Plasma Proteomic Analysis Identified Proteins Associated with Faulty Neutrophils Functionality in Decompensated Cirrhosis Patients with Sepsis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Patients and Blood Sampling
- (a)
- Temperature >38.0 °C or <36.0 °C
- (b)
- TLC (Total leukocyte count) < 4 or >12 × 109/L
- (c)
- Pulse > 90 beats/minutes
- (d)
- RR (Respiratory rate) >20 or <32 beats/minutes
2.2. Blood Sampling
2.3. Endotoxin Estimation
2.4. Multi-Parametric Whole Blood Immunophenotyping
2.5. Phagocytic Activity
2.6. Oxidative Burst Activity
2.7. Expression of Neutrophilic CD64 and Monocytic HLA-DR (nCD64-mHLADR Index)
2.8. Isolation of Neutrophils
2.9. NETosis
2.10. Estimation of Cell Free DNA
2.11. Cytokine Multiplex Bead Array Assay
2.12. Plasma Proteomic Analysis
2.13. Quantitative RT-PCR Analysis
2.14. Statistical Analysis
3. Results
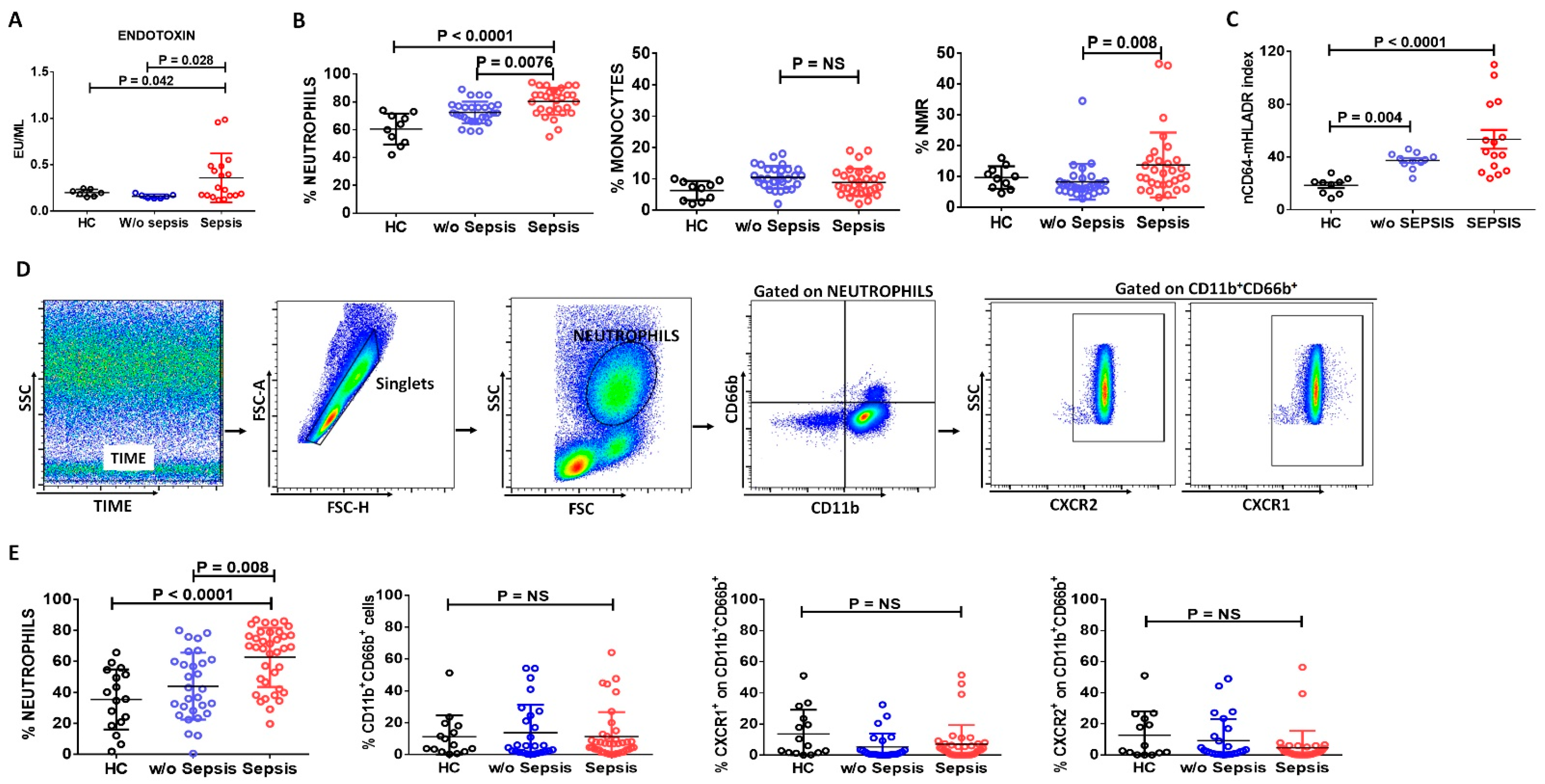
3.1. Neutrophils in Sepsis
3.1.1. Increase in Neutrophils and Neutrophil-to-Monocyte Ratio in Sepsis Patients
3.1.2. Assessment of Neutrophilic CD64 and Monocytic HLADR Expression in Sepsis
3.1.3. Decreased Neutrophils Chemoattractant Receptors in Sepsis Patients
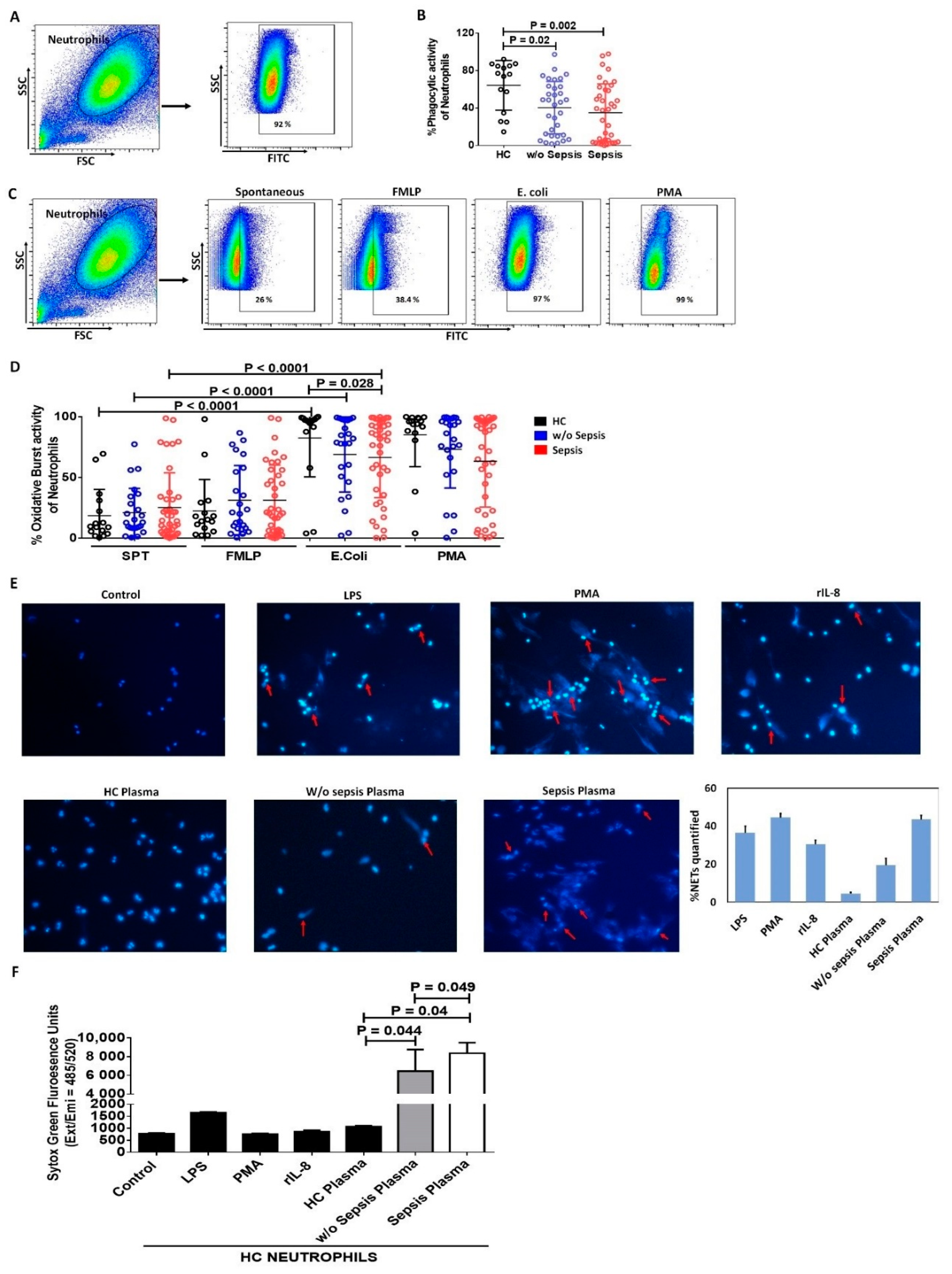
3.1.4. Decreased Phagocytic Activity of Neutrophils in Sepsis
3.1.5. Increase in Neutrophil Extracellular Traps and Cell Free DNA Concentration in Sepsis Plasma
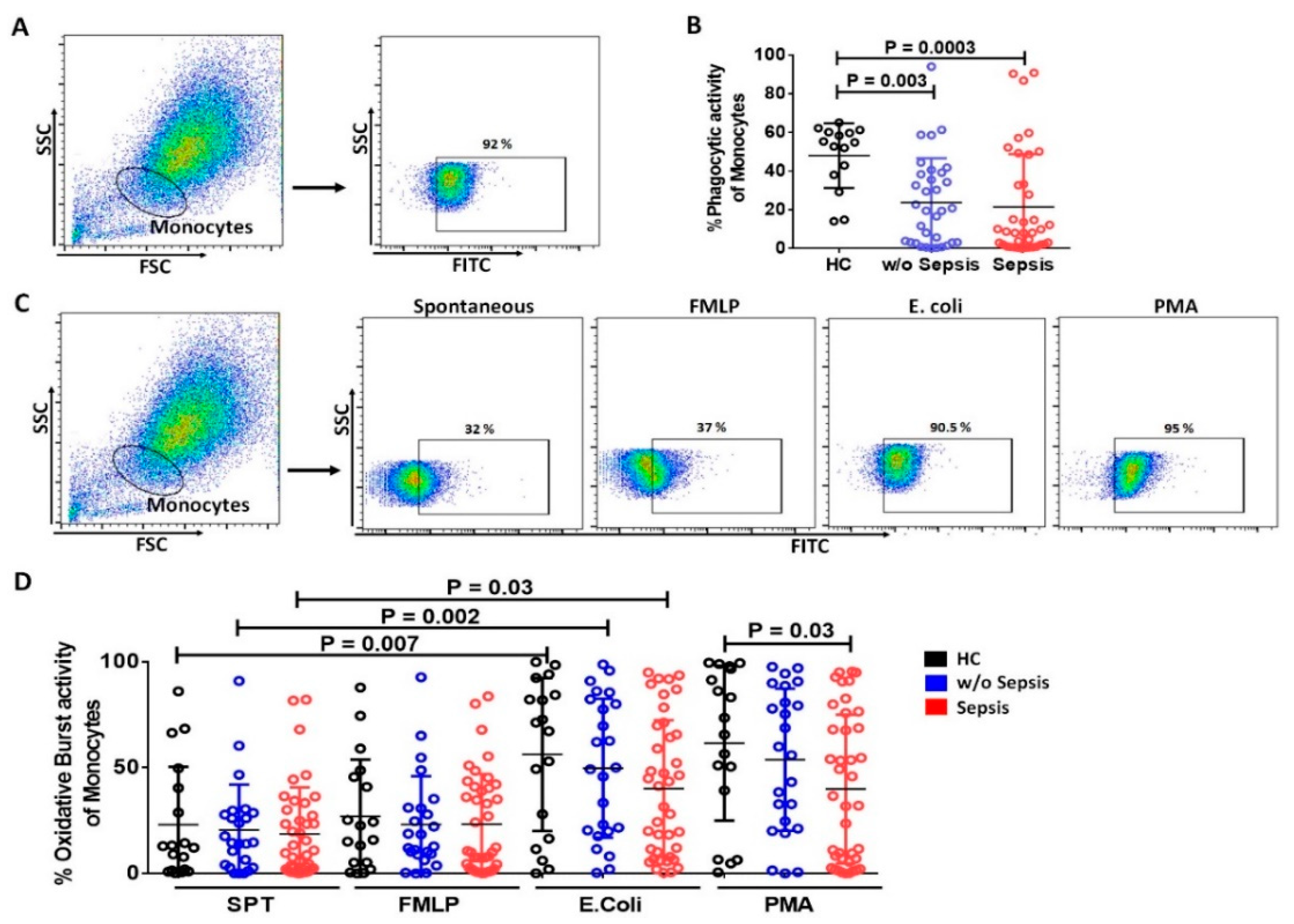
3.2. Monocytes in Sepsis
3.2.1. Reduced mHLA-DR in Sepsis Patients
3.2.2. Decrease in Phagocytic and Oxidative Burst Activity of Monocytes in Sepsis
3.3. Relation of Soluble Factors in Sepsis
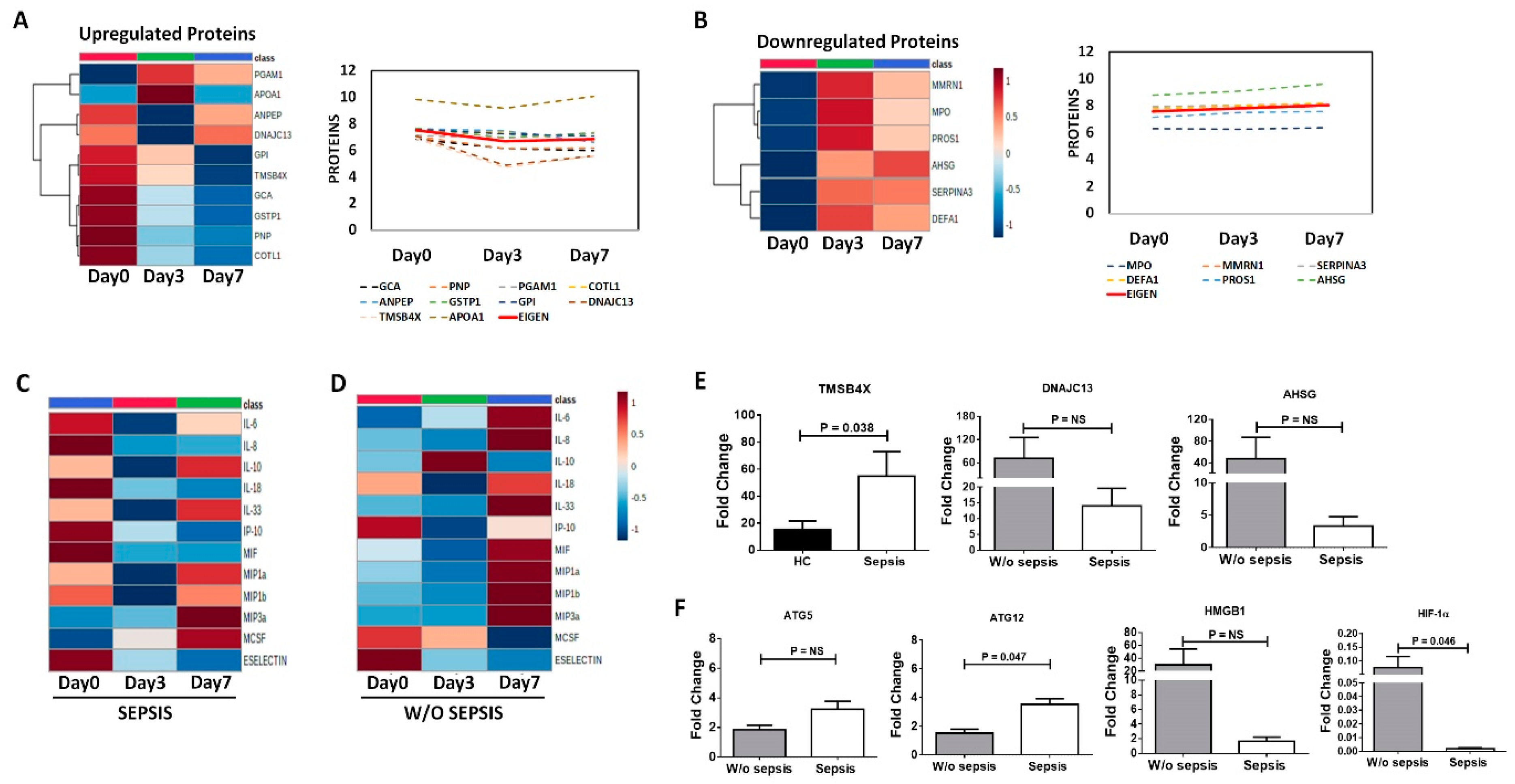
3.3.1. Neutrophils Specific Plasma Proteins in Sepsis
3.3.2. Increased Cytokines in Sepsis
3.3.3. Decrease Cytokines in Sepsis
3.3.4. Correlation of Neutrophils Functionality with Plasma Proteins and Cytokines in Sepsis
3.3.5. Differential Expression of Proteins and Cytokines at Follow-Up Time Points in Sepsis Patients
3.3.6. QRT-PCR Analysis for Validation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schuppan, D.; Afdhal, N.H. Liver Cirrhosis. Lancet 2008, 371, 838–851. [Google Scholar] [CrossRef]
