Complex Role of Microbiome in Pancreatic Tumorigenesis: Potential Therapeutic Implications
Abstract
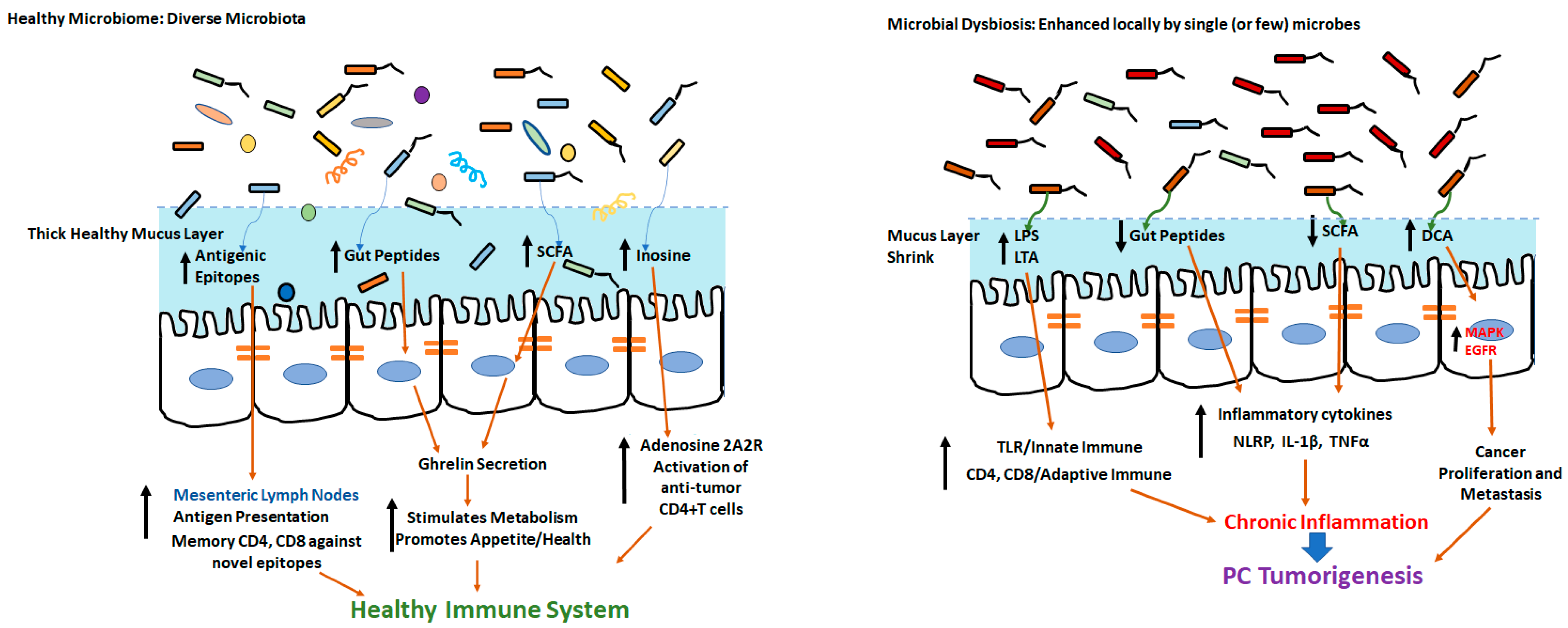
:1. Introduction
2. Pancreatic Microbiome
3. Role of Microbiome in Pancreatic Carcinogenesis
3.1. Alterations of Microbial Metabolites
3.2. Microbiota-Mediated Immunoregulation
4. Secondary Impact of Lifestyle Co-Morbidities on Microbiome-Modulated Carcinogenesis
5. Microbiome-Specific Biomarkers
6. Microbiome in Cancer Therapy
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- American Cancer Society. Cancer Facts & Figures 2022. Atlanta Am. Cancer Soc. 2022. [Google Scholar]
- Andersson, R.; Haglund, C.; Seppanen, H.; Ansari, D. Pancreatic cancer—The past, the present, and the future. Scand. J. Gastroenterol. 2022, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kindler, H.L. A Glimmer of Hope for Pancreatic Cancer. N. Engl. J. Med. 2018, 379, 2463–2464. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.P.; Hong, T.S.; Bardeesy, N. Pancreatic adenocarcinoma. N. Engl. J. Med. 2014, 371, 1039. [Google Scholar] [CrossRef] [PubMed]
- Lopez, N.E.; Prendergast, C.; Lowy, A.M. Borderline resectable pancreatic cancer: Definitions and management. World J. Gastroenterol. 2014, 20, 10740–10751. [Google Scholar] [CrossRef]
- Sender, R.; Fuchs, S.; Milo, R. Revised Estimates for the Number of Human and Bacteria Cells in the Body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, D.; Leung, R.K.; Guan, W.; Au, W.W. Involvement of gut microbiome in human health and disease: Brief overview, knowledge gaps and research opportunities. Gut Pathog. 2018, 10, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helmink, B.A.; Khan, M.A.W.; Hermann, A.; Gopalakrishnan, V.; Wargo, J.A. The microbiome, cancer, and cancer therapy. Nat. Med. 2019, 25, 377. [Google Scholar] [CrossRef]
- Vandeven, N.; Nghiem, P. Pathogen-driven cancers and emerging immune therapeutic strategies. Cancer Immunol. Res. 2014, 2, 9–14. [Google Scholar] [CrossRef] [Green Version]
- Nejman, D.; Livyatan, I.; Fuks, G.; Gavert, N.; Zwang, Y.; Geller, L.T.; Rotter-Maskowitz, A.; Weiser, R.; Mallel, G.; Gigi, E.; et al. The human tumor microbiome is composed of tumor type-specific intracellular bacteria. Science 2020, 368, 973–980. [Google Scholar] [CrossRef] [PubMed]
- Pushalkar, S.; Hundeyin, M.; Daley, D.; Zambirinis, C.P.; Kurz, E.; Mishra, A.; Mohan, N.; Aykut, B.; Usyk, M.; Torres, L.E.; et al. The Pancreatic Cancer Microbiome Promotes Oncogenesis by Induction of Innate and Adaptive Immune Suppression. Cancer Discov. 2018, 8, 403–416. [Google Scholar] [CrossRef] [Green Version]
- Jeong, J.Y.; Kim, T.B.; Kim, J.; Choi, H.W.; Kim, E.J.; Yoo, H.J.; Lee, S.; Jun, H.R.; Yoo, W.; Kim, S.; et al. Diversity in the Extracellular Vesicle-Derived Microbiome of Tissues According to Tumor Progression in Pancreatic Cancer. Cancers 2020, 12, 2346. [Google Scholar] [CrossRef] [PubMed]
- Aykut, B.; Pushalkar, S.; Chen, R.; Li, Q.; Abengozar, R.; Kim, J.I.; Shadaloey, S.A.; Wu, D.; Preiss, P.; Verma, N.; et al. The fungal mycobiome promotes pancreatic oncogenesis via activation of MBL. Nature 2019, 574, 264–267. [Google Scholar] [CrossRef] [PubMed]
- Rogers, M.B.; Aveson, V.; Firek, B.; Yeh, A.; Brooks, B.; Brower-Sinning, R.; Steve, J.; Banfield, J.F.; Zureikat, A.; Hogg, M.; et al. Disturbances of the Perioperative Microbiome Across Multiple Body Sites in Patients Undergoing Pancreaticoduodenectomy. Pancreas 2017, 46, 260–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, Z.; Jiang, J.; Xie, H.; Li, A.; Lu, H.; Xu, S.; Zhou, L.; Zhang, H.; Cui, G.; Chen, X.; et al. Gut microbial profile analysis by MiSeq sequencing of pancreatic carcinoma patients in China. Oncotarget 2017, 8, 95176–95191. [Google Scholar] [CrossRef] [Green Version]
- Kartal, E.; Schmidt, T.S.B.; Molina-Montes, E.; Rodriguez-Perales, S.; Wirbel, J.; Maistrenko, O.M.; Akanni, W.A.; Alashkar Alhamwe, B.; Alves, R.J.; Carrato, A.; et al. A faecal microbiota signature with high specificity for pancreatic cancer. Gut 2022, 71, 1359–1372. [Google Scholar] [CrossRef] [PubMed]
