Regulation of the Intestinal Extra-Adrenal Steroidogenic Pathway Component LRH-1 by Glucocorticoids in Ulcerative Colitis
Abstract
1. Introduction
2. Materials and Methods
2.1. Human Samples
2.2. Reagents
2.3. Ex Vivo Culture of Intestinal Biopsies
2.4. Human Colonic Organoids
2.5. Cell Line In Vitro Experiments
2.6. RT-qPCR Assays
2.7. Immunohistochemistry (IHC)
2.8. Animal Experimentation
2.9. Immunofluorescence of Slides from Patient’ Biopsies, Murine DSS-Induced Models and Organoid Experiments
2.10. Transcriptome Array
2.11. Cortisol Measurement
2.12. Cytometric Bead Array (CBA)
2.13. In Silico Analysis of GREs in Human NR5A2 Gene, Immunoprecipitation of Chromatin Coupled to Real Time PCR and Formaldehyde-Assisted Isolation of Regulatory Elements (FAIRE) Analysis
2.14. Western Blotting
2.15. Statistical Analysis
3. Results
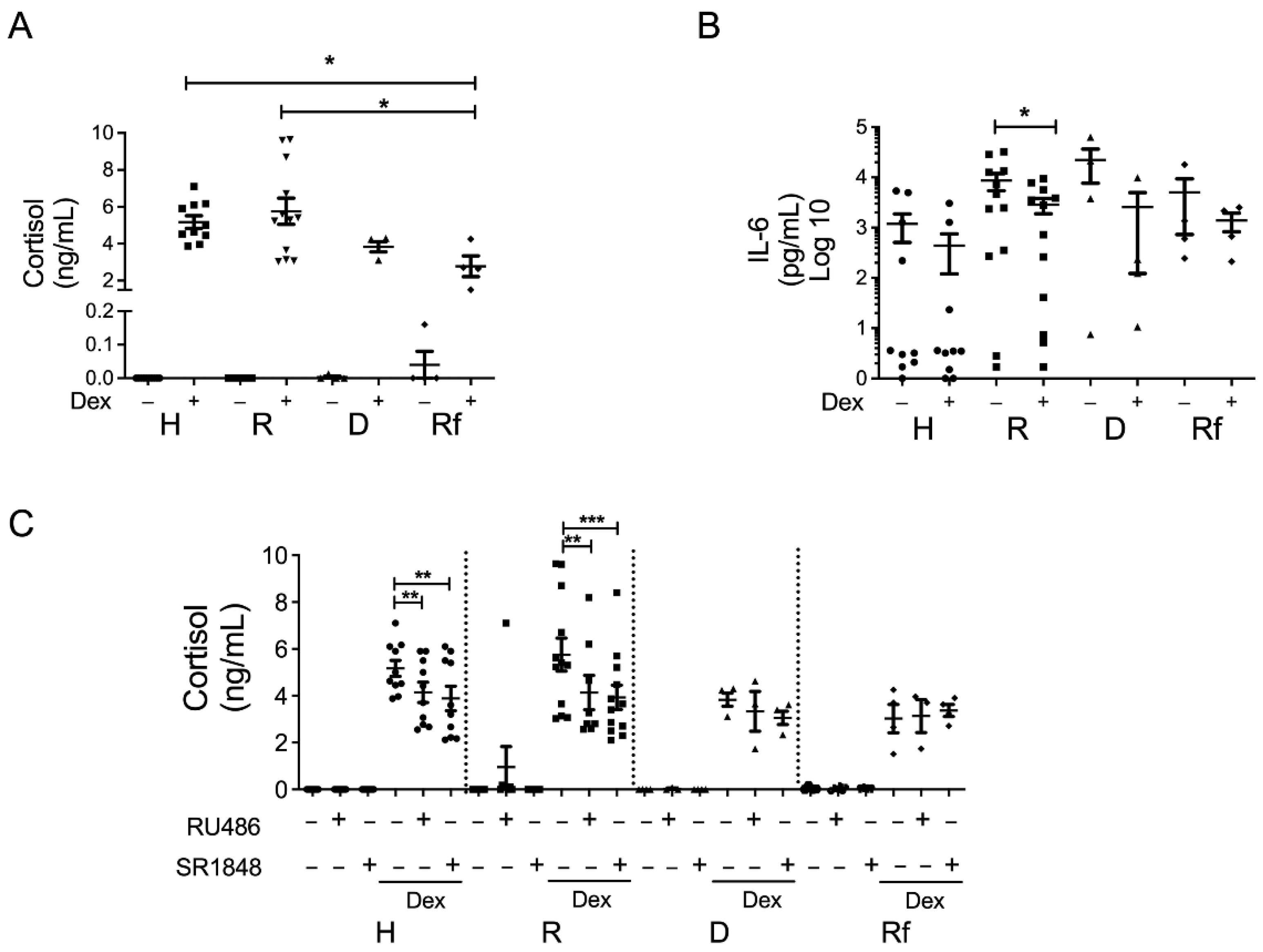
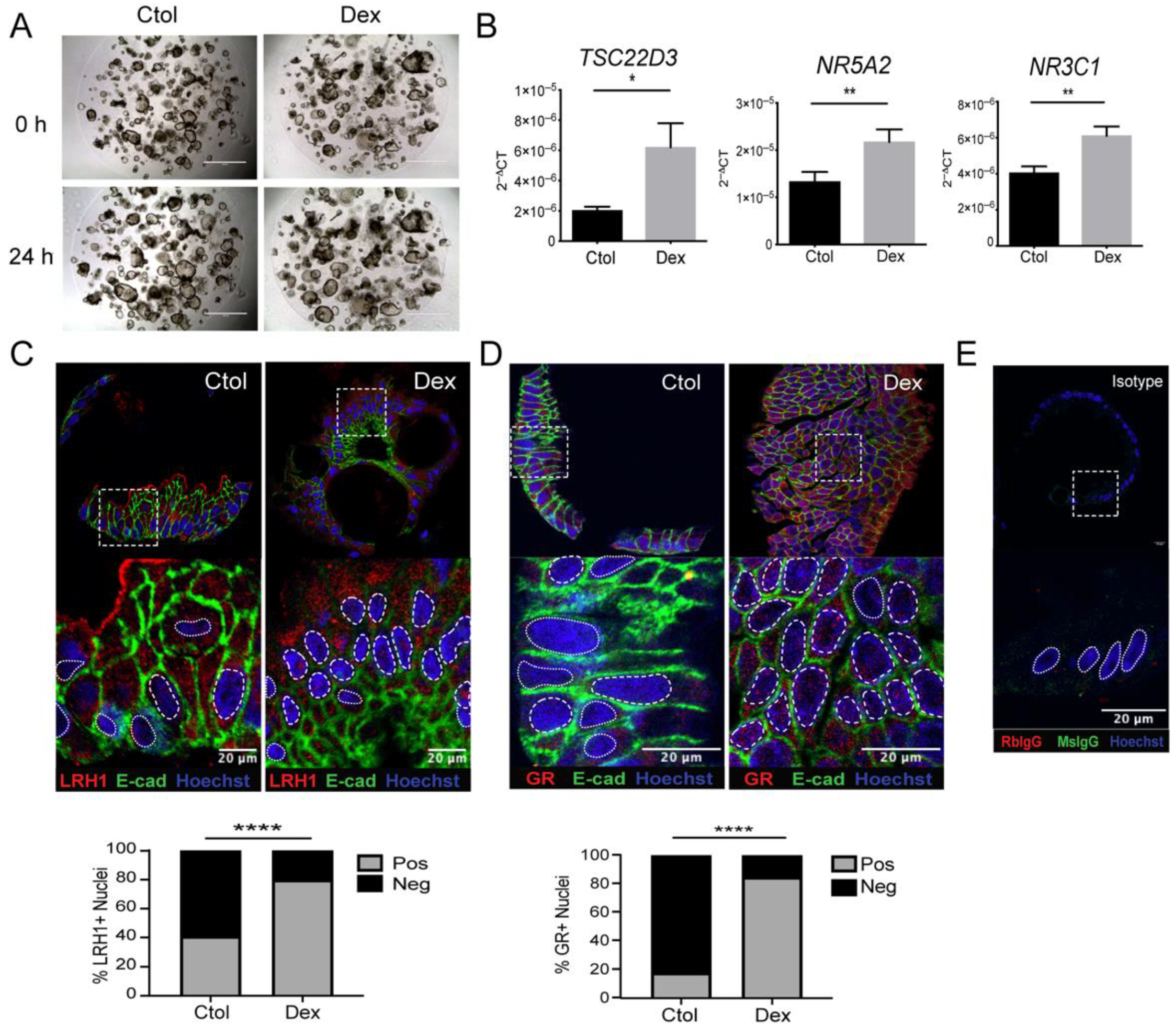
3.1. Dexamethasone Regulates Steroidogenesis in Colonic Mucosa from Steroid-Responder UC Patients
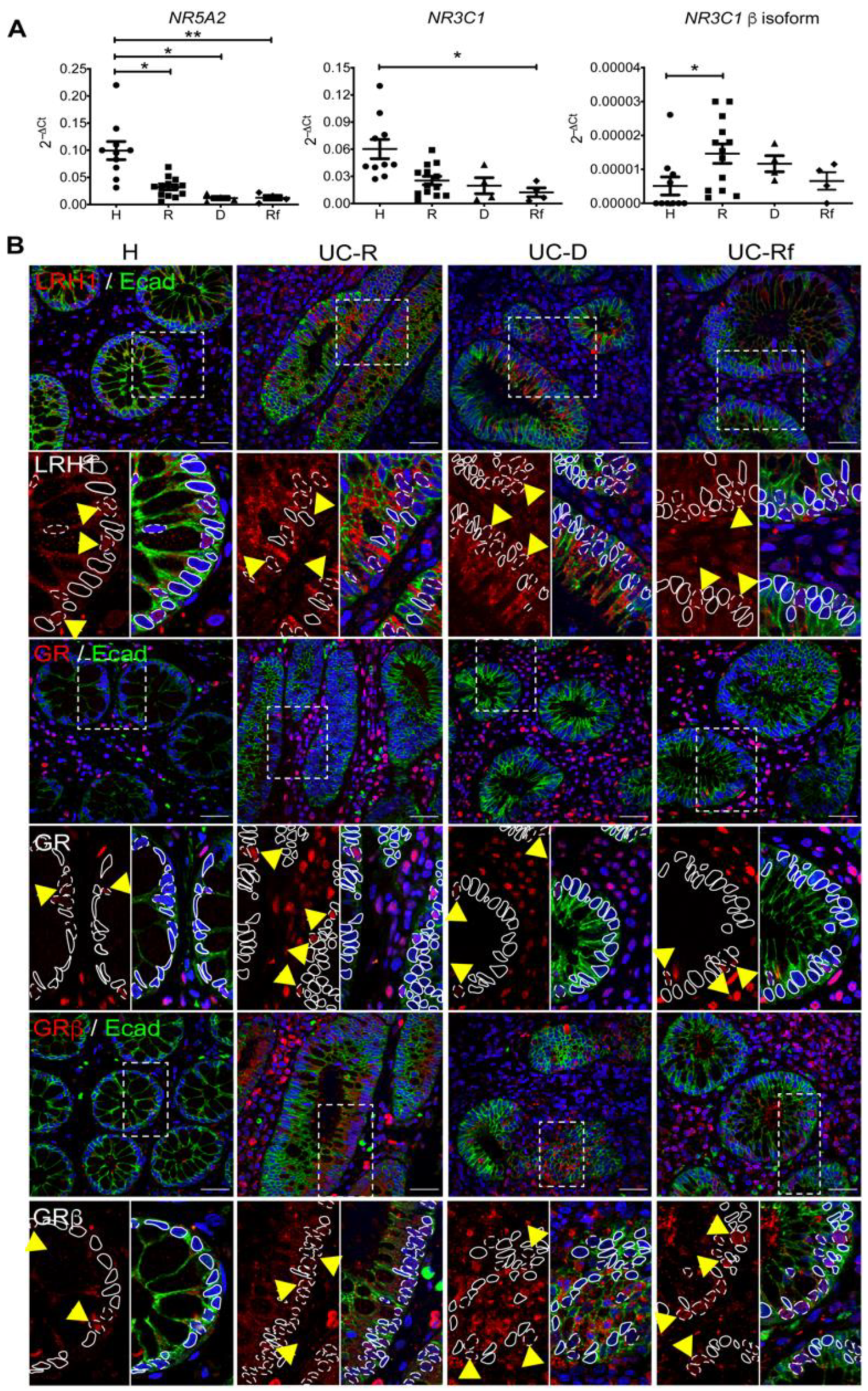
3.2. Steroidogenesis Pathway Components in Intestinal Mucosa of UC Patients