- Tsao, G.G.; Abraldes, J.G.; Berzigotti, A.; Bosch, J. Portal Hypertensive Bleeding in Cirrhosis: Risk Stratification, Diagnosis, and Management: 2016 Practice Guidance by the American Association for the Study of Liver Diseases. Hepatology 2017, 65, 310–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kovach, M.A.; Standiford, T.J. The function of neutrophils in sepsis. Curr. Opin. Infect. Dis. 2012, 25, 321–327. [Google Scholar] [CrossRef]
- Mookerjee, R.P.; Stadlbauer, V.; Lidder, S.; Wright, G.A.; Hodges, S.J.; Davies, N.A.; Jalan, R. Neutrophil dysfunction in alcoholic hepatitis superimposed on cirrhosis is reversible and predicts the outcome. Hepatology 2007, 46, 831–840. [Google Scholar] [CrossRef] [PubMed]
- Xing, T.; Li, L.; Cao, H.; Huang, J. Altered immune function of monocytes in different stages of patients with acute on chronic liver failure. Clin. Exp. Immunol. 2007, 147, 184–188. [Google Scholar] [CrossRef]
- Riordan, S.M.; Skinner, N.; Nagree, A.; McCallum, H.; McIver, C.J.; Kurtovic, J.; A Hamilton, J.; Bengmark, S.; Williams, R.; Visvanathan, K. Peripheral blood mononuclear cell expression of Toll-like receptors and relation to cytokine levels in cirrhosis. Hepatology 2003, 37, 1154–1164. [Google Scholar] [CrossRef] [PubMed]
- Albillos, A.; Lario, M.; Mon, M.A. Cirrhosis-associated immune dysfunction: Distinctive features and clinical relevance. J. Hepatol. 2014, 61, 1385–1396. [Google Scholar] [CrossRef] [Green Version]
- Almeida, J.; Galhenage, S.; Yu, J.; Kurtovic, J.; Riordan, S.M. Gut flora and bacterial translocation in chronic liver disease. World J. Gastroenterol. 2006, 12, 1493–1502. [Google Scholar] [CrossRef]
- Bal, C.K.; Daman, K.; Bhatia, V. Predictors of fifty days in-hospital mortality in decompensated cirrhosis patients with spontaneous bacterial peritonitis. World J. Hepatol. 2016, 8, 566–572. [Google Scholar] [CrossRef]
- Kobayashi, Y. The role of chemokines in neutrophil biology. Front. Biosci. 2008, 13, 2400–2407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ley, K.; Hoffman, H.M.; Paul, K.; Cassatella, M.Z.; Zychlinsky, A.; Hedrick, C.C.; Catz, S.D. Neutrophils: New insights and open questions. Sci. Immunol. 2018, 3, eaat4579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pradhan, R.; Jain, P.; Paria, A.; Saha, A.; Sahoo, J.; Sen, A.; Mukherjee, S.; Som, T.; Hazra, A.; Warner, N.; et al. Ratio of neutrophilic CD64 and monocytic HLA-DR: A novel parameter in diagnosis and prognostication of neonatal sepsis. Cytom. B Clin. Cytom. 2016, 90, 295–302. [Google Scholar] [CrossRef] [PubMed]
