulin-stimulated Glut4 trafficking to the plasma membrane via the exocyst and the motor protein Myo1c. Dev. Cell 2007, 13, 391–404. [Google Scholar] [CrossRef]
- Skorobogatko, Y.; Dragan, M.; Cordon, C.; Reilly, S.M.; Hung, C.W.; Xia, W.; Zhao, P.; Wallace, M.; Lackey, D.E.; Chen, X.W.; et al. RalA controls glucose homeostasis by regulating glucose uptake in brown fat. Proc. Natl. Acad. Sci. USA 2018, 115, 7819–7824. [Google Scholar] [CrossRef] [Green Version]
- Chang, Y.C.; Chi, L.H.; Chang, W.M.; Su, C.Y.; Lin, Y.F.; Chen, C.L.; Chen, M.H.; Chang, P.M.; Wu, A.T.; Hsiao, M. Glucose transporter 4 promotes head and neck squamous cell carcinoma metastasis through the TRIM24-DDX58 axis. J. Hematol. Oncol. 2017, 10, 11. [Google Scholar] [CrossRef] [Green Version]
- Gu, C.J.; Xie, F.; Zhang, B.; Yang, H.L.; Cheng, J.; He, Y.Y.; Zhu, X.Y.; Li, D.J.; Li, M.Q. High Glucose Promotes Epithelial-Mesenchymal Transition of Uterus Endometrial Cancer Cells by Increasing ER/GLUT4-Mediated VEGF Secretion. Cell Physiol. Biochem. 2018, 50, 706–720. [Google Scholar] [CrossRef] [PubMed]
- Shipitsin, M.; Feig, L.A. RalA but not RalB enhances polarized delivery of membrane proteins to the basolateral surface of epithelial cells. Mol. Cell Biol. 2004, 24, 5746–5756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, B.; Bogatcheva, G.; Washington, M.K.; Coffey, R.J. Transformation of polarized epithelial cells by apical mistrafficking of epiregulin. Proc. Natl. Acad. Sci. USA 2013, 110, 8960–8965. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.W.; Inoue, M.; Hsu, S.C.; Saltiel, A.R. RalA-exocyst-dependent recycling endosome trafficking is required for the completion of cytokinesis. J. Biol. Chem. 2006, 281, 38609–38616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holly, R.M.; Mavor, L.M.; Zuo, Z.; Blankenship, J.T. A rapid, membrane-dependent pathway directs furrow formation through RalA in the early Drosophila embryo. Development 2015, 142, 2316–2328. [Google Scholar] [CrossRef]
- Cascone, I.; Selimoglu, R.; Ozdemir, C.; Del Nery, E.; Yeaman, C.; White, M.; Camonis, J. Distinct roles of RalA and RalB in the progression of cytokinesis are supported by distinct RalGEFs. EMBO J. 2008, 27, 2375–2387. [Google Scholar] [CrossRef] [Green Version]
- Ganem, N.J.; Godinho, S.A.; Pellman, D. A mechanism linking extra centrosomes to chromosomal instability. Nature 2009, 460, 278–282. [Google Scholar] [CrossRef] [Green Version]
- Silkworth, W.T.; Nardi, I.K.; Scholl, L.M.; Cimini, D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE 2009, 4, e6564. [Google Scholar] [CrossRef] [Green Version]
- Storchova, Z.; Pellman, D. From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell Biol. 2004, 5, 45–54. [Google Scholar] [CrossRef]
- Fujiwara, T.; Bandi, M.; Nitta, M.; Ivanova, E.V.; Bronson, R.T.; Pellman, D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature 2005, 437, 1043–1047. [Google Scholar] [CrossRef]
- Zhang, X.; Chen, F.; Huang, P.; Wang, X.; Zhou, K.; Zhou, C.; Yu, L.; Peng, Y.; Fan, J.; Zhou, J.; et al. Exosome-depleted MiR-148a-3p derived from Hepatic Stellate Cells Promotes Tumor Progression via ITGA5/PI3K/Akt Axis in Hepatocellular Carcinoma. Int. J. Biol. Sci. 2022, 18, 2249–2260. [Google Scholar] [CrossRef] [PubMed]