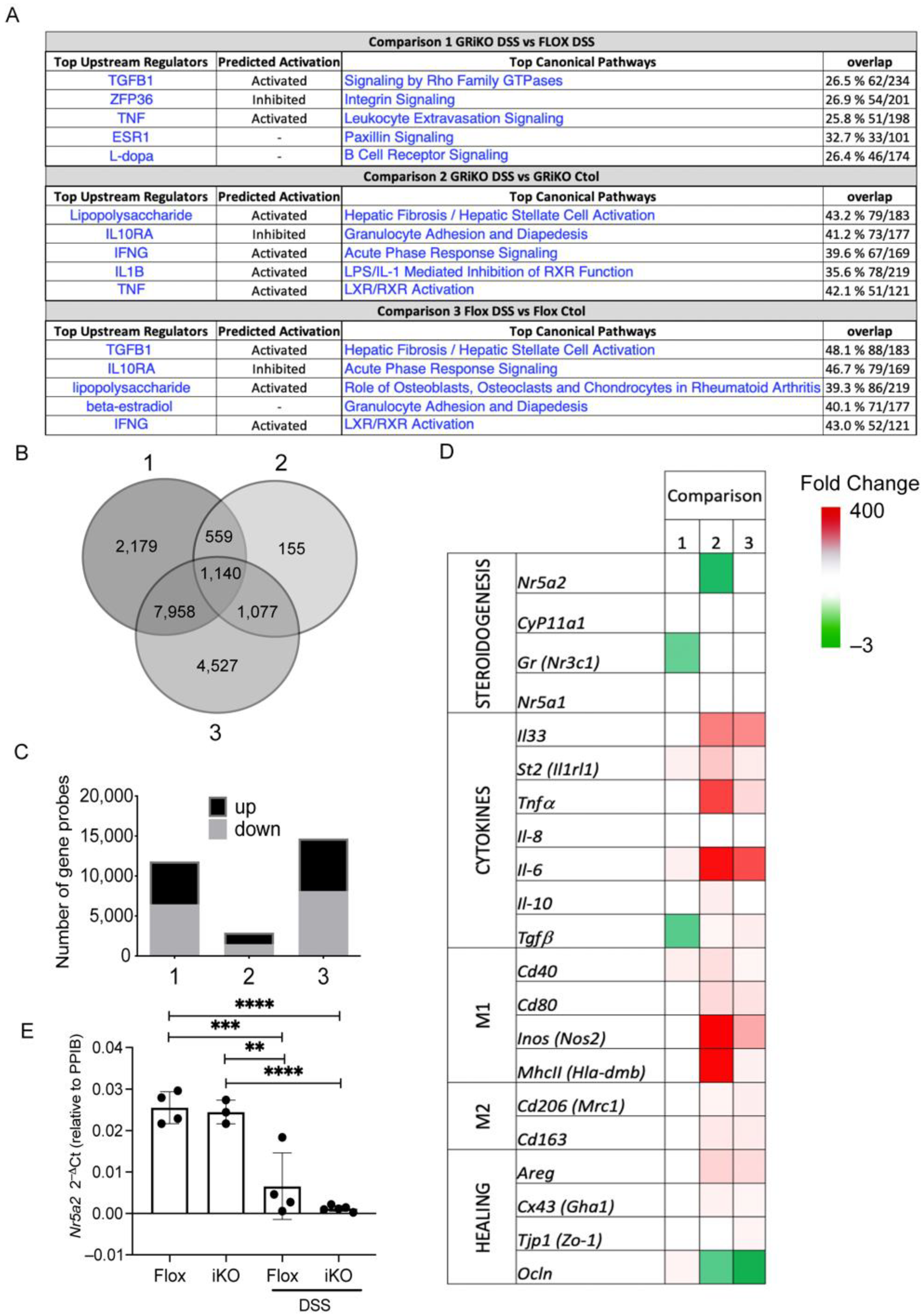
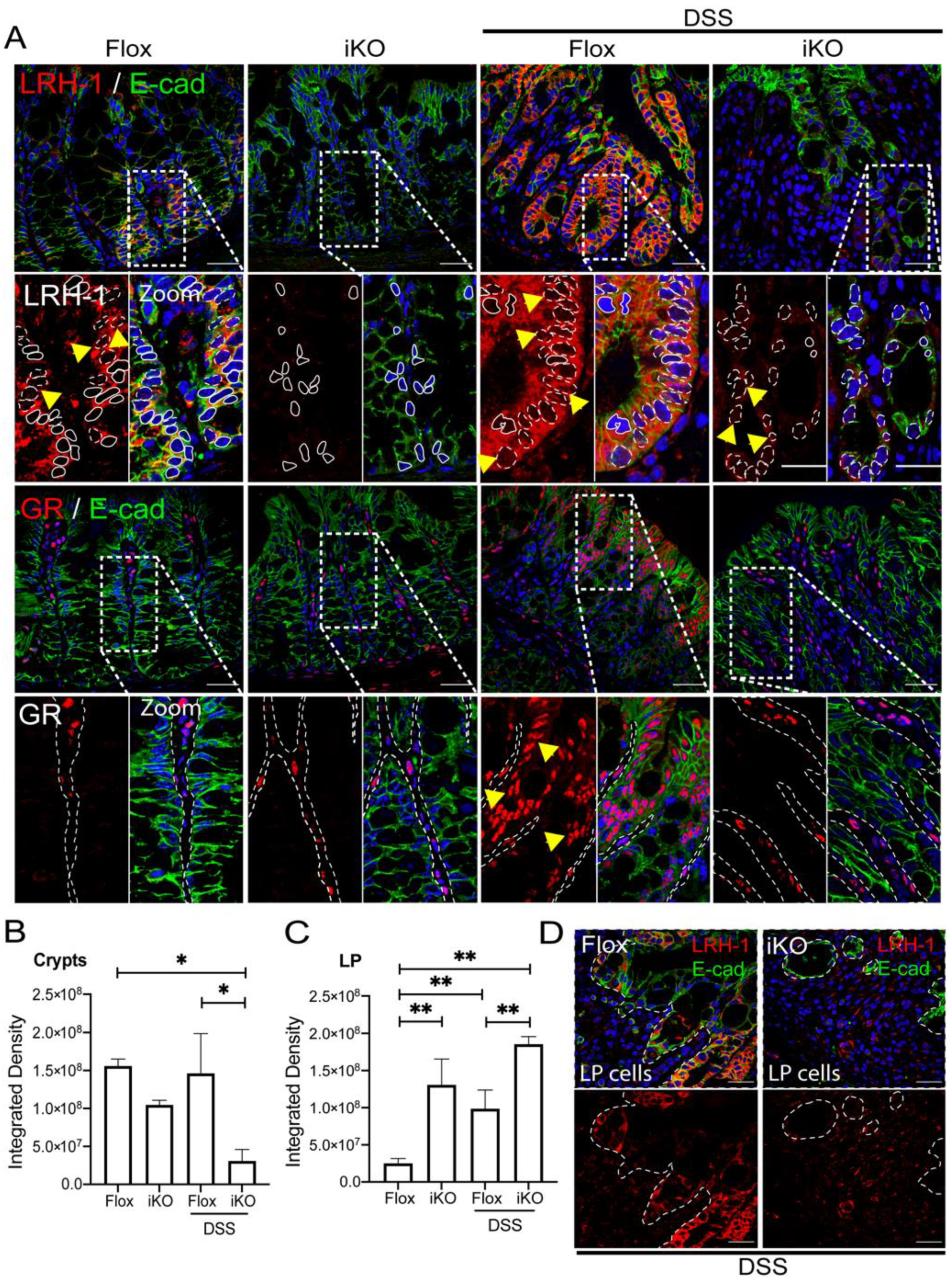
3.3. GRiKO DSS Mouse Model Shows Strong Upregulation of Inflammatory Mediators and Spatially Distinct LRH-1 Expression in Intestinal Mucosa
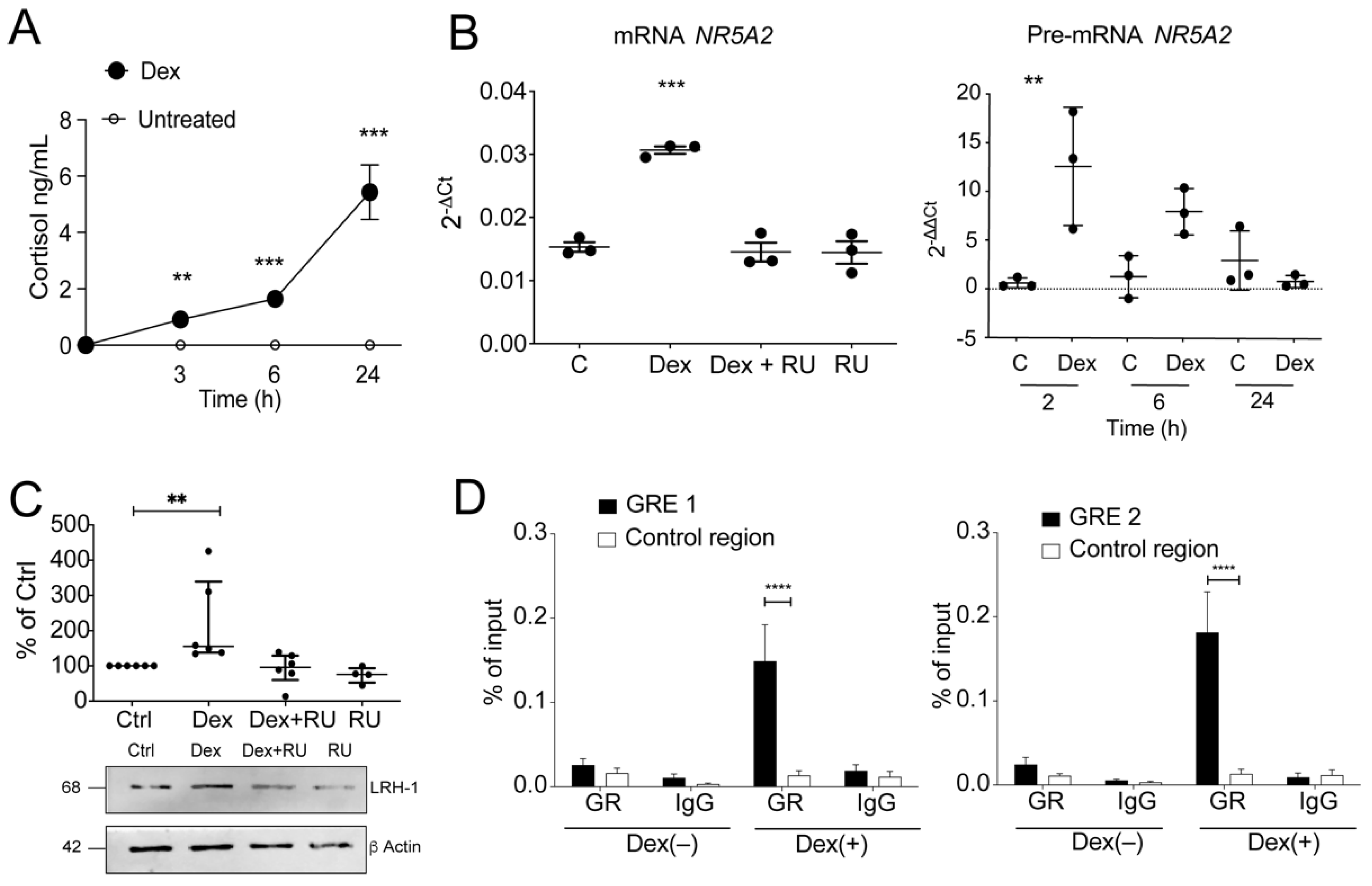
3.4. GR-Dependent Activation of Steroidogenesis in Human Colonocytes
3.5. Glucocorticoids Regulate LRH-1 Expression by Binding to GREs in Regulatory Regions of the LRH-1/NR5A2 Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rubin, D.T.; Ananthakrishnan, A.N.; Siegel, C.A.; Sauer, B.G.; Long, M.D. ACG Clinical Guideline: Ulcerative colitis in adults. Am. J. Gastroenterol. 2019, 114, 384–413. [Google Scholar] [CrossRef] [PubMed]
- Sands, B.E. Medical Therapy of Steroid-Resistant Crohn’s Disease. Can. J. Gastroenterol. 2000, 14, 33C–37C. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lichtenstein, G.R. Approach to corticosteroid-dependent and corticosteroid-refractory Crohn’s disease. Inflamm. Bowel Dis. 2001, 7, S23–S29. [Google Scholar] [CrossRef] [PubMed]