- Pires, R.H.; Felix, S.B.; Delcea, M. The architecture of neutrophil extracellular traps investigated by atomic force microscopy. Nanoscale 2016, 8, 14193–14202. [Google Scholar] [CrossRef] [PubMed]
- Boussif, A.; Rolas, L.; Weiss, E.; Bouriche, H.; Moreau, R.; Périanin, A. Impaired intracellular signaling, myeloperoxidase release and bactericidal activity of neutrophils from patients with alcoholic cirrhosis. J. Hepatol. 2016, 64, 1041–1048. [Google Scholar] [CrossRef]
- Lominadze, G.; Powell, D.W.; Luerman, G.C.; Link, A.J.; Ward, R.A.; McLeish, K.R. Proteomic analysis of human neutrophil granules. Mol. Cell Proteom. 2005, 4, 1503–1521. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ravenhill, B.J.; Soday, L.; Houghton, J.; Antrobus, R.; Weekes, M.P. Comprehensive cell surface proteomics defines markers of classical, intermediate and non-classical monocytes. Sci. Rep. 2020, 10, 4560. [Google Scholar] [CrossRef]
- Tomazella, G.G.; Silva, I.D.; Laure, H.J.; Rosa, J.C.; Chammas, R.; Wiker, H.G.; A de Souza, G.; Greene, L.J. Proteomic analysis of total cellular proteins of human neutrophils. Proteome Sci. 2009, 7, 32. [Google Scholar] [CrossRef] [Green Version]
- Yin, X.; Xin, H.; Mao, S.; Wu, G.; Guo, L. The Role of Autophagy in Sepsis: Protection and Injury to Organs. Front Physiol. 2019, 10, 1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ho, J.; Yu, J.; Wong, S.H.; Zhang, L.; Liu, X.; Wong, W.T.; Leung, C.C.H.; Choi, G.; Wang, M.H.T.; Gin, T.; et al. Autophagy in sepsis: Degradation into exhaustion? Autophagy 2016, 12, 1073–1082. [Google Scholar] [CrossRef] [Green Version]
- Shrestha, S.; Lee, J.M.; Hong, C.W. Autophagy in neutrophils. Korean J. Physiol. Pharm. 2020, 24, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, R.K.; Burns, B. Systemic Inflammatory Response Syndrome. 2021.7.28. In StatPearls [Internet]; StatPearls Publishing: Treasure Island, FL, USA, 2022. [Google Scholar]