- Machado, E.; White-Gilbertson, S.; van de Vlekkert, D.; Janke, L.; Moshiach, S.; Campos, Y.; Finkelstein, D.; Gomero, E.; Mosca, R.; Qiu, X.; et al. Regulated lysosomal exocytosis mediates cancer progression. Sci. Adv. 2015, 1, e1500603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Wang, D.; Ding, D.; Feng, Y.; Hou, R.; Liu, D.; Lin, C.; Gao, Y. The Role and Application of Exosomes in Gastric and Colorectal Cancer. Front. Pharmacol. 2021, 12, 825475. [Google Scholar] [CrossRef] [PubMed]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell. Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef]
- Gu, P.; Sun, M.; Li, L.; Yang, Y.; Jiang, Z.; Ge, Y.; Wang, W.; Mu, W.; Wang, H. Breast Tumor-Derived Exosomal MicroRNA-200b-3p Promotes Specific Organ Metastasis Through Regulating CCL2 Expression in Lung Epithelial Cells. Front. Cell Dev. Biol. 2021, 9, 657158. [Google Scholar] [CrossRef]
- Wang, M.; Qin, Z.; Wan, J.; Yan, Y.; Duan, X.; Yao, X.; Jiang, Z.; Li, W. Tumor-derived exosomes drive pre-metastatic niche formation in lung via modulating CCL1(+) fibroblast and CCR8(+) Treg cell interactions. Cancer Immunol. Immunother. 2022. [Google Scholar] [CrossRef]
- Fong, M.Y.; Zhou, W.; Liu, L.; Alontaga, A.Y.; Chandra, M.; Ashby, J.; Chow, A.; O’Connor, S.T.; Li, S.; Chin, A.R.; et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat. Cell Biol. 2015, 17, 183–194. [Google Scholar] [CrossRef] [Green Version]
- Webber, J.; Steadman, R.; Mason, M.D.; Tabi, Z.; Clayton, A. Cancer exosomes trigger fibroblast to myofibroblast differentiation. Cancer Res. 2010, 70, 9621–9630. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Deng, T.; Liu, R.; Bai, M.; Zhou, L.; Wang, X.; Li, S.; Yang, H.; Li, J.; Ning, T.; et al. Exosome-delivered EGFR regulates liver microenvironment to promote gastric cancer liver metastasis. Nat. Commun. 2017, 8, 15016. [Google Scholar] [CrossRef] [Green Version]
- Hyenne, V.; Apaydin, A.; Rodriguez, D.; Spiegelhalter, C.; Hoff-Yoessle, S.; Diem, M.; Tak, S.; Lefebvre, O.; Schwab, Y.; Goetz, J.G.; et al. RAL-1 controls multivesicular body biogenesis and exosome secretion. J. Cell Biol. 2015, 211, 27–37. [Google Scholar] [CrossRef] [Green Version]
- Hyenne, V.; Labouesse, M.; Goetz, J.G. The Small GTPase Ral orchestrates MVB biogenesis and exosome secretion. Small GTPases 2018, 9, 445–451. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vitale, N.; Mawet, J.; Camonis, J.; Regazzi, R.; Bader, M.F.; Chasserot-Golaz, S. The Small GTPase RalA controls exocytosis of large dense core secretory granules by interacting with ARF6-dependent phospholipase D1. J. Biol. Chem. 2005, 280, 29921–29928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, S.C.; Oxford, G.; Baras, A.S.; Owens, C.; Havaleshko, D.; Brautigan, D.L.; Safo, M.K.; Theodorescu, D. Expression of Ral GTPases, Their Effectors, and Activators in Human Bladder Cancer. Clin. Cancer Res. 2007, 13, 3803–3813. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Li, Y.; Luo, X.; Fei, J. Inhibition of small GTPase RalA regulates growth and arsenic-induced apoptosis in chronic myeloid leukemia (CML) cells. Cell. Signal. 2012, 24, 1134–1140. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Feng, M.; Yin, Z.; Luo, X.; Yang, J.; Li, Y.; Li, T.; Wang, R.; Fei, J. RalA, a GTPase targeted by miR-181a, promotes transformation and progression by activating the Ras-related signaling pathway in chronic myelogenous leukemia. Oncotarget 2016, 7, 20561–20573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eckfeldt, C.E.; Pomeroy, E.J.; Lee, R.D.W.; Hazen, K.S.; Lee, L.A.; Moriarity, B.S.; Largaespada, D.A. RALB provides critical survival signals downstream of Ras in acute myeloid leukemia. Oncotarget 2016, 7, 65147–65156. [Google Scholar] [CrossRef] [Green Version]