- Kohi, S.; Macgregor-Das, A.; Dbouk, M.; Yoshida, T.; Chuidian, M.; Abe, T.; Borges, M.; Lennon, A.M.; Shin, E.J.; Canto, M.I.; et al. Alterations in the Duodenal Fluid Microbiome of Patients with Pancreatic Cancer. Clin. Gastroenterol. Hepatol. 2022, 20, e196–e227. [Google Scholar] [CrossRef] [PubMed]
- Lu, H.; Ren, Z.; Li, A.; Li, J.; Xu, S.; Zhang, H.; Jiang, J.; Yang, J.; Luo, Q.; Zhou, K.; et al. Tongue coating microbiome data distinguish patients with pancreatic head cancer from healthy controls. J. Oral. Microbiol. 2019, 11, 1563409. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Alekseyenko, A.V.; Wu, J.; Peters, B.A.; Jacobs, E.J.; Gapstur, S.M.; Purdue, M.P.; Abnet, C.C.; Stolzenberg-Solomon, R.; Miller, G.; et al. Human oral microbiome and prospective risk for pancreatic cancer: A population-based nested case-control study. Gut 2018, 67, 120–127. [Google Scholar] [CrossRef] [Green Version]
- Wei, A.L.; Li, M.; Li, G.Q.; Wang, X.; Hu, W.M.; Li, Z.L.; Yuan, J.; Liu, H.Y.; Zhou, L.L.; Li, K.; et al. Oral microbiome and pancreatic cancer. World J. Gastroenterol. 2020, 26, 7679–7692. [Google Scholar] [CrossRef] [PubMed]
- Mitsuhashi, K.; Nosho, K.; Sukawa, Y.; Matsunaga, Y.; Ito, M.; Kurihara, H.; Kanno, S.; Igarashi, H.; Naito, T.; Adachi, Y.; et al. Association of Fusobacterium species in pancreatic cancer tissues with molecular features and prognosis. Oncotarget 2015, 6, 7209–7220. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Del Castillo, E.; Meier, R.; Chung, M.; Koestler, D.C.; Chen, T.; Paster, B.J.; Charpentier, K.P.; Kelsey, K.T.; Izard, J.; Michaud, D.S. The Microbiomes of Pancreatic and Duodenum Tissue Overlap and Are Highly Subject Specific but Differ between Pancreatic Cancer and Noncancer Subjects. Cancer Epidemiol. Biomark. Prev. 2019, 28, 370–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alkharaan, H.; Lu, L.; Gabarrini, G.; Halimi, A.; Ateeb, Z.; Sobkowiak, M.J.; Davanian, H.; Fernandez Moro, C.; Jansson, L.; Del Chiaro, M.; et al. Circulating and Salivary Antibodies to Fusobacterium nucleatum Are Associated with Cystic Pancreatic Neoplasm Malignancy. Front. Immunol. 2020, 11, 2003. [Google Scholar] [CrossRef] [PubMed]
- Michaud, D.S.; Izard, J.; Wilhelm-Benartzi, C.S.; You, D.H.; Grote, V.A.; Tjonneland, A.; Dahm, C.C.; Overvad, K.; Jenab, M.; Fedirko, V.; et al. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut 2013, 62, 1764–1770. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Liu, W.; Wu, J. Helicobacter pylori infection and pancreatic cancer risk: A meta-analysis. J. Cancer Res. Ther. 2016, 12, 229–232. [Google Scholar]
- Trikudanathan, G.; Philip, A.; Dasanu, C.A.; Baker, W.L. Association between Helicobacter pylori infection and pancreatic cancer. A cumulative meta-analysis. JOP J. Pancreas 2011, 12, 26–31. [Google Scholar]
- Schulte, A.; Pandeya, N.; Fawcett, J.; Fritschi, L.; Risch, H.A.; Webb, P.M.; Whiteman, D.C.; Neale, R.E. Association between Helicobacter pylori and pancreatic cancer risk: A meta-analysis. Cancer Causes Control 2015, 26, 1027. [Google Scholar] [CrossRef]
- Chakladar, J.; Kuo, S.Z.; Castaneda, G.; Li, W.T.; Gnanasekar, A.; Yu, M.A.; Chang, E.Y.; Wang, X.Q.; Ongkeko, W.M. The Pancreatic Microbiome is Associated with Carcinogenesis and Worse Prognosis in Males and Smokers. Cancers 2020, 12, 2672. [Google Scholar] [CrossRef] [PubMed]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156. [Google Scholar] [CrossRef] [Green Version]
- Thomas, R.M.; Gharaibeh, R.Z.; Gauthier, J.; Beveridge, M.; Pope, J.L.; Guijarro, M.V.; Yu, Q.; He, Z.; Ohland, C.; Newsome, R.; et al. Intestinal microbiota enhances pancreatic carcinogenesis in preclinical models. Carcinogenesis 2018, 39, 1068–1078. [Google Scholar] [CrossRef]
- Diehl, G.E.; Longman, R.S.; Zhang, J.X.; Breart, B.; Galan, C.; Cuesta, A.; Schwab, S.R.; Littman, D.R. Microbiota restricts trafficking of bacteria to mesenteric lymph nodes by CX(3)CR1(hi) cells. Nature 2013, 494, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Aldars-Garcia, L.; Chaparro, M.; Gisbert, J.P. Systematic Review: The Gut Microbiome and Its Potential Clinical Application in Inflammatory Bowel Disease. Microorganisms 2021, 9, 977. [Google Scholar] [CrossRef] [PubMed]