- Tripathi, G.; Sharma, N.; Bindal, V.; Yadav, M.; Mathew, B.; Sharma, S.; Gupta, E.; Maras, J.S.; Sarin, S.K. Protocol for global proteome, virome, and metaproteome profiling of respiratory specimen (VTM) in COVID-19 patient by LC-MS/MS-based analysis. STAR Protoc. 2022, 3, 101045. [Google Scholar] [CrossRef] [PubMed]
- Singer, M.; Deutschman, C.S.; Seymour, C.W.; Shankar-Hari, M.; Annane, D.; Bauer, M.; Bellomo, R.; Bernard, G.R.; Chiche, J.-D.; Coopersmith, C.M.; et al. The Third International Consensus Definitions for Sepsis and Septic Shock (Sepsis-3). JAMA 2016, 315, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Faivre, V.; Lukaszewicz, A.C.; Payen, D. Downregulation of Blood Monocyte HLA-DR in ICU Patients Is Also Present in Bone Marrow Cells. PLoS ONE 2016, 11, e0164489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Li, L.; Shi, S. Expression of monocyte HLA-DR and blood lactic acid level in patients with sepsis and correlation with prognosis. Int. J. Clin. Exp. Med. 2018, 11, 4905–4911. [Google Scholar]
- Taneja, R.; Sharma, A.P.; Hallett, M.B.; Findlay, G.P.; Morris, M.R. Immature circulating neutrophils in sepsis have impaired phagocytosis and calcium signaling. Shock 2008, 30, 618–622. [Google Scholar] [CrossRef] [PubMed]
- Wilson, M.E. Effects of bacterial endotoxins on neutrophil function. Rev. Infect. Dis. 1985, 7, 404–418. [Google Scholar] [CrossRef] [PubMed]
- Hiki, N.; Berger, D.; Prigl, C.; Boelke, E.; Wiedeck, H.; Seidelmann, M.; Staib, L.; Kaminishi, M.; Oohara, T.; Beger, H.G. Endotoxin binding and elimination by monocytes: Secretion of soluble CD14 represents an inducible mechanism counteracting reduced expression of membrane CD14 in patients with sepsis and in a patient with paroxysmal nocturnal hemoglobinuria. Infect. Immun. 1998, 66, 1135–1141. [Google Scholar] [CrossRef] [Green Version]
- Patel, J.M.; Sapey, E.; Parekh, D.; Scott, A.; Dosanjh, D.; Gao, F.; Thickett, D.R. Sepsis Induces a Dysregulated Neutrophil Phenotype that is Associated with Increased Mortality. Mediat. Inflamm. 2018, 11, 4065362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cid, J.; Aguinaco, R.; Sánchez, R.; García-Pardo, G.; Llorente, A. Neutrophil CD64 expression as marker of bacterial infection: A systematic review and meta-analysis. J. Infect. 