Effect of Thrombin on the Metabolism and Function of Murine Macrophages
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Monocyte Isolation and Culture
2.3. Macrophage Polarization
2.4. Immunoblotting
2.5. RT-qPCR
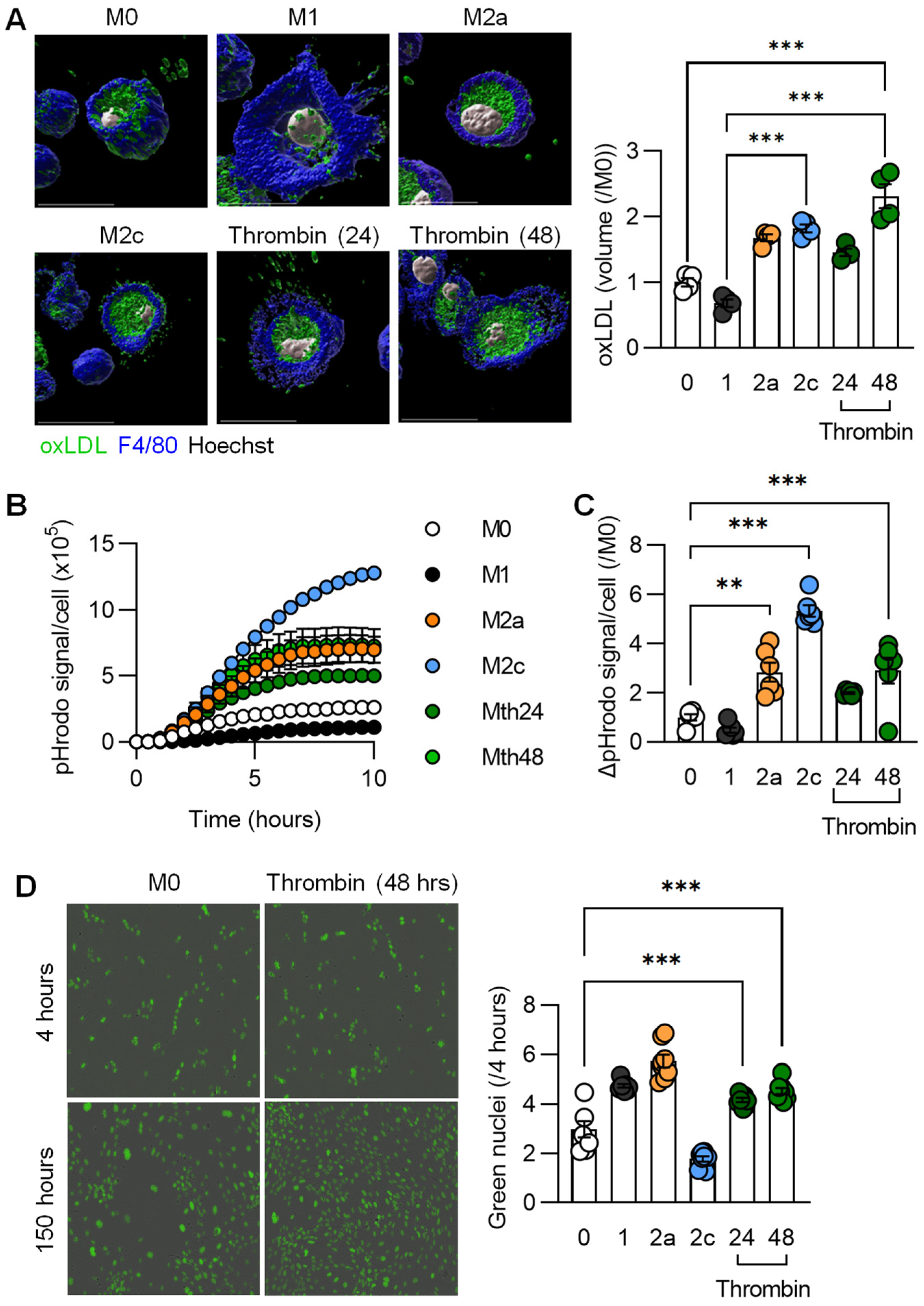
2.6. Phagocytosis Assays
2.7. FACS Analysis
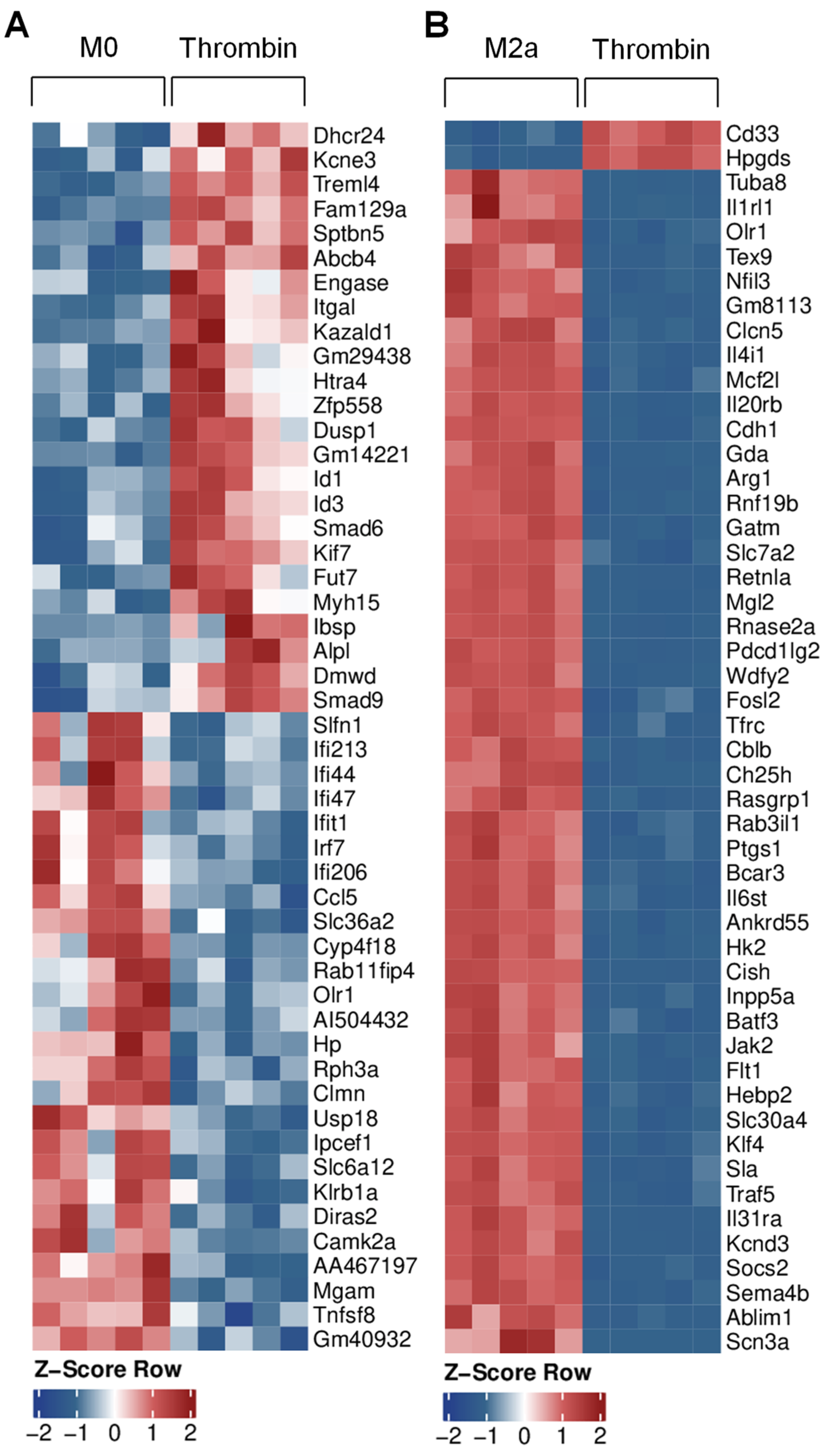
2.8. RNA Sequencing
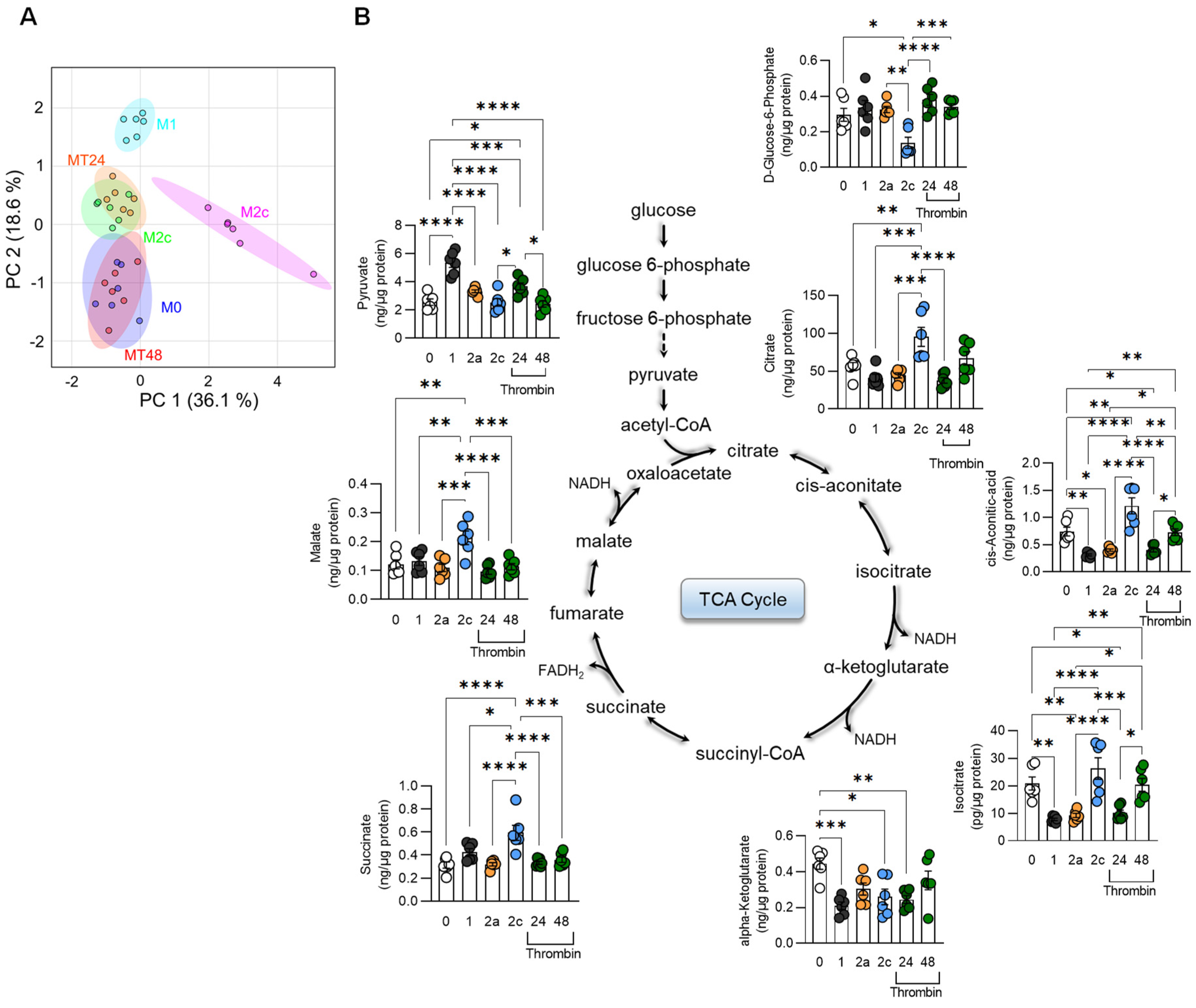
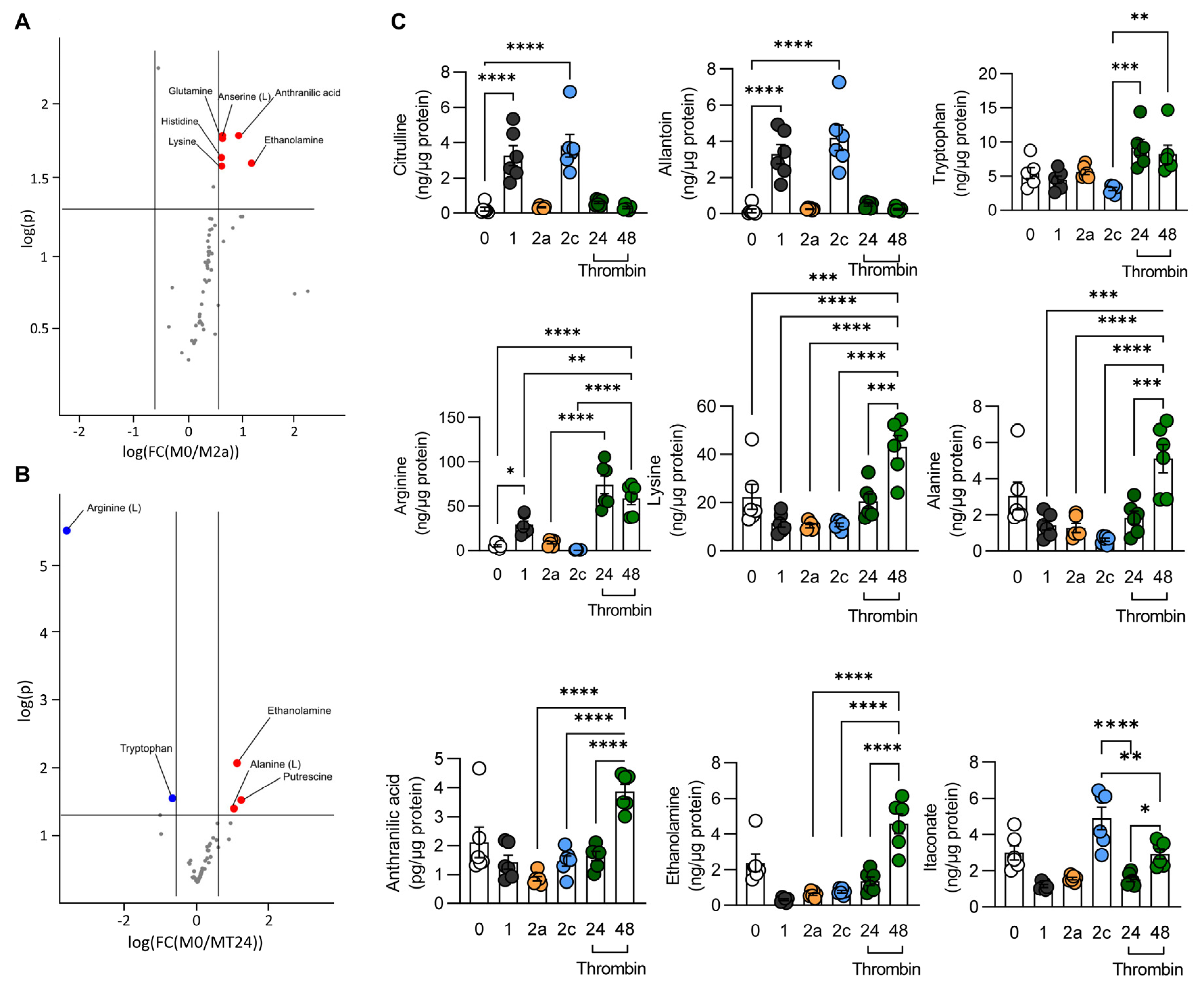
2.9. Metabolomics
2.9.1. Amino Acid Analyses
2.9.2. TCA Cycle
2.10. Immunofluorescence
2.11. Endothelial Cell Proliferation
2.12. Statistical Analyses
3. Results
3.1. Effects of Thrombin on Macrophage Gene Expression
3.2. Metabolic Characterization of Thrombin-Polarized Macrophages
3.3. Functional Characterization of Thrombin-Polarized Macrophages
3.4. Transcriptional Characterization of Thrombin-Polarized Macrophages
3.5. Impact of SMOC1 on Thrombin Polarized Macrophages
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Popović, M.; Smiljanić, K.; Dobutović, B.; Syrovets, T.; Simmet, T.; Isenović, E.R. Thrombin and vascular inflammation. Mol. Cell. Biochem. 2012, 359, 301–313. [Google Scholar] [CrossRef] [PubMed]
- Hirano, K. The roles of proteinase-activated receptors in the vascular physiology and pathophysiology. Arter. Thromb. Vasc. Biol. 2007, 27, 27–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Posma, J.J.N.; Posthuma, J.J.; Spronk, H.M.H. Coagulation and non-coagulation effects of thrombin. J. Thromb. Haemost. 2016, 14, 1908–1916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Candia, E.; Hall, S.W.; Rutella, S.; Landolfi, R.; Andrews, R.K.; de Cristofaro, R. Binding of thrombin to glycoprotein Ib accelerates the hydrolysis of Par-1 on intact platelets. J. Biol. Chem. 2001, 276, 4692–4698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogatcheva, N.V.; Garcia, J.G.N.; Verin, A.D. Molecular mechanisms of thrombin-induced endothelial cell permeability. Biochemistry 2002, 67, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Kyselova, A.; Elgheznawy, A.; Wittig, I.; Heidler, J.; Mann, A.W.; Ruf, W.; Fleming, I.; Randriamboavonjy, V. Platelet-derived calpain cleaves the endothelial protease-activated receptor 1 to induce vascular inflammation in diabetes. Basic. Res. Cardiol. 2020, 115, 75. [Google Scholar] [CrossRef]
- Szaba, F.M.; Smiley, S.T. Roles for thrombin and fibrin(ogen) in cytokine/chemokine production and macrophage adhesion in vivo. Blood 2002, 99, 1053–1059. [Google Scholar] [CrossRef] [Green Version]