- Fujishima, S.; Takeda, H.; Kawata, S.; Yamakawa, M. The relationship between the expression of the glucocorticoid receptor in biopsied colonic mucosa and the glucocorticoid responsiveness of ulcerative colitis patients. Clin. Immunol. 2009, 133, 208–217. [Google Scholar] [CrossRef]
- Chen, H.-L.; Li, L.-R. Glucocorticoid Receptor Gene Polymorphisms and Glucocorticoid Resistance in Inflammatory Bowel Disease: A Meta-Analysis. Dig. Dis. Sci. 2012, 57, 3065–3075. [Google Scholar] [CrossRef]
- Taves, M.D.; Gomez-Sanchez, C.E.; Soma, K.K. Extra-adrenal glucocorticoids and mineralocorticoids: Evidence for local synthesis, regulation, and function. Am. J. Physiol.-Endocrinol. Metab. 2011, 301, E11–E24. [Google Scholar] [CrossRef]
- Asser, L.; Hescot, S.; Viengchareun, S.; Delemer, B.; Trabado, S.; Lombès, M. Autocrine positive regulatory feedback of glucocorticoid secretion: Glucocorticoid receptor directly impacts H295R human adrenocortical cell function. Mol. Cell. Endocrinol. 2014, 395, 1–9. [Google Scholar] [CrossRef]