2010, 60, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Ueda, T.; Aoyama-Ishikawa, M.; Nakao, A.; Yamada, T.; Usami, M.; Kotani, J. A simple scoring system based on neutrophil count in sepsis patients. Med. Hypotheses 2014, 82, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Pilsczek, H.F.; Salina, D.; Poon, K.K.H.; Fahey, C.; Yipp, B.G.; Sibley, C.D.; Robbins, S.M.; Green, F.H.Y.; Surette, M.G.; Sugai, M.; et al. A novel mechanism of rapid nuclear neutrophil extracellular trap formation in response to Staphylococcus aureus. J. Immunol. 2010, 185, 7413–7425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jorch, S.K.; Kubes, P. An emerging role for neutrophil extracellular traps in noninfectious disease. Nat. Med. 2017, 23, 279–287. [Google Scholar] [CrossRef] [PubMed]
- Honda, M.; Kubes, P. Neutrophils and neutrophil extracellular traps in the liver and gastrointestinal system. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 206–221. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.O.; Kim, H.-S.; Youn, J.-C.; Shin, E.-C.; Park, S. Serum cytokine profiles in healthy young and elderly population assessed using multiplexed bead-based immunoassays. J. Transl. Med. 2011, 9, 113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fiddler, C.A.; Parfrey, H.; Cowburn, A.S.; Luo, D.; Nash, G.B.; Murphy, G.; Chilvers, E.R. The aminopeptidase CD13 induces homotypic aggregation in neutrophils and impairs collagen invasion. PLoS ONE 2016, 11, e0160108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cowburn, A.S.; Sobolewski, A.; Reed, B.J.; Deighton, J.; Murray, J.; Cadwallader, K.A.; Bradley, J.R.; Chilvers, E.R. Aminopeptidase N (CD13) regulates tumor necrosis factor-alpha-induced apoptosis in human neutrophils. J. Biol. Chem. 2006, 281, 12458–12467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhattacharya, C.; Manna, S.K.; Samanta, S.; Gupta, S.K.; Samanta, A.K. An aminopeptidase regulates LPS stimulated interleukin-8 receptor on the surface of human neutrophils. Indian J. Biochem. Biophys. 1997, 34, 214–219. [Google Scholar] [PubMed]