- Cantrell, R.; Palumbo, J.S. The thrombin–inflammation axis in cancer progression. Thromb. Res. 2020, 191, S117–S122. [Google Scholar] [CrossRef]
- Snyder, K.M.; Kessler, C.M. The pivotal role of thrombin in cancer biology and tumorigenesis. Semin. Thromb. Hemost. 2008, 34, 734–741. [Google Scholar] [CrossRef] [Green Version]
- Hara, T.; Phuong, P.T.; Fukuda, D.; Yamaguchi, K.; Murata, C.; Nishimoto, S.; Yagi, S.; Kusunose, K.; Yamada, H.; Soeki, T.; et al. Protease-activated receptor-2 plays a critical role in vascular inflammation and atherosclerosis in apolipoprotein E-deficient mice. Circulation 2018, 138, 1706–1719. [Google Scholar] [CrossRef]
- Jones, S.M.; Mann, A.; Conrad, K.; Saum, K.; Hall, D.E.; McKinney, L.M.; Robbins, N.; Thompson, J.; Peairs, A.D.; Camerer, E.; et al. PAR2 (protease-activated receptor 2) deficiency attenuates atherosclerosis in mice. Arter. Thromb. Vasc. Biol. 2018, 38, 1271–1282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Zambrano, M.; Rodriguez-Montesinos, J.; Crespo-Avilan, G.E.; Muñoz-Vega, M.; Preissner, K.T. Thrombin promotes macrophage polarization into M1-like phenotype to induce inflammatory Responses. Thromb. Haemost. 2020, 120, 658–670. [Google Scholar] [CrossRef]
- Chen, L.; Gao, B.; Zhang, Y.; Lu, H.; Li, X.; Pan, L.; Yin, L.; Zhi, X. PAR2 promotes M1 macrophage polarization and inflammation via FOXO1 pathway. J. Cell. Biochem. 2019, 120, 9799–9809. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, H.; Leonard, H.; Chen, D.; Lawrence, T.; Robson, M.; Goossens, P.; McVey, J.H.; Dorling, A. PAR-1 signaling on macrophages is required for effective in vivo delayed-type hypersensitivity responses. iScience 2021, 24, 101981. [Google Scholar] [CrossRef] [PubMed]
- García-González, G.; Sánchez-González, A.; Hernández-Bello, R.; González, G.M.; Franco-Molina, M.A.; Coronado-Cerda, E.E.; Palma-Nicolás, J.P. Triggering of protease-activated receptors (PARs) induces alternative M2 macrophage polarization with impaired plasticity. Mol. Immunol. 2019, 114, 278–288. [Google Scholar] [CrossRef]
- White, M.J.V.; Gomer, R.H. Trypsin, tryptase, and thrombin polarize macrophages towards a pro-fibrotic M2a phenotype. PLoS ONE 2015, 10, e0138748. [Google Scholar] [CrossRef] [Green Version]
- Delgado Lagos, F.; Elgheznawy, A.; Kyselova, A.; Meyer Zu Heringdorf, D.; Ratiu, C.; Randriamboavonjy, V.; Mann, A.W.; Fisslthaler, B.; Siragusa, M.; Fleming, I. Secreted modular calcium-binding protein 1 binds and activates thrombin to account for platelet hyperreactivity in diabetes. Blood 2021, 137, 1641–1651. [Google Scholar] [CrossRef]
- Fleming, I.; Fisslthaler, B.; Dixit, M.; Busse, R. Role of PECAM-1 in the shear-stress-induced activation of Akt and the endothelial nitric oxide synthase (eNOS) in endothelial cells. J. Cell Sci. 2005, 118, 4103–4111. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- UniProt Consortium. Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res. 2014, 42, D191–D198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, T.; Gan, S.; Zhu, Q.; Dai, D.; Li, N.; Wang, H.; Chen, X.; Hou, D.; Wang, Y.; Pan, Q.; et al. Modulation of M2 macrophage polarization by the crosstalk between Stat6 and Trim24. Nat. Commun. 2019, 10, 4353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mantovani, A.; Sica, A.; Sozzani, S.; Allavena, P.; Vecchi, A.; Locati, M. The chemokine system in diverse forms of macrophage activation and polarization. Trends. Immunol. 2004, 25, 677–686. [Google Scholar] [CrossRef]
- Yao, Y.; Xu, X.-H.; Jin, L. Macrophage polarization in physiological and pathological pregnancy. Front. Immunol. 2019, 10, 792. [Google Scholar] [CrossRef]
- Martinez, F.O.; Sica, A.; Mantovani, A.; Locati, M. Macrophage activation and polarization. Front. Biosci. 2008, 13, 453–461. [Google Scholar] [CrossRef] [Green Version]
- Ferrante, C.J.; Pinhal-Enfield, G.; Elson, G.; Cronstein, B.N.; Hasko, G.; Outram, S.; Leibovich, S.J. The adenosine-dependent angiogenic switch of macrophages to an M2-like phenotype is independent of interleukin-4 receptor alpha (IL-4Rα) signaling. Inflammation 2013, 36, 921–931. [Google Scholar] [CrossRef]
- Boada-Romero, E.; Martinez, J.; Heckmann, B.L.; Green, D.R. The clearance of dead cells by efferocytosis. Nat. Rev. Mol. Cell Biol. 2020, 21, 398–414. [Google Scholar] [CrossRef]
- Zippel, N.; Malik, R.A.; Frömel, T.; Popp, R.; Bess, E.; Strilic, B.; Wettschureck, N.; Fleming, I.; Fisslthaler, B. Transforming growth factor-β-activated kinase 1 regulates angiogenesis via AMP-activated protein kinase-α1 and redox balance in endothelial cells. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 2792–2799. [Google Scholar] [CrossRef] [Green Version]
- Abouzeid, H.; Boisset, G.; Favez, T.; Youssef, M.; Marzouk, I.; Shakankiry, N.; Bayoumi, N.; Descombes, P.; Agosti, C.; Munier, F.L.; et al. Mutations in the SPARC-related modular calcium-binding protein 1 gene, SMOC1, cause Waardenburg anophthalmia syndrome. Am. J. Hum. Genet. 2011, 88, 92–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okada, I.; Hamanoue, H.; Terada, K.; Tohma, T.; Megarbane, A.; Chouery, E.; Abou-Ghoch, J.; Jalkh, N.; Cogulu, O.; Ozkinay, F.; et al. SMOC1 is essential for ocular and limb development in humans and mice. Am. J. Hum. Genet. 2011, 88, 30–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-X.; Zhang, S.-X.; Wu, H.-J.; Rong, X.-L.; Guo, J. M2b macrophage polarization and its roles in diseases. J. Leukoc. Biol. 2019, 106, 345–358. [Google Scholar] [CrossRef] [Green Version]
- Orecchioni, M.; Ghosheh, Y.; Pramod, A.B.; Ley, K. Macrophage polarization: Different gene signatures in M1(LPS+) vs. classically and M2(LPS-) vs. alternatively activated macrophages. Front. Immunol. 2019, 10, 1084. [Google Scholar] [CrossRef]
- Collot-Teixeira, S.; Martin, J.; McDermott-Roe, C.; Poston, R.; McGregor, J.L. CD36 and macrophages in atherosclerosis. Cardiovasc. Res. 2007, 75, 468–477. [Google Scholar] [CrossRef]
- Tian, K.; Du, G.; Wang, X.; Wu, X.; Li, L.; Liu, W.; Wu, R. MMP-9 secreted by M2-type macrophages promotes Wilms’ tumour metastasis through the PI3K/AKT pathway. Mol. Biol. Rep. 2022, 1–12. [Google Scholar] [CrossRef]
- Saha, S.; Shalova, I.N.; Biswas, S.K. Metabolic regulation of macrophage phenotype and function. Immunol. Rev. 2017, 280, 102–111. [Google Scholar] [CrossRef]
- Yamashita, A.; Hayashi, Y.; Nemoto-Sasaki, Y.; Ito, M.; Oka, S.; Tanikawa, T.; Waku, K.; Sugiura, T. Acyltransferases and transacylases that determine the fatty acid composition of glycerolipids and the metabolism of bioactive lipid mediators in mammalian cells and model organisms. Prog. Lipid. Res. 2014, 53, 18–81. [Google Scholar] [CrossRef]
- Rubio, J.M.; Astudillo, A.M.; Casas, J.; Balboa, M.A.; Balsinde, J. Regulation of phagocytosis in macrophages by membrane ethanolamine plasmalogens. Front. Immunol. 2018, 9, 1723. [Google Scholar] [CrossRef]