- Alamri, A. Diversity of Microbial Signatures in Asthmatic Airways. Int. J. Gen. Med. 2021, 14, 1367–1378. [Google Scholar] [CrossRef]
- Hrncir, T.; Hrncirova, L.; Kverka, M.; Hromadka, R.; Machova, V.; Trckova, E.; Kostovcikova, K.; Kralickova, P.; Krejsek, J.; Tlaskalova-Hogenova, H. Gut Microbiota and NAFLD: Pathogenetic Mechanisms, Microbiota Signatures, and Therapeutic Interventions. Microorganisms 2021, 9, 957. [Google Scholar] [CrossRef]
- Kunovsky, L.; Dite, P.; Jabandziev, P.; Dolina, J.; Vaculova, J.; Blaho, M.; Bojkova, M.; Dvorackova, J.; Uvirova, M.; Kala, Z.; et al. Helicobacter pylori infection and other bacteria in pancreatic cancer and autoimmune pancreatitis. World J. Gastrointest. Oncol. 2021, 13, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Binder Gallimidi, A.; Fischman, S.; Revach, B.; Bulvik, R.; Maliutina, A.; Rubinstein, A.M.; Nussbaum, G.; Elkin, M. Periodontal pathogens Porphyromonas gingivalis and Fusobacterium nucleatum promote tumor progression in an oral-specific chemical carcinogenesis model. Oncotarget 2015, 6, 22613–22623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gnanasekaran, J.; Binder Gallimidi, A.; Saba, E.; Pandi, K.; Eli Berchoer, L.; Hermano, E.; Angabo, S.; Makkawi, H.A.; Khashan, A.; Daoud, A.; et al. Intracellular Porphyromonas gingivalis Promotes the Tumorigenic Behavior of Pancreatic Carcinoma Cells. Cancers 2020, 12, 2331. [Google Scholar] [CrossRef] [PubMed]
- Sethi, V.; Kurtom, S.; Tarique, M.; Lavania, S.; Malchiodi, Z.; Hellmund, L.; Zhang, L.; Sharma, U.; Giri, B.; Garg, B.; et al. Gut Microbiota Promotes Tumor Growth in Mice by Modulating Immune Response. Gastroenterology 2018, 155, 33–37.e6. [Google Scholar] [CrossRef] [PubMed]
- Pinato, D.J.; Howlett, S.; Ottaviani, D.; Urus, H.; Patel, A.; Mineo, T.; Brock, C.; Power, D.; Hatcher, O.; Falconer, A.; et al. Association of Prior Antibiotic Treatment with Survival and Response to Immune Checkpoint Inhibitor Therapy in Patients with Cancer. JAMA Oncol. 2019, 5, 1774. [Google Scholar] [CrossRef] [PubMed]
- Hakozaki, T.; Richard, C.; Elkrief, A.; Hosomi, Y.; Benlaifaoui, M.; Mimpen, I.; Terrisse, S.; Derosa, L.; Zitvogel, L.; Routy, B.; et al. The Gut Microbiome Associates with Immune Checkpoint Inhibition Outcomes in Patients with Advanced Non-Small Cell Lung Cancer. Cancer Immunol. Res. 2020, 8, 1243–1250. [Google Scholar] [CrossRef]
- Jin, Y.; Dong, H.; Xia, L.; Yang, Y.; Zhu, Y.; Shen, Y.; Zheng, H.; Yao, C.; Wang, Y.; Lu, S. The Diversity of Gut Microbiome is Associated with Favorable Responses to Anti-Programmed Death 1 Immunotherapy in Chinese Patients with NSCLC. J. Thorac. Oncol. 2019, 14, 1378. [Google Scholar] [CrossRef] [PubMed]
- Cani, P.D.; Jordan, B.F. Gut microbiota-mediated inflammation in obesity: A link with gastrointestinal cancer. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Sivaprakasam, S.; Gurav, A.; Paschall, A.V.; Coe, G.L.; Chaudhary, K.; Cai, Y.; Kolhe, R.; Martin, P.; Browning, D.; Huang, L.; et al. An essential role of Ffar2 (Gpr43) in dietary fibre-mediated promotion of healthy composition of gut microbiota and suppression of intestinal carcinogenesis. Oncogenesis 2016, 5, e238. [Google Scholar] [CrossRef] [Green Version]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of Gpr109a, receptor for niacin and the commensal metabolite butyrate, suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.; Kwon, O.; Ryu, T.Y.; Jung, C.R.; Kim, J.; Min, J.K.; Kim, D.S.; Son, M.Y.; Cho, H.S. Propionate of a microbiota metabolite induces cell apoptosis and cell cycle arrest in lung cancer. Mol. Med. Rep. 2019, 20, 1569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohara, T.; Mori, T. Antiproliferative Effects of Short-chain Fatty Acids on Human Colorectal Cancer Cells via Gene Expression Inhibition. Anticancer Res. 2019, 39, 4659–4666. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coutzac, C.; Jouniaux, J.M.; Paci, A.; Schmidt, J.; Mallardo, D.; Seck, A.; Asvatourian, V.; Cassard, L.; Saulnier, P.; Lacroix, L.; et al. Systemic short chain fatty acids limit antitumor effect of CTLA-4 blockade in hosts with cancer. Nat. Commun. 2020, 11, 2168. [Google Scholar] [CrossRef]