- Xu, P.; Roes, J.; Segal, A.W.; Radulovic, M. The role of grancalcin in adhesion of neutrophils. Cell. Immunol. 2006, 240, 116–121. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Shinomiya, H.; Kirikae, T.; Hirata, H.; Asano, Y. Characterization of Murine Grancalcin Specifically Expressed in Leukocytes and Its Possible Role in Host Defense against Bacterial Infection. Biosci. Biotechnol. Biochem. 2004, 68, 894–902. [Google Scholar] [CrossRef]
- Blackburn, W.D.; Dohlman, J.G.; Venkatachalapathi, Y.V.; Pillion, D.J.; Koopman, W.J.; Segrest, J.P.; Anantharamaiah, G.M. Apolipoprotein A-I decreases neutrophil degranulation and superoxide production. J. Lipid Res. 1991, 32, 1911–1918. [Google Scholar] [CrossRef]
- Sharifov, O.F.; Xu, X.; Gaggar, A.; Grizzle, W.E.; Mishra, V.K.; Honavar, J.; Litovsky, S.H.; Palgunachari, M.N.; White, C.R.; Anantharamaiah, G.M.; et al. Anti-inflammatory mechanisms of apolipoprotein A-I mimetic peptide in acute respiratory distress syndrome secondary to sepsis. PLoS ONE 2013, 8, e64486. [Google Scholar] [CrossRef] [PubMed]
- Aratani, Y. Myeloperoxidase: Its Role for Host Defense, Inflammation, and Neutrophil Function. Arch. Biochem. Biophys. 2018, 640, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, G.T.; Green, E.R.; Mecsas, J. Neutrophils to the ROScue: Mechanisms of NADPH Oxidase Activation and Bacterial Resistance. Front. Cell. Infect. Microbiol. 2017, 7, 373. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, Y.; Ouchi, Y. Antimicrobial peptide defensin identification of novel isoforms and the characterization of their physiological roles and their significance in the pathogenesis of diseases. Proc. Jpn. Acad. B Phys. Biol. Sci. 2012, 88, 152–166. [Google Scholar] [CrossRef] [Green Version]
- Brook, M.; Tomlinson, G.H.; Miles, K.; Smith, R.W.O.; Rossi, A.G.; Hiemstra, P.S.; Wout, E.F.A.V.; Dean, J.L.E.; Gray, N.K.; Lu, W.; et al. Neutrophil-derived alpha defensins control inflammation by inhibiting macrophage mRNA translation. Proc. Natl. Acad. Sci. USA 2016, 113, 4350–4355. [Google Scholar] [CrossRef] [Green Version]