- O’Neill, L.A.J.; Artyomov, M.N. Itaconate: The poster child of metabolic reprogramming in macrophage function. Nat. Rev. Immunol. 2019, 19, 273–281. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Alexander, P.B.; Wang, X.-F. TGF-β family signaling in the control of cell proliferation and survival. Cold Spring Harb. Perspect. Biol. 2017, 9, a022145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imamura, T.; Takase, M.; Nishihara, A.; Oeda, E.; Hanai, J.; Kawabata, M.; Miyazono, K. Smad6 inhibits signalling by the TGF-beta superfamily. Nature 1997, 389, 622–626. [Google Scholar] [CrossRef] [PubMed]
- Tsukamoto, S.; Mizuta, T.; Fujimoto, M.; Ohte, S.; Osawa, K.; Miyamoto, A.; Yoneyama, K.; Murata, E.; Machiya, A.; Jimi, E.; et al. Smad9 is a new type of transcriptional regulator in bone morphogenetic protein signaling. Sci. Rep. 2014, 4, 7596. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Ming, X.-F. Functions of arginase isoforms in macrophage inflammatory responses: Impact on cardiovascular diseases and metabolic disorders. Front. Immunol. 2014, 5, 533. [Google Scholar] [CrossRef] [Green Version]
- Yue, Y.; Huang, W.; Liang, J.; Guo, J.; Ji, J.; Yao, Y.; Zheng, M.; Cai, Z.; Lu, L.; Wang, J. IL4I1 is a novel regulator of M2 macrophage polarization that can inhibit T cell activation via L-tryptophan and arginine depletion and IL-10 production. PLoS ONE 2015, 10, e0142979. [Google Scholar] [CrossRef]
- Liao, X.; Sharma, N.; Kapadia, F.; Zhou, G.; Lu, Y.; Hong, H.; Paruchuri, K.; Mahabeleshwar, G.H.; Dalmas, E.; Venteclef, N.; et al. Krüppel-like factor 4 regulates macrophage polarization. J. Clin. Investig. 2011, 121, 2736–2749. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Li, M.; Sanchez, E.; Soof, C.M.; Bujarski, S.; Ng, N.; Cao, J.; Hekmati, T.; Zahab, B.; Nosrati, J.D.; et al. JAK1/2 pathway inhibition suppresses M2 polarization and overcomes resistance of myeloma to lenalidomide by reducing TRIB1, MUC1, CD44, CXCL12, and CXCR4 expression. Br. J. Haematol. 2020, 188, 283–294. [Google Scholar] [CrossRef]
- Wilson, H.M. SOCS proteins in macrophage polarization and function. Front. Immunol. 2014, 5, 357. [Google Scholar] [CrossRef] [Green Version]
- Gersdorff, N.; Muller, M.; Schall, A.; Miosge, N. Secreted modular calcium-binding protein-1 localization during mouse embryogenesis. Histochem. Cell. Biol. 2006, 126, 705–712. [Google Scholar] [CrossRef]