- Pomeroy, E.J.; Lee, L.A.; Lee, R.D.W.; Schirm, D.K.; Temiz, N.A.; Ma, J.; Gruber, T.A.; Diaz-Flores, E.; Moriarity, B.S.; Downing, J.R.; et al. Ras oncogene-independent activation of RALB signaling is a targetable mechanism of escape from NRAS(V12) oncogene addiction in acute myeloid leukemia. Oncogene 2017, 36, 3263–3273. [Google Scholar] [CrossRef] [Green Version]
- Seibold, M.; Stühmer, T.; Kremer, N.; Mottok, A.; Scholz, C.-J.; Schlosser, A.; Leich, E.; Holzgrabe, U.; Brünnert, D.; Barrio, S.; et al. RAL GTPases mediate multiple myeloma cell survival and are activated independently of oncogenic RAS. Haematologica 2019, 105, 2316–2326. [Google Scholar] [CrossRef]
- Gotoh, T.; Cai, D.; Tian, X.; Feig, L.A.; Lerner, A. p130Cas Regulates the Activity of AND-34, a Novel Ral, Rap1, and R-Ras Guanine Nucleotide Exchange Factor. J. Biol. Chem. 2000, 275, 30118–30123. [Google Scholar] [CrossRef] [Green Version]
- Yu, Y.; Feig, L.A. Involvement of R-Ras and Ral GTPases in estrogen-independent proliferation of breast cancer cells. Oncogene 2002, 21, 7557–7568. [Google Scholar] [CrossRef] [Green Version]
- Martin, T.D.; Samuel, J.C.; Routh, E.D.; Der, C.J.; Yeh, J.J. Activation and involvement of Ral GTPases in colorectal cancer. Cancer Res. 2011, 71, 206–215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khawaja, H.; Campbell, A.; Roberts, J.Z.; Javadi, A.; O’Reilly, P.; McArt, D.; Allen, W.L.; Majkut, J.; Rehm, M.; Bardelli, A.; et al. RALB GTPase: A critical regulator of DR5 expression and TRAIL sensitivity in KRAS mutant colorectal cancer. Cell Death Dis. 2020, 11, 930. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Zhang, W.; Wang, L.; Liang, W.; Cai, A.; Gao, Y.; Chen, L. RCC2 Interacts with Small GTPase RalA and Regulates Cell Proliferation and Motility in Gastric Cancer. Onco. Targets Ther. 2020, 13, 3093–3103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nanami, T.; Hoshino, I.; Ito, M.; Yajima, S.; Oshima, Y.; Suzuki, T.; Shiratori, F.; Nabeya, Y.; Funahashi, K.; Shimada, H. Prevalence of autoantibodies against Ras-like GTPases, RalA, in patients with gastric cancer. Mol. Clin. Oncol. 2020, 13, 28. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Chen, Y.; Liu, S.; Qiu, S.; Gao, S.; Huang, X.; Zhang, J.; Peng, X.; Qiani, W.; Zhang, J.Y. Immunogenicity of Ra1A and its Tissue-Specific Expression in Hepatocellular Carcinoma. Int. J. Immunopathol. Pharmacol. 2009, 22, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Ezzeldin, M.; Borrego-Diaz, E.; Taha, M.; Esfandyari, T.; Wise, A.L.; Peng, W.; Rouyanian, A.; Asvadi Kermani, A.; Soleimani, M.; Patrad, E.; et al. RalA signaling pathway as a therapeutic target in hepatocellular carcinoma (HCC). Mol. Oncol. 2014, 8, 1043–1053. [Google Scholar] [CrossRef]
- Male, H.; Patel, V.; Jacob, M.A.; Borrego-Diaz, E.; Wang, K.; Young, D.A.; Wise, A.L.; Huang, C.; Van Veldhuizen, P.; O’Brien-Ladner, A.; et al. Inhibition of RalA signaling pathway in treatment of non-small cell lung cancer. Lung Cancer 2012, 77, 252–259. [Google Scholar] [CrossRef]
- Peschard, P.; McCarthy, A.; Leblanc-Dominguez, V.; Yeo, M.; Guichard, S.; Stamp, G.; Marshall, C.J. Genetic deletion of RALA and RALB small GTPases reveals redundant functions in development and tumorigenesis. Curr. Biol. 2012, 22, 2063–2068. [Google Scholar] [CrossRef] [Green Version]
- Biondini, M.; Duclos, G.; Meyer-Schaller, N.; Silberzan, P.; Camonis, J.; Parrini, M.C. RalB regulates contractility-driven cancer dissemination upon TGFβ stimulation via the RhoGEF GEF-H1. Sci. Rep. 2015, 5, 11759. [Google Scholar] [CrossRef] [Green Version]