- Maekawa, T.; Fukaya, R.; Takamatsu, S.; Itoyama, S.; Fukuoka, T.; Yamada, M.; Hata, T.; Nagaoka, S.; Kawamoto, K.; Eguchi, H.; et al. Possible involvement of Enterococcus infection in the pathogenesis of chronic pancreatitis and cancer. Biochem. Biophys. Res. Commun. 2018, 506, 962–969. [Google Scholar] [CrossRef]
- Cao, H.; Xu, M.; Dong, W.; Deng, B.; Wang, S.; Zhang, Y.; Wang, S.; Luo, S.; Wang, W.; Qi, Y.; et al. Secondary bile acid-induced dysbiosis promotes intestinal carcinogenesis. Int. J. Cancer 2017, 140, 2545–2556. [Google Scholar] [CrossRef] [Green Version]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef]
- Wang, S.; Dong, W.; Liu, L.; Xu, M.; Wang, Y.; Liu, T.; Zhang, Y.; Wang, B.; Cao, H. Interplay between bile acids and the gut microbiota promotes intestinal carcinogenesis. Mol. Carcinog. 2019, 58, 1155–1167. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagathihalli, N.S.; Beesetty, Y.; Lee, W.; Washington, M.K.; Chen, X.; Lockhart, A.C.; Merchant, N.B. Novel mechanistic insights into ectodomain shedding of EGFR Ligands Amphiregulin and TGF-alpha: Impact on gastrointestinal cancers driven by secondary bile acids. Cancer Res. 2014, 74, 2062–2072. [Google Scholar] [CrossRef] [Green Version]
- Halimi, A.; Gabarrini, G.; Sobkowiak, M.J.; Ateeb, Z.; Davanian, H.; Gaiser, R.A.; Arnelo, U.; Valente, R.; Wong, A.Y.W.; Moro, C.F.; et al. Isolation of pancreatic microbiota from cystic precursors of pancreatic cancer with intracellular growth and DNA damaging properties. Gut Microbes 2021, 13, 1983101. [Google Scholar] [CrossRef]
- Cani, P.D. Human gut microbiome: Hopes, threats and promises. Gut 2018, 67, 1716–1725. [Google Scholar] [CrossRef] [PubMed]
- Cougnoux, A.; Dalmasso, G.; Martinez, R.; Buc, E.; Delmas, J.; Gibold, L.; Sauvanet, P.; Darcha, C.; Dechelotte, P.; Bonnet, M.; et al. Bacterial genotoxin colibactin promotes colon tumour growth by inducing a senescence-associated secretory phenotype. Gut 2014, 63, 1932–1942. [Google Scholar] [CrossRef]
- Nougayrede, J.P.; Taieb, F.; De Rycke, J.; Oswald, E. Cyclomodulins: Bacterial effectors that modulate the eukaryotic cell cycle. Trends Microbiol. 2005, 13, 103–110. [Google Scholar] [CrossRef]
- Mowat, A.M.; Agace, W.W. Regional specialization within the intestinal immune system. Nat. Rev. Immunol. 2014, 14, 667. [Google Scholar] [CrossRef]
- Fulde, M.; Hornef, M.W. Maturation of the enteric mucosal innate immune system during the postnatal period. Immunol. Rev. 2014, 260, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Kau, A.L.; Ahern, P.P.; Griffin, N.W.; Goodman, A.L.; Gordon, J.I. Human nutrition, the gut microbiome and the immune system. Nature 2011, 474, 327–336. [Google Scholar] [CrossRef] [Green Version]
- Schwabe, R.F.; Jobin, C. The microbiome and cancer. Nat. Rev. Cancer 2013, 13, 800. [Google Scholar] [CrossRef] [Green Version]
- Rakoff-Nahoum, S.; Medzhitov, R. Toll-like receptors and cancer. Nat. Rev. Cancer 2009, 9, 57–63. [Google Scholar] [CrossRef] [PubMed]
- Ochi, A.; Graffeo, C.S.; Zambirinis, C.P.; Rehman, A.; Hackman, M.; Fallon, N.; Barilla, R.M.; Henning, J.R.; Jamal, M.; Rao, R.; et al. Toll-like receptor 7 regulates pancreatic carcinogenesis in mice and humans. J. Clin. Investig. 2012, 122, 4118–4129. [Google Scholar] [CrossRef] [PubMed]
- Martinon, F.; Mayor, A.; Tschopp, J. The inflammasomes: Guardians of the body. Annu. Rev. Immunol. 2009, 27, 229–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, S.; Shapiro, B.; Vucic, E.A.; Vogt, S.; Bar-Sagi, D. Tumor Cell-Derived IL1beta Promotes Desmoplasia and Immune Suppression in Pancreatic Cancer. Cancer Res. 2020, 80, 1088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Incio, J.; Liu, H.; Suboj, P.; Chin, S.M.; Chen, I.X.; Pinter, M.; Ng, M.R.; Nia, H.T.; Grahovac, J.; Kao, S.; et al. Obesity-Induced Inflammation and Desmoplasia Promote Pancreatic Cancer Progression and Resistance to Chemotherapy. Cancer Discov. 2016, 6, 852. [Google Scholar] [CrossRef] [Green Version]
- Moreira, L.O.; Zamboni, D.S. NOD1 and NOD2 Signaling in Infection and Inflammation. Front. Immunol. 2012, 3, 328. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, T.; Hovingh, E.S.; Foerster, E.G.; Abdel-Nour, M.; Philpott, D.J.; Girardin, S.E. NOD1 and NOD2 in inflammation, immunity and disease. Arch. Biochem. Biophys. 2019, 670, 69–81. [Google Scholar] [CrossRef]