- Vannahme, C.; Smyth, N.; Miosge, N.; Gosling, S.; Frie, C.; Paulsson, M.; Maurer, P.; Hartmann, U. Characterization of SMOC-1, a novel modular calcium-binding protein in basement membranes. J. Biol. Chem. 2002, 277, 37977–37986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradshaw, A.D. Diverse biological functions of the SPARC family of proteins. Int. J. Biochem. Cell Biol. 2012, 44, 480–488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, Y.A.; Lim, J.; Kim, K.M.; Acharya, B.; Cho, J.Y.; Bae, Y.C.; Shin, H.I.; Kim, S.Y.; Park, E.K. Secretome analysis of human BMSCs and identification of SMOC1 as an important ECM protein in osteoblast differentiation. J. Proteome. Res. 2010, 9, 2946–2956. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Haque, R.U.; Dammer, E.B.; Duong, D.M.; Ping, L.; Johnson, E.C.B.; Lah, J.J.; Levey, A.I.; Seyfried, N.T. Targeted mass spectrometry to quantify brain-derived cerebrospinal fluid biomarkers in Alzheimer’s disease. Clin. Proteom. 2020, 17, 19. [Google Scholar] [CrossRef] [PubMed]
- Sathe, G.; Albert, M.; Darrow, J.; Saito, A.; Troncoso, J.; Pandey, A.; Moghekar, A. Quantitative proteomic analysis of the frontal cortex in Alzheimer’s disease. J. Neurochem. 2020, 156, 988–1002. [Google Scholar] [CrossRef]
- Bai, B.; Wang, X.; Li, Y.; Chen, P.-C.; Yu, K.; Dey, K.K.; Yarbro, J.M.; Han, X.; Lutz, B.M.; Rao, S.; et al. Deep multilayer brain proteomics identifies molecular networks in Alzheimer’s disease progression. Neuron 2020, 105, 975–991. [Google Scholar] [CrossRef]
- Montgomery, M.K.; Bayliss, J.; Devereux, C.; Bezawork-Geleta, A.; Roberts, D.; Huang, C.; Schittenhelm, R.B.; Ryan, A.; Townley, S.L.; Selth, L.A.; et al. SMOC1 is a glucose-responsive hepatokine and therapeutic target for glycemic control. Sci. Transl. Med. 2020, 12, eaaz8048. [Google Scholar] [CrossRef]
- Ghodsian, N.; Gagnon, E.; Bourgault, J.; Gobeil, É.; Manikpurage, H.D.; Perrot, N.; Girard, A.; Mitchell, P.L.; Arsenault, B.J. Blood levels of the SMOC1 hepatokine are not causally linked with type 2 diabetes: A bidirectional mendelian randomization study. Nutrients 2021, 13, 4208. [Google Scholar] [CrossRef]
- Awwad, K.; Hu, J.; Shi, L.; Mangels, N.; Abdel Malik, R.; Zippel, N.; Fisslthaler, B.; Eble, J.A.; Pfeilschifter, J.; Popp, R.; et al. Role of secreted modular calcium binding protein 1 (SMOC1) in transforming growth factor β signaling and angiogenesis. Cardiovasc. Res. 2015, 106, 284–294. [Google Scholar] [CrossRef] [Green Version]
- You, D.; Qiao, Q.; Ono, K.; Wei, M.; Tan, W.; Wang, C.; Liu, Y.; Liu, G.; Zheng, M. miR-223-3p inhibits the progression of atherosclerosis via down-regulating the activation of MEK1/ERK1/2 in macrophages. Aging 2022, 14, 1865–1878. [Google Scholar] [CrossRef]