- Zipfel, P.A.; Brady, D.C.; Kashatus, D.F.; Ancrile, B.D.; Tyler, D.S.; Counter, C.M. Ral activation promotes melanomagenesis. Oncogene 2010, 29, 4859–4864. [Google Scholar] [CrossRef] [Green Version]
- Bodempudi, V.; Yamoutpoor, F.; Pan, W.; Dudek, A.Z.; Esfandyari, T.; Piedra, M.; Babovick-Vuksanovic, D.; Woo, R.A.; Mautner, V.F.; Kluwe, L.; et al. Ral Overactivation in Malignant Peripheral Nerve Sheath Tumors. Mol. Cell. Biol. 2009, 29, 3964–3974. [Google Scholar] [CrossRef] [Green Version]
- Ganapathy, S.; Fagman, J.B.; Shen, L.; Yu, T.; Zhou, X.; Dai, W.; Makriyannis, A.; Chen, C. Ral A, via activating the mitotic checkpoint, sensitizes cells lacking a functional Nf1 to apoptosis in the absence of protein kinase C. Oncotarget 2016, 7, 84326–84337. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Terai, K.; Peng, W.; Rouyanian, A.; Liu, J.; Roby, K.F.; Wise, A.L.; Ezzeldin, M.; Larson, J.; Woo, R.A. The role of RalA in biology and therapy of ovarian cancer. Oncotarget 2013, 5, 1–14. [Google Scholar]
- Gong, S.; Chen, Y.; Meng, F.; Zhang, Y.; Wu, H.; Li, C.; Zhang, G. RCC2, a regulator of the RalA signaling pathway, is identified as a novel therapeutic target in cisplatin-resistant ovarian cancer. FASEB J. 2019, 33, 5350–5365. [Google Scholar] [CrossRef] [PubMed]
- Beel, S.; Kolloch, L.; Apken, L.H.; Jürgens, L.; Bolle, A.; Sudhof, N.; Ghosh, S.; Wardelmann, E.; Meisterernst, M.; Steinestel, K.; et al. κB-Ras and Ral GTPases regulate acinar to ductal metaplasia during pancreatic adenocarcinoma development and pancreatitis. Nat. Commun. 2020, 11, 3409. [Google Scholar] [CrossRef]
- Lim, K.-H.; O’Hayer, K.; Adam, S.J.; Kendall, S.D.; Campbell, P.M.; Der, C.J.; Counter, C.M. Divergent Roles for RalA and RalB in Malignant Growth of Human Pancreatic Carcinoma Cells. Curr. Biol. 2006, 16, 2385–2394. [Google Scholar] [CrossRef] [Green Version]
- Seguin, L.; Kato, S.; Franovic, A.; Camargo, M.F.; Lesperance, J.; Elliott, K.C.; Yebra, M.; Mielgo, A.; Lowy, A.M.; Husain, H.; et al. An integrin β3–KRAS–RalB complex drives tumour stemness and resistance to EGFR inhibition. Nat. Cell Biol. 2014, 16, 457–468. [Google Scholar] [CrossRef] [Green Version]
- Oeckinghaus, A.; Postler, T.S.; Rao, P.; Schmitt, H.; Schmitt, V.; Grinberg-Bleyer, Y.; Kühn, L.I.; Gruber, C.W.; Lienhard, G.E.; Ghosh, S. κB-Ras proteins regulate both NF-κB-dependent inflammation and Ral-dependent proliferation. Cell Rep. 2014, 8, 1793–1807. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Zheng, Y.; Chen, Q.; Dai, Y.; Li, T. MicroRNA-139 inhibits pancreatic-cancer carcinogenesis by suppressing RalB via the Ral/RAC/PI3K pathway. Arch. Biochem. Biophys. 2021, 704, 108719. [Google Scholar] [CrossRef]
- Yin, J.; Pollock, C.; Tracy, K.; Chock, M.; Martin, P.; Oberst, M.; Kelly, K. Activation of the RalGEF/Ral Pathway Promotes Prostate Cancer Metastasis to Bone. Mol. Cell. Biol. 2007, 27, 7538–7550. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Lang, C.; Wu, Z.; Dai, Y.; He, S.; Guo, W.; Huang, S.; Du, H.; Ren, D.; Peng, X. MAZ promotes prostate cancer bone metastasis through transcriptionally activating the KRas-dependent RalGEFs pathway. J. Exp. Clin. Cancer Res. 2019, 38, 391. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Muders, M.H.; Li, J.; Rinaldo, F.; Tindall, D.J.; Datta, K. Loss of NKX3.1 Favors Vascular Endothelial Growth Factor-C Expression in Prostate Cancer. Cancer Res. 2008, 68, 8770–8778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinaldo, F.; Li, J.; Wang, E.; Muders, M.; Datta, K. RalA regulates vascular endothelial growth factor-C (VEGF-C) synthesis in prostate cancer cells during androgen ablation. Oncogene 2007, 26, 1731–1738. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Dai, L.; Lei, N.; Xing, M.; Li, P.; Luo, C.; Casiano, C.A.; Zhang, J.-Y. Evaluation and characterization of anti-RalA autoantibody as a potential serum biomarker in human prostate cancer. Oncotarget 2016, 7, 43546–43556. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhu, L.; Zong, H.; Song, X.; Wang, L.; Wang, X.; Yang, D.; Wang, J. Subcellular localization of aquaporin 3 in prostate cancer is regulated by RalA. Oncol. Rep. 2018, 39, 2171–2177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Zhang, Y.; Kim, W.J.; Daaka, Y. PGE2 promotes renal carcinoma cell invasion through activated RalA. Oncogene 2013, 32, 1408–1415. [Google Scholar] [CrossRef] [Green Version]
- Zuo, X.; Kwon, S.H.; Janech, M.G.; Dang, Y.; Lauzon, S.D.; Fogelgren, B.; Polgar, N.; Lipschutz, J.H. Primary cilia and the exocyst are linked to urinary extracellular vesicle production and content. J. Biol. Chem. 2019, 294, 19099–19110. [Google Scholar] [CrossRef]
- Ward, Y.; Wang, W.; Woodhouse, E.; Linnoila, I.; Liotta, L.; Kelly, K. Signal Pathways Which Promote Invasion and Metastasis: Critical and Distinct Contributions of Extracellular Signal-Regulated Kinase and Ral-Specific Guanine Exchange Factor Pathways. Mol. Cell. Biol. 2001, 21, 5958–5969. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Chan, L.; Fiji, H.D.; Dahl, R.; Kwon, O.; Tamanoi, F. In vivo antitumor effect of a novel inhibitor of protein geranylgeranyltransferase-I. Mol. Cancer Ther. 2009, 8, 1218–1226. [Google Scholar] [CrossRef] [Green Version]
- Hamada, M.; Miki, T.; Iwai, S.; Shimizu, H.; Yura, Y. Involvement of RhoA and RalB in geranylgeranyltransferase I inhibitor-mediated inhibition of proliferation and migration of human oral squamous cell carcinoma cells. Cancer. Chemother. Pharmacol. 2011, 68, 559–569. [Google Scholar] [CrossRef]
- Ginn, K.F.; Fangman, B.; Terai, K.; Wise, A.; Ziazadeh, D.; Shah, K.; Gartrell, R.; Ricke, B.; Kimura, K.; Mathur, S.; et al. RalA is overactivated in medulloblastoma. J. Neurooncol. 2016, 130, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Dadon, T.; Chenna, V.; Yabuuchi, S.; Bannerji, R.; Booher, R.; Strack, P.; Azad, N.; Nelkin, B.D.; Maitra, A. Combined Inhibition of Cyclin-Dependent Kinases (Dinaciclib) and AKT (MK-2206) Blocks Pancreatic Tumor Growth and Metastases in Patient-Derived Xenograft Models. Mol. Cancer Ther. 2015, 14, 1532–1539. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Murphy, A.G.; Zahurak, M.; Shah, M.; Weekes, C.D.; Hansen, A.; Siu, L.L.; Spreafico, A.; LoConte, N.; Anders, N.M.; Miles, T.; et al. A Phase I Study of Dinaciclib in Combination with MK-2206 in Patients with Advanced Pancreatic Cancer. Clin. Transl. Sci. 2020, 13, 1178–1188. [Google Scholar] [CrossRef] [PubMed]
- Tomita, M.; Mori, N. Aurora A selective inhibitor MLN8237 suppresses the growth and survival of HTLV-1-infected T-cells in vitro. Cancer Sci. 2010, 101, 1204–1211. [Google Scholar] [CrossRef]
- Görgün, G.; Calabrese, E.; Hideshima, T.; Ecsedy, J.; Perrone, G.; Mani, M.; Ikeda, H.; Bianchi, G.; Hu, Y.; Cirstea, D.; et al. A novel Aurora-A kinase inhibitor MLN8237 induces cytotoxicity and cell-cycle arrest in multiple myeloma. Blood 2010, 115, 5202–5213. [Google Scholar] [CrossRef] [Green Version]
- Asteriti, I.A.; Giubettini, M.; Lavia, P.; Guarguaglini, G. Aurora-A inactivation causes mitotic spindle pole fragmentation by unbalancing microtubule-generated forces. Mol. Cancer 2011, 10, 131. [Google Scholar] [CrossRef] [Green Version]