- Kamada, N.; Chen, G.Y.; Inohara, N.; Nunez, G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 2013, 14, 685–690. [Google Scholar] [CrossRef]
- Ijssennagger, N.; Belzer, C.; Hooiveld, G.J.; Dekker, J.; van Mil, S.W.; Muller, M.; Kleerebezem, M.; van der Meer, R. Gut microbiota facilitates dietary heme-induced epithelial hyperproliferation by opening the mucus barrier in colon. Proc. Natl. Acad. Sci. USA 2015, 112, 10038–10043. [Google Scholar] [CrossRef] [Green Version]
- Reinhardt, C.; Bergentall, M.; Greiner, T.U.; Schaffner, F.; Ostergren-Lunden, G.; Petersen, L.C.; Ruf, W.; Backhed, F. Tissue factor and PAR1 promote microbiota-induced intestinal vascular remodelling. Nature 2012, 483, 627–631. [Google Scholar] [CrossRef] [Green Version]
- Riquelme, E.; Zhang, Y.; Zhang, L.; Montiel, M.; Zoltan, M.; Dong, W.; Quesada, P.; Sahin, I.; Chandra, V.; San Lucas, A.; et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell 2019, 178, 795–806.e12. [Google Scholar] [CrossRef] [PubMed]
- Rogers, C.J.; Prabhu, K.S.; Vijay-Kumar, M. The microbiome and obesity-an established risk for certain types of cancer. Cancer J. 2014, 20, 176. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Morris, J.S.; Liu, J.; Hassan, M.M.; Day, R.S.; Bondy, M.L.; Abbruzzese, J.L. Body mass index and risk, age of onset, and survival in patients with pancreatic cancer. Jama 2009, 301, 2553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkegard, J.; Mortensen, F.V.; Cronin-Fenton, D. Chronic Pancreatitis and Pancreatic Cancer Risk: A Systematic Review and Meta-analysis. Am. J. Gastroenterol. 2017, 112, 1366–1372. [Google Scholar] [CrossRef] [Green Version]
- Tomita, Y.; Azuma, K.; Nonaka, Y.; Kamada, Y.; Tomoeda, M.; Kishida, M.; Tanemura, M.; Miyoshi, E. Pancreatic fatty degeneration and fibrosis as predisposing factors for the development of pancreatic ductal adenocarcinoma. Pancreas 2014, 43, 1032–1041. [Google Scholar] [CrossRef]
- Kumar, K.; Ghoshal, U.C.; Srivastava, D.; Misra, A.; Mohindra, S. Small intestinal bacterial overgrowth is common both among patients with alcoholic and idiopathic chronic pancreatitis. Pancreatology 2014, 14, 280–283. [Google Scholar] [CrossRef]
- El Kurdi, B.; Babar, S.; El Iskandarani, M.; Bataineh, A.; Lerch, M.M.; Young, M.; Singh, V.P. Factors That Affect Prevalence of Small Intestinal Bacterial Overgrowth in Chronic Pancreatitis: A Systematic Review, Meta-Analysis, and Meta-Regression. Clin. Transl. Gastroenterol. 2019, 10, e00072. [Google Scholar] [CrossRef] [Green Version]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef]
- Scara, S.; Bottoni, P.; Scatena, R. CA 19-9: Biochemical and Clinical Aspects. Adv. Exp. Med. Biol. 2015, 867, 247–260. [Google Scholar]
- Farrell, J.J.; Zhang, L.; Zhou, H.; Chia, D.; Elashoff, D.; Akin, D.; Paster, B.J.; Joshipura, K.; Wong, D.T. Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 2012, 61, 582–588. [Google Scholar] [CrossRef]
- Sun, H.; Zhao, X.; Zhou, Y.; Wang, J.; Ma, R.; Ren, X.; Wang, H.; Zou, L. Characterization of Oral Microbiome and Exploration of Potential Biomarkers in Patients with Pancreatic Cancer. Biomed. Res. Int. 2020, 2020, 4712498. [Google Scholar] [CrossRef] [PubMed]
- Weniger, M.; Hank, T.; Qadan, M.; Ciprani, D.; Michelakos, T.; Niess, H.; Heiliger, C.; Ilmer, M.; D’Haese, J.G.; Ferrone, C.R.; et al. Influence of Klebsiella pneumoniae and quinolone treatment on prognosis in patients with pancreatic cancer. Br. J. Surg. 2021, 108, 709. [Google Scholar] [PubMed]
- Mathijssen, R.H.; van Alphen, R.J.; Verweij, J.; Loos, W.J.; Nooter, K.; Stoter, G.; Sparreboom, A. Clinical pharmacokinetics and metabolism of irinotecan (CPT-11). Clin. Cancer Res. 2001, 7, 2182. [Google Scholar]