- Wang, J.; Bai, X.; Song, Q.; Fan, F.; Hu, Z.; Cheng, G.; Zhang, Y. miR-223 inhibits lipid deposition and inflammation by suppressing Toll-like receptor 4 signaling in macrophages. Int. J. Mol. Sci. 2015, 16, 24965–24982. [Google Scholar] [CrossRef] [PubMed]
- Long, F.-Q.; Kou, C.-X.; Li, K.; Wu, J.; Wang, Q.-Q. MiR-223-3p inhibits rTp17-induced inflammasome activation and pyroptosis by targeting NLRP3. J. Cell Mol. Med. 2020, 24, 14405–14414. [Google Scholar] [CrossRef] [PubMed]
- Favero, A.; Segatto, I.; Perin, T.; Belletti, B. The many facets of miR-223 in cancer: Oncosuppressor, oncogenic driver, therapeutic target, and biomarker of response. Wiley Interdiscip Rev. RNA 2021, 12, e1659. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, J.; Zhou, W.; Hsu, A.Y.; Deng, Q. miRNA-223 at the crossroads of inflammation and cancer. Cancer Lett. 2019, 451, 136–141. [Google Scholar] [CrossRef] [PubMed]
- Jaberi, N.; Soleimani, A.; Pashirzad, M.; Abdeahad, H.; Mohammadi, F.; Khoshakhlagh, M.; Khazaei, M.; Ferns, G.A.; Avan, A.; Hassanian, S.M. Role of thrombin in the pathogenesis of atherosclerosis. J. Cell Biochem. 2019, 120, 4757–4765. [Google Scholar] [CrossRef]
- Gray, E.; Hogwood, J.; Mulloy, B. The anticoagulant and antithrombotic mechanisms of heparin. Handb. Exp. Pharm. 2012, 207, 43–61. [Google Scholar] [CrossRef]
- Schweickert, P.G.; Yang, Y.; White, E.E.; Cresswell, G.M.; Elzey, B.D.; Ratliff, T.L.; Arumugam, P.; Antoniak, S.; Mackman, N.; Flick, M.J.; et al. Thrombin-PAR1 signaling in pancreatic cancer promotes an immunosuppressive microenvironment. J. Thromb. Haemost. 2021, 19, 161–172. [Google Scholar] [CrossRef]
- Zhong, Y.-C.; Zhang, T.; Di, W.; Li, W.-P. Thrombin promotes epithelial ovarian cancer cell invasion by inducing epithelial-mesenchymal transition. J. Gynecol. Oncol. 2013, 24, 265–272. [Google Scholar] [CrossRef] [Green Version]
- Otsuki, T.; Fujimoto, D.; Hirono, Y.; Goi, T.; Yamaguchi, A. Thrombin conducts epithelial-mesenchymal transition via protease-activated receptor-1 in human gastric cancer. Int. J. Oncol. 2014, 45, 2287–2294. [Google Scholar] [CrossRef] [Green Version]
- Brellier, F.; Ruggiero, S.; Zwolanek, D.; Martina, E.; Hess, D.; Brown-Luedi, M.; Hartmann, U.; Koch, M.; Merlo, A.; Lino, M.; et al. SMOC1 is a tenascin-C interacting protein over-expressed in brain tumors. Matrix Biol. 2011, 30, 225–233. [Google Scholar] [CrossRef]