- Mueller, M.; Atanasov, A.; Cima, I.; Corazza, N.; Schoonjans, K.; Brunner, T. Differential regulation of glucocorticoid synthesis in murine intestinal epithelial versus adrenocortical cell lines. Endocrinology 2007, 148, 1445–1453. [Google Scholar] [CrossRef]
- Slominski, R.M.; Tuckey, R.C.; Manna, P.R.; Jetten, A.M.; Postlethwaite, A.; Raman, C.; Slominski, A.T. Extra-adrenal glucocorticoid biosynthesis: Implications for autoimmune and inflammatory disorders. Genes Immun. 2020, 21, 150–168. [Google Scholar] [CrossRef]
- Cima, I.; Corazza, N.; Dick, B.; Fuhrer, A.; Herren, S.; Jakob, S.; Ayuni, E.; Mueller, C.; Brunner, T. Intestinal epithelial cells synthesize glucocorticoids and regulate T cell activation. J. Exp. Med. 2004, 200, 1635–1646. [Google Scholar] [CrossRef]
- Bayrer, J.R.; Wang, H.; Nattiv, R.; Suzawa, M.; Escusa, H.S.; Fletterick, R.J.; Klein, O.D.; Moore, D.D.; Ingraham, H.A. LRH-1 mitigates intestinal inflammatory disease by maintaining epithelial homeostasis and cell survival. Nat. Commun. 2018, 9, 4055. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Marcos, P.J.; Auwerx, J.; Schoonjans, K. Emerging actions of the nuclear receptor LRH-1 in the gut. Biochim. Biophys. Acta -Mol. Basis Dis. 2011, 1812, 947–955. [Google Scholar] [CrossRef] [PubMed]