- Stringer, A.M.; Gibson, R.J.; Logan, R.M.; Bowen, J.M.; Yeoh, A.S.; Keefe, D.M. Faecal microflora and beta-glucuronidase expression are altered in an irinotecan-induced diarrhea model in rats. Cancer Biol. Ther. 2008, 7, 1919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vande Voorde, J.; Sabuncuoglu, S.; Noppen, S.; Hofer, A.; Ranjbarian, F.; Fieuws, S.; Balzarini, J.; Liekens, S. Nucleoside-catabolizing enzymes in mycoplasma-infected tumor cell cultures compromise the cytostatic activity of the anticancer drug gemcitabine. J. Biol. Chem. 2014, 289, 13054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.M.; Hsu, L.J.; Lee, H.L.; Lin, C.P.; Huang, S.W.; Lai, C.J.; Lin, C.W.; Chen, W.T.; Chen, Y.J.; Lin, Y.C.; et al. Lactobacillus Attenuate the Progression of Pancreatic Cancer Promoted by Porphyromonas Gingivalis in K-ras(G12D) Transgenic Mice. Cancers 2020, 12, 3522. [Google Scholar] [CrossRef]
- Kita, A.; Fujiya, M.; Konishi, H.; Tanaka, H.; Kashima, S.; Iwama, T.; Ijiri, M.; Murakami, Y.; Takauji, S.; Goto, T.; et al. Probioticderived ferrichrome inhibits the growth of refractory pancreatic cancer cells. Int. J. Oncol. 2020, 57, 721–732. [Google Scholar] [CrossRef]
- Viaud, S.; Saccheri, F.; Mignot, G.; Yamazaki, T.; Daillere, R.; Hannani, D.; Enot, D.P.; Pfirschke, C.; Engblom, C.; Pittet, M.J.; et al. The intestinal microbiota modulates the anticancer immune effects of cyclophosphamide. Science 2013, 342, 971–976. [Google Scholar] [CrossRef] [Green Version]
- Iida, N.; Dzutsev, A.; Stewart, C.A.; Smith, L.; Bouladoux, N.; Weingarten, R.A.; Molina, D.A.; Salcedo, R.; Back, T.; Cramer, S.; et al. Commensal bacteria control cancer response to therapy by modulating the tumor microenvironment. Science 2013, 342, 967–970. [Google Scholar] [CrossRef]
- Round, J.L.; Mazmanian, S.K. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc. Natl. Acad. Sci USA 2010, 107, 12204–12209. [Google Scholar] [CrossRef] [Green Version]
- Balachandran, V.P.; Beatty, G.L.; Dougan, S.K. Broadening the Impact of Immunotherapy to Pancreatic Cancer: Challenges and Opportunities. Gastroenterology 2019, 156, 2056–2072. [Google Scholar] [CrossRef]
- Upadhrasta, S.; Zheng, L. Strategies in Developing Immunotherapy for Pancreatic Cancer: Recognizing and Correcting Multiple Immune “defects” in the Tumor Microenvironment. J. Clin. Med. 2019, 8, 1472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feig, C.; Jones, J.O.; Kraman, M.; Wells, R.J.; Deonarine, A.; Chan, D.S.; Connell, C.M.; Roberts, E.W.; Zhao, Q.; Caballero, O.L.; et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 20212–20217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Byrne, K.T.; Yan, F.; Yamazoe, T.; Chen, Z.; Baslan, T.; Richman, L.P.; Lin, J.H.; Sun, Y.H.; Rech, A.J.; et al. Tumor Cell-Intrinsic Factors Underlie Heterogeneity of Immune Cell Infiltration and Response to Immunotherapy. Immunity 2018, 49, 178–193.e7. [Google Scholar] [CrossRef] [Green Version]
- Sivan, A.; Corrales, L.; Hubert, N.; Williams, J.B.; Aquino-Michaels, K.; Earley, Z.M.; Benyamin, F.W.; Lei, Y.M.; Jabri, B.; Alegre, M.L.; et al. Commensal Bifidobacterium promotes antitumor immunity and facilitates anti-PD-L1 efficacy. Science 2015, 350, 1084–1089. [Google Scholar] [CrossRef] [Green Version]
- Mager, L.F.; Burkhard, R.; Pett, N.; Cooke, N.C.A.; Brown, K.; Ramay, H.; Paik, S.; Stagg, J.; Groves, R.A.; Gallo, M.; et al. Microbiome-derived inosine modulates response to checkpoint inhibitor immunotherapy. Science 2020, 369, 1481–1489. [Google Scholar] [CrossRef] [PubMed]
- Baruch, E.N.; Youngster, I.; Ben-Betzalel, G.; Ortenberg, R.; Lahat, A.; Katz, L.; Adler, K.; Dick-Necula, D.; Raskin, S.; Bloch, N.; et al. Fecal microbiota transplant promotes response in immunotherapy-refractory melanoma patients. Science 2021, 371, 602–609. [Google Scholar] [CrossRef]
- Davar, D.; Dzutsev, A.K.; McCulloch, J.A.; Rodrigues, R.R.; Chauvin, J.M.; Morrison, R.M.; Deblasio, R.N.; Menna, C.; Ding, Q.; Pagliano, O.; et al. Fecal microbiota transplant overcomes resistance to anti-PD-1 therapy in melanoma patients. Science 2021, 371, 595–602. [Google Scholar] [CrossRef]
- O’Keefe, S.J.; Li, J.V.; Lahti, L.; Ou, J.; Carbonero, F.; Mohammed, K.; Posma, J.M.; Kinross, J.; Wahl, E.; Ruder, E.; et al. Fat, fibre and cancer risk in African Americans and rural Africans. Nat. Commun. 2015, 6, 6342. [Google Scholar] [CrossRef] [Green Version]