- Inchanalkar, S.; Deshpande, N.U.; Kasherwal, V.; Jayakannan, M.; Balasubramanian, N. Polymer Nanovesicle-Mediated Delivery of MLN8237 Preferentially Inhibits Aurora Kinase A To Target RalA and Anchorage-Independent Growth in Breast Cancer Cells. Mol. Pharm. 2018, 15, 3046–3059. [Google Scholar] [CrossRef]
- Yan, C.; Liu, D.; Li, L.; Wempe, M.F.; Guin, S.; Khanna, M.; Meier, J.; Hoffman, B.; Owens, C.; Wysoczynski, C.L.; et al. Discovery and characterization of small molecules that target the GTPase Ral. Nature 2014, 515, 443–447. [Google Scholar] [CrossRef] [Green Version]
| Cancer Subtype | RALA Role | RALB Role |
|---|---|---|
| Bladder cancer | RALA inhibits migration [97] | Phosphorylation of RALB is required for tumor growth and metastasis [23] and migration [97] |
| Blood Cancers | Upregulated and supports proliferation in CML [154] | May promote AML colony formation [156], regulates migration in MM [99] |
| Breast cancer | Involved in tamoxifen resistance [159], required for tumor growth, invasion, and metastasis [22] | Opposes proliferation and tumor growth [22], promotes metastasis [103] |
| Colorectal cancer | Stable RALA knockdown reduced colony formation via Exo84 [161] | Stable RALB knockdown increased colony formation via SEC5 [161], transient knockdown increased cell death and RALB is upregulated in the CRIS-B subtype [162] |
| Gastric cancer | Interacts with RCC2 and the MAPK/JNK pathway [163] | Limited data |
| Hepatic Cancer | Upregulated and activated in cancer tissue [47,66,174], increases migration and stemness [166] | Limited data, does not appear to be upregulated [166] |
| Pancreatic cancer | Required for tumor establish- ment [176], involved in anchorage-independent growth [84] | Involved in invasion [91] |
| Lung cancer | Important for tumorigenesis, proliferation, and invasion [167] | Important for invasion [188], cell cycle regulation [56], and migration [169] |
| Melanoma | Disruption of either RALA or RALB decreases tumor growth [170] | Disruption of either RALA or RALB decreases tumor growth [170] |
| Nerve sheath tumors | Associated with invasion [171] and apoptosis [172] | Limited data, upstream inhibition of RALB induces apoptosis when NF1 is decreased [172] |
| Ovarian cancer | GTP-RALA is overexpressed in cancerous cells, important in growth and invasion [173] | Limited data, GTP-RALB is not overexpressed in cancerous cells [173] |
| Prostate cancer | Central to invasion and migration [93], promotes prostate to bone metastasis [180,181] | Limited data |
| Renal cancer | Required for invasion [186] | Limited data, not required for invasion [186] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Richardson, D.S.; Spehar, J.M.; Han, D.T.; Chakravarthy, P.A.; Sizemore, S.T. The RAL Enigma: Distinct Roles of RALA and RALB in Cancer. Cells 2022, 11, 1645. https://doi.org/10.3390/cells11101645
Richardson DS, Spehar JM, Han DT, Chakravarthy PA, Sizemore ST. The RAL Enigma: Distinct Roles of RALA and RALB in Cancer. Cells. 2022; 11(10):1645. https://doi.org/10.3390/cells11101645
Chicago/Turabian StyleRichardson, Dillon S., Jonathan M. Spehar, David T. Han, Prathik A. Chakravarthy, and Steven T. Sizemore. 2022. "The RAL Enigma: Distinct Roles of RALA and RALB in Cancer" Cells 11, no. 10: 1645. https://doi.org/10.3390/cells11101645
APA StyleRichardson, D. S., Spehar, J. M., Han, D. T., Chakravarthy, P. A., & Sizemore, S. T. (2022). The RAL Enigma: Distinct Roles of RALA and RALB in Cancer. Cells, 11(10), 1645. https://doi.org/10.3390/cells11101645