- Sidler, D.; Renzulli, P.; Schnoz, C.; Berger, B.; Schneider-Jakob, S.; Flück, C.; Inderbitzin, D.; Corazza, N.; Candinas, D.; Brunner, T. Colon cancer cells produce immunoregulatory glucocorticoids. Oncogene 2011, 30, 2411–2419. [Google Scholar] [CrossRef] [PubMed]
- Noti, M.; Corazza, N.; Tuffin, G.; Schoonjans, K.; Brunner, T. Lipopolysaccharide induces intestinal glucocorticoid synthesis in a TNFalpha-dependent manner. FASEB J. 2010, 24, 1340–1346. [Google Scholar] [CrossRef]
- Mahata, B.; Pramanik, J.; van der Weyden, L.; Polanski, K.; Kar, G.; Riedel, A.; Chen, X.; Fonseca, N.A.; Kundu, K.; Campos, L.S.; et al. Tumors induce de novo steroid biosynthesis in T cells to evade immunity. Nat. Commun. 2020, 11, 3588. [Google Scholar] [CrossRef]
- Schwaderer, J.; Gaiser, A.-K.; Phan, T.S.; Delgado, M.E.; Brunner, T. Liver receptor homolog-1 (NR5a2) regulates CD95/Fas ligand transcription and associated T-cell effector functions. Cell Death Dis. 2017, 8, e2745. [Google Scholar] [CrossRef]