- Leschner, S.; Weiss, S. Salmonella-allies in the fight against cancer. J. Mol. Med. 2010, 88, 763–773. [Google Scholar] [CrossRef]
- Tan, W.; Duong, M.T.; Zuo, C.; Qin, Y.; Zhang, Y.; Guo, Y.; Hong, Y.; Zheng, J.H.; Min, J.J. Targeting of pancreatic cancer cells and stromal cells using engineered oncolytic Salmonella typhimurium. Mol. Ther. 2022, 30, 662–671. [Google Scholar] [CrossRef] [PubMed]
- Ebelt, N.D.; Zamloot, V.; Zuniga, E.; Passi, K.B.; Sobocinski, L.J.; Young, C.A.; Blazar, B.R.; Manuel, E.R. Collagenase-Expressing Salmonella Targets Major Collagens in Pancreatic Cancer Leading to Reductions in Immunosuppressive Subsets and Tumor Growth. Cancers 2021, 13, 3565. [Google Scholar] [CrossRef]
- Ebelt, N.D.; Zuniga, E.; Passi, K.B.; Sobocinski, L.J.; Manuel, E.R. Hyaluronidase-Expressing Salmonella Effectively Targets Tumor-Associated Hyaluronic Acid in Pancreatic Ductal Adenocarcinoma. Mol. Cancer Ther. 2020, 19, 706–716. [Google Scholar] [CrossRef] [Green Version]
- Chavez-Arroyo, A.; Portnoy, D.A. Why is Listeria monocytogenes such a potent inducer of CD8+ T-cells? Cell Microbiol. 2020, 22, e13175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flickinger, J.C., Jr.; Rodeck, U.; Snook, A.E. Listeria monocytogenes as a Vector for Cancer Immunotherapy: Current Understanding and Progress. Vaccines 2018, 6, 48. [Google Scholar] [CrossRef] [Green Version]
- Salas-Benito, D.; Melero, I.; Ponz-Sarvise, M. Vaccination for Pancreatic Ductal Adenocarcinoma: A Hard Nut to Crack. Clin. Cancer Res. 2019, 25, 5435–5437. [Google Scholar] [CrossRef] [PubMed]
- Kim, V.M.; Blair, A.B.; Lauer, P.; Foley, K.; Che, X.; Soares, K.; Xia, T.; Muth, S.T.; Kleponis, J.; Armstrong, T.D.; et al. Anti-pancreatic tumor efficacy of a Listeria-based, Annexin A2-targeting immunotherapy in combination with anti-PD-1 antibodies. J. Immunother. Cancer 2019, 7, 132. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.; Lira, V.; Hudson, T.E.; Lemmens, E.E.; Hanson, W.G.; Flores, R.; Barajas, G.; Katibah, G.E.; Desbien, A.L.; Lauer, P.; et al. Recombinant Listeria promotes tumor rejection by CD8(+) T cell-dependent remodeling of the tumor microenvironment. Proc. Natl. Acad. Sci. USA 2018, 115, 8179. [Google Scholar] [CrossRef] [Green Version]
- Brockstedt, D.G.; Giedlin, M.A.; Leong, M.L.; Bahjat, K.S.; Gao, Y.; Luckett, W.; Liu, W.; Cook, D.N.; Portnoy, D.A.; Dubensky, T.W., Jr. Listeria-based cancer vaccines that segregate immunogenicity from toxicity. Proc. Natl. Acad. Sci. USA 2004, 101, 13832–13837. [Google Scholar] [CrossRef] [Green Version]
- Le, D.T.; Picozzi, V.J.; Ko, A.H.; Wainberg, Z.A.; Kindler, H.; Wang-Gillam, A.; Oberstein, P.; Morse, M.A.; Zeh, H.J., 3rd; Weekes, C.; et al. Results from a Phase IIb, Randomized, Multicenter Study of GVAX Pancreas and CRS-207 Compared with Chemotherapy in Adults with Previously Treated Metastatic Pancreatic Adenocarcinoma (ECLIPSE Study). Clin. Cancer Res. 2019, 25, 5493–5502. [Google Scholar] [CrossRef]
- Le, D.T.; Wang-Gillam, A.; Picozzi, V.; Greten, T.F.; Crocenzi, T.; Springett, G.; Morse, M.; Zeh, H.; Cohen, D.; Fine, R.L.; et al. Safety and survival with GVAX pancreas prime and Listeria Monocytogenes-expressing mesothelin (CRS-207) boost vaccines for metastatic pancreatic cancer. J. Clin. Oncol. 2015, 33, 1325–1333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Microbial Taxa | Impact of PC Tumorigenesis | Model/Methodology | |
|---|---|---|---|
| Oral Microbiota | Leptotrichia | Enhances [18,20]; Inhibits [19] | Ref#[18,19,20] Human—16S rRNA gene sequencing |
| Fusobacterium | Enhances [21,22,23] | Ref#[21] Human—TaqMan gene expression Assay for Fusobacterium (RT-qPCR); Ref#[22] Human—16S rRNA gene sequencing; Ref#[23] Human—ELISA and RT-qPCR on saliva | |
| Haemophilus | Inhibits [18] | Ref#[18] Human—16S rRNA gene sequencing | |
| Porphyromonas gingivalis | Enhances [19,24] | Ref#[19] Human—16S rRNA gene sequencing; Ref#[24] Human—serology/plasma antibodies | |
| Aggregatibacter | Enhances [19] | Ref#[19] Human—16S rRNA gene sequencing | |
| Gut Microbiota/Mycobiota | Helicobacter pylori | Enhances [25,26,27] | Ref#[25,26,27] Human—meta-analysis |
| Proteobacteria | Inhibits [28] | Ref#[28] Human—meta-analysis RNA-sequencing data from The Cancer Genome Atlas (TCGA) | |