- Winkler, I.G.; Hendy, J.; Coughlin, P.; Horvath, A.; Lévesque, J.P. Serine protease inhibitors serpina1 and serpina3 are downregulated in bone marrow during hematopoietic progenitor mobilization. J. Exp. Med. 2005, 201, 1077–1088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Besemer, A.S.; Maus, J.; Ax, M.D.A.; Stein, A.; Vo, S.; Freese, C.; Nalbach, K.; von Hilchen, C.; Pfalzgraf, I.F.; Koziollek-Drechsler, I.; et al. Receptor-mediated endocytosis 8 (RME-8)/DNAJC13 is a novel positive modulator of autophagy and stabilizes cellular protein homeostasis. Cell. Mol. Life Sci. 2021, 78, 645–660. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Zhu, S.; Li, J.; Huang, Y.; Zhou, R.; Fan, X.; Yang, H.; Gong, X.; Eissa, N.T.; Jahnen-Dechent, W.; et al. A hepatic protein, fetuin-A, occupies a protective role in lethal systemic inflammation. PLoS ONE 2011, 6, e16945. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Jung, Y. Potential role of thymosin Beta 4 in liver fibrosis. Int. J. Mol. Sci. 2015, 16, 10624–10635. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Badamchian, M.; Fagarasan, M.O.; Danner, R.L.; Suffredini, A.F.; Damavandy, A.; Goldstein, A.L. Thymosin beta(4) reduces lethality and down-regulates inflammatory mediators in endotoxin-induced septic shock. Int. Immunopharmacol. 2003, 3, 1225–1233. [Google Scholar] [CrossRef]
- Xing, Z.; Li, S.; Liu, Z.; Zhang, C.; Bai, Z. CircSERPINA3 regulates SERPINA3-mediated apoptosis, autophagy and aerobic glycolysis of prostate cancer cells by competitively binding to MiR-653-5p and recruiting BUD13. J. Transl. Med. 2021, 19, 492. [Google Scholar] [CrossRef] [PubMed]
- Duan, F.; Mei, C.; Yang, L.; Zheng, J.; Lu, H.; Xia, Y.; Hsu, S. Vitamin K2 promotes PI3K/AKT/HIF-1α-mediated glycolysis that leads to AMPK-dependent autophagic cell death in bladder cancer cells. Sci. Rep. 2020, 10, 7714. [Google Scholar] [CrossRef] [PubMed]
- Sun, Q.; Li, S.; Wang, Y.; Peng, H.; Zhang, X.; Zheng, Y.; Li, C. Phosphoglyceric acid mutase-1 contributes to oncogenic mTOR-mediated tumor growth and confers non-small cell lung cancer patients with poor prognosis. Cell Death Differ. 2018, 25, 1160–1173. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| MEDIAN and RANGE | Healthy Control (HC, n = 15) | DC w/o Sepsis (n = 30) | DC with Sepsis (n = 40) | p Value–w/o and with Sepsis |
|---|---|---|---|---|
| Age | 32 (20–40) | 47 (22–60) | 43 (29–60) | 0.02 |
| Male: Female | 11:4 | 25:5 | 38:2 | - |