- Coste, A.; Dubuquoy, L.; Barnouin, R.; Annicotte, J.; Magnier, B.; Notti, M.; Corazza, N.; Antal, M.C.; Metzger, D.; Desreumaux, P.; et al. LRH-1-mediated glucocorticoid synthesis in enterocytes protects against inflammatory bowel disease. Proc. Natl. Acad. Sci. USA 2007, 104, 13098–13103. [Google Scholar] [CrossRef]
- Huang, S.C.; Lee, C.T.; Chung, B.C. Tumor necrosis factor suppresses NR5A2 activity and intestinal glucocorticoid synthesis to sustain chronic colitis. Sci. Signal. 2014, 7, ra20. [Google Scholar] [CrossRef]
- Olivares-Morales, M.J.; De La Fuente, M.K.; Dubois-Camacho, K.; Parada, D.; Diaz-Jiménez, D.; Torres-Riquelme, A.; Xu, X.; Chamorro-Veloso, N.; Naves, R.; Gonzalez, M.-J.; et al. Glucocorticoids Impair Phagocytosis and Inflammatory Response Against Crohn’s Disease-Associated Adherent-Invasive Escherichia coli. Front. Immunol. 2018, 9, 1026. [Google Scholar] [CrossRef]
- Lamb, C.A.; Kennedy, N.A.; Raine, T.; Hendy, P.A.; Smith, P.J.; Limdi, J.K.; Hayee, B.; Lomer, M.C.E.; Parkes, G.C.; Selinger, C.; et al. British Society of Gastroenterology consensus guidelines on the management of inflammatory bowel disease in adults. Gut 2019, 68, s1–s106. [Google Scholar] [CrossRef]