| Bifidobacterium | Enhances [11] | Ref#[11] Human—16S rRNA gene sequencing | |
| Ascomycota (fungi) | Enhances [13] | Ref#[13] Human- MiSeq, alpha and beta diversity indices (Chao1, Shannon and Simpson indices) | |
| Ascomycota (fungi) | Enhances [13] | Ref#[13] Human—MiSeq, alpha and beta diversity indices (Chao1, Shannon and Simpson indices) | |
| Intratumoral Microbiota/Mycobiota | Proteobacteria | Enhances [11] | Ref#[11] Human—16S rRNA gene sequencing |
| Enterobacteriaceae | Enhances [29] | Ref#[29] Human—rRNA fluorescence in situ hybridization | |
| Pseudomonadaceae | Enhances [11,29] | Ref#[11] Human—16S rRNA gene sequencing | |
| Bacteroidetes | Enhances [16] | Ref#[16] Human—shotgun metagenomic and 16S rRNA amplicon sequencing | |
| Malassezia (fungi) | Enhances [13] | Ref#[13] Human—MiSeq, alpha and beta diversity indices (Chao1, Shannon and Simpson indices); |
| Intervention | Identifier# | Title | Tumor Type | Status |
|---|---|---|---|---|
| 16S rRNA gene sequencing assay | NCT03302637 | Oral Microbiome and Pancreatic Cancer | Pancreatic Cancer | Completed |
| Diagnostic test: microbiome evaluation | NCT04274972 | The Microbiome of Pancreatic Cancer: „PANDEMIC” Study (PANDEMIC) | Pancreas Cancer | Unknown |
| Diagnostic test: dental plaque sampling (qPCR) | NCT04993846 | Pancreatic Cancer and Oral Microbiome | Pancreas Cancer | Recruiting |
| Other: oral and rectal swabs for microbiome sequencing | NCT04922515 | Pancreatic Ductal Adenocarcinoma—Microbiome as Predictor of Subtypes (PDA-MAPS) | Pancreatic Cancer | Recruiting |
| Phase 4 Drug: pembrolizumab, ciprofloxacin and metronidazole | NCT03891979 | Gut Microbiome Modulation to Enable Efficacy of Checkpoint-based Immunotherapy in Pancreatic Adenocarcinoma | Pancreatic Cancer | Withdrawn |
| Drug: immunotherapy and chemotherapeutic agent microbiome analysis | NCT04638751 | ARGONAUT: Stool and Blood Sample Bank for Cancer Patients | Non-Small Cell Lung Cancer, Colorectal Cancer, Triple Negative Breast Cancer, Pancreas Cancer | Recruiting |
| Microbiome analysis | NCT04189393 | Microbiome Analysis in esoPhageal, PancreatIc and Colorectal CaNcer Patients Undergoing Gastrointestinal Surgery (MA-PPING) | Gastrointestinal Cancer, Colorectal Cancer, Pancreatic Cancer, Esophageal Cancer | Unknown |
| Microbiota analysis | NCT04931069 | Correlation between Complications after Pancreaticoduodenectomy and Microbiota (COMPAMIC) | Pancreatic Cancer | Recruiting |
| Phase 1 Drug: MRx0518 Radiation: hypofractionated pre-operative radiation | NCT04193904 | A Study of Live Biotherapeutic Product MRx0518 With Hypofractionated Radiation Therapy in Resectable Pancreatic Cancer | Pancreatic Cancer | Recruiting |
| phase 1 and 2 Drug: durvalumab Radiation: stereotactic ablative body radiotherapy (SABR) Microbiome analysis | NCT03245541 | Radiation Therapy in Combination With Durvalumab for People With Pancreatic Cancer | Pancreatic Adenocarcinoma | Recruiting |
| Observational study only microbiome analysis | NCT04476082 | Nutrition in Gastrointestinal Tumors (NutriGIT) | Pancreatic Cancer, Oesophageal Cancer, Colon Cancer, Liver Cancer, Rectal Cancer, Bile Duct Cancer | Recruiting |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amara, S.; Yang, L.V.; Tiriveedhi, V.; Muzaffar, M. Complex Role of Microbiome in Pancreatic Tumorigenesis: Potential Therapeutic Implications. Cells 2022, 11, 1900. https://doi.org/10.3390/cells11121900
Amara S, Yang LV, Tiriveedhi V, Muzaffar M. Complex Role of Microbiome in Pancreatic Tumorigenesis: Potential Therapeutic Implications. Cells. 2022; 11(12):1900. https://doi.org/10.3390/cells11121900
Chicago/Turabian StyleAmara, Suneetha, Li V. Yang, Venkataswarup Tiriveedhi, and Mahvish Muzaffar. 2022. "Complex Role of Microbiome in Pancreatic Tumorigenesis: Potential Therapeutic Implications" Cells 11, no. 12: 1900. https://doi.org/10.3390/cells11121900
APA StyleAmara, S., Yang, L. V., Tiriveedhi, V., & Muzaffar, M. (2022). Complex Role of Microbiome in Pancreatic Tumorigenesis: Potential Therapeutic Implications. Cells, 11(12), 1900. https://doi.org/10.3390/cells11121900