| Total Bilirubin (mg/dL) | 1 (0.3–1.5) | 6.8 (1.6–22) | 17.9 (2.2–31.7) | <0.00 |
| AST (IU/mL) | 20 (5–40) | 62 (31–510) | 118 (35–1037) | 0.04 |
| ALT (IU/mL) | 25 (10–40) | 37 (20–634) | 46 (11–233) | NS |
| INR (s) | 1 (0.8–1.2) | 1.9 (1.1–3.3) | 2.7 (1.3–6.75) | <0.00 |
| PCT (ng/mL) | 0.8 (0.2–2) | 0.4 (0.04–3.25) | 3 (0.07–88) | 0.04 |
| MELD Na | 8 (6–10) | 26 (10–37) | 34 (29–40) | <0.00 |
| Lactate (mmol/L) | 1.5 (1–2) | 1.4 (0.6–5.2) | 2.1 (0.2–13.3) | 0.02 |
| Sodium (mmol/L) | 140 (136–145) | 133 (124.3–142.7) | 129 (119–150) | NS |
| Creatinine (mg/mL) | 0.6 (0.2–1) | 0.86 (0.3–2.9) | 1.57 (0.3–4.78) | 0.03 |
| SOFA score | - | 7 (4–11) | 12 (10–16) | <0.00 |
| SIRS Criteria | ||||
| TLC (109 L) | 6 (4–11) | 6.6 (3.1–19.8) | 14 (2.7–43.6) | 0.01 |
| PULSE (/minute) | 70 (60–100) | 76 (60–110) | 94 (62–132) | 0.01 |
| RR (/minute) | 14 (12–16) | 20 (16–24) | 22 (14–34) | NS |
| TEMPERATURE | 98 (97–99) | 98.4 (97–98.9) | 98.4 (96–100) | NS |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sehgal, R.; Kaur, N.; Maiwall, R.; Ramakrishna, G.; Maras, J.S.; Trehanpati, N. Plasma Proteomic Analysis Identified Proteins Associated with Faulty Neutrophils Functionality in Decompensated Cirrhosis Patients with Sepsis. Cells 2022, 11, 1745. https://doi.org/10.3390/cells11111745
Sehgal R, Kaur N, Maiwall R, Ramakrishna G, Maras JS, Trehanpati N. Plasma Proteomic Analysis Identified Proteins Associated with Faulty Neutrophils Functionality in Decompensated Cirrhosis Patients with Sepsis. Cells. 2022; 11(11):1745. https://doi.org/10.3390/cells11111745
Chicago/Turabian StyleSehgal, Rashi, Navkiran Kaur, Rakhi Maiwall, Gayatri Ramakrishna, Jaswinder Singh Maras, and Nirupma Trehanpati. 2022. "Plasma Proteomic Analysis Identified Proteins Associated with Faulty Neutrophils Functionality in Decompensated Cirrhosis Patients with Sepsis" Cells 11, no. 11: 1745. https://doi.org/10.3390/cells11111745
APA StyleSehgal, R., Kaur, N., Maiwall, R., Ramakrishna, G., Maras, J. S., & Trehanpati, N. (2022). Plasma Proteomic Analysis Identified Proteins Associated with Faulty Neutrophils Functionality in Decompensated Cirrhosis Patients with Sepsis. Cells, 11(11), 1745. https://doi.org/10.3390/cells11111745