- Dignass, A.; Eliakim, R.; Magro, F.; Maaser, C.; Chowers, Y.; Geboes, K.; Mantzaris, G.; Reinisch, W.; Colombel, J.F.; Vermeire, S.; et al. Second European evidence-based consensus on the diagnosis and management of ulcerative colitis Part 1: Definitions and diagnosis. J. Crohn’s Colitis 2012, 6, 965–990. [Google Scholar] [CrossRef]
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.J.; van Es, J.H.; van den Brink, S.; van Houdt, W.J.; Pronk, A.; van Gorp, J.; Siersema, P.D.; et al. Long-term Expansion of Epithelial Organoids From Human Colon, Adenoma, Adenocarcinoma, and Barrett’s Epithelium. Gastroenterology 2011, 141, 1762–1772. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Vries, R.G.; Snippert, H.J.; Van De Wetering, M.; Barker, N.; Stange, D.E.; Van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Fujii, M.; Matano, M.; Toshimitsu, K.; Takano, A.; Mikami, Y.; Nishikori, S.; Sugimoto, S.; Sato, T. Human Intestinal Organoids Maintain Self-Renewal Capacity and Cellular Diversity in Niche-Inspired Culture Condition. Cell Stem Cell 2018, 23, 787–793.e6. [Google Scholar] [CrossRef] [PubMed]