- Gong, X.; Liu, L.; Xiong, J.; Li, X.; Xu, J.; Xiao, Y.; Li, J.; Luo, X.; Mao, D.; Liu, L. Construction of a prognostic gene signature associated with immune infiltration in glioma: A comprehensive analysis based on the CGGA. J. Oncol. 2021, 2021, 6620159. [Google Scholar] [CrossRef] [PubMed]
- Walline, H.M.; Komarck, C.M.; McHugh, J.B.; Bellile, E.L.; Brenner, J.C.; Prince, M.E.; McKean, E.L.; Chepeha, D.B.; Wolf, G.T.; Worden, F.P.; et al. Genomic integration of high risk HPV alters gene expression in oropharyngeal squamous cell carcinoma. Mol. Cancer Res. 2016, 14, 941–952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peraramelli, S.; Zhou, Q.; Zhou, Q.; Wanko, B.; Zhao, L.; Nishimura, T.; Leung, T.H.; Mizuno, S.; Ito, M.; Myles, T.; et al. Thrombin cleavage of osteopontin initiates osteopontin’s tumor-promoting activity. J. Thromb. Haemost. 2022, 20, 1256–1270. [Google Scholar] [CrossRef] [PubMed]


| Gene | Forward | Reverse |
|---|---|---|
| NOS | GTGGTGACAAGCACATTTGG | GTTCGTCCCCTTCTCCTGTT |
| TNF-α | GGCCTTCCTACCTTCAGACC | CCGGCCTTCCAAATAAATAC |
| FIZZ-1 | CCCTTCTCATCTGCATCTCC | CAGTAGCAGTCATCCCAGCA |
| CD206 | TGGATGGATGGGAGCAAAGT | GCTGCTGTTATGTCTCTGGC |
| MMP9 | GAAGGCAAACCCTGTGTGTT | AGAGTACTGCTTGCCCAGGA |
| CD36 | AAACCCAGATGACGTGGCAA | AAGATGGCTCCATTGGGCTG |
| EF2 | GACATCACCAAGGGTGTGCAG | GCGGTCAGCACACTGGCATA |
| 18S | CTTTGGTCGCTCGCTCCTC | CTGACCGGGTTGGTTTTGAT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ukan, Ü.; Delgado Lagos, F.; Kempf, S.; Günther, S.; Siragusa, M.; Fisslthaler, B.; Fleming, I. Effect of Thrombin on the Metabolism and Function of Murine Macrophages. Cells 2022, 11, 1718. https://doi.org/10.3390/cells11101718
Ukan Ü, Delgado Lagos F, Kempf S, Günther S, Siragusa M, Fisslthaler B, Fleming I. Effect of Thrombin on the Metabolism and Function of Murine Macrophages. Cells. 2022; 11(10):1718. https://doi.org/10.3390/cells11101718
Chicago/Turabian StyleUkan, Ürün, Fredy Delgado Lagos, Sebastian Kempf, Stefan Günther, Mauro Siragusa, Beate Fisslthaler, and Ingrid Fleming. 2022. "Effect of Thrombin on the Metabolism and Function of Murine Macrophages" Cells 11, no. 10: 1718. https://doi.org/10.3390/cells11101718
APA StyleUkan, Ü., Delgado Lagos, F., Kempf, S., Günther, S., Siragusa, M., Fisslthaler, B., & Fleming, I. (2022). Effect of Thrombin on the Metabolism and Function of Murine Macrophages. Cells, 11(10), 1718. https://doi.org/10.3390/cells11101718