- Oakley, R.H.; Cruz-Topete, D.; He, B.; Foley, J.F.; Myers, P.H.; Xu, X.; Gomez-Sanchez, C.E.; Chambon, P.; Willis, M.S.; Cidlowski, J.A. Cardiomyocyte glucocorticoid and mineralocorticoid receptors directly and antagonistically regulate heart disease in mice. Sci. Signal. 2019, 12, eaau9685. [Google Scholar] [CrossRef]
- Oakley, R.H.; Ren, R.; Cruz-Topete, D.; Bird, G.S.; Myers, P.H.; Boyle, M.C.; Schneider, M.D.; Willis, M.S.; Cidlowski, J.A. Essential role of stress hormone signaling in cardiomyocytes for the prevention of heart disease. Proc. Natl. Acad. Sci. USA 2013, 110, 17035–17040. [Google Scholar] [CrossRef]
- Tang, S.; Zhang, Z.; Oakley, R.H.; Li, W.; He, W.; Xu, X.; Ji, M.; Xu, Q.; Chen, L.; Wellman, A.S.; et al. Intestinal epithelial glucocorticoid receptor promotes chronic inflammation–associated colorectal cancer. JCI Insight 2021, 6, e151815. [Google Scholar] [CrossRef]
- Shichijo, K.; Matuu, M.; Ikeda, Y.; Naito, S.; Ito, M.; Sekine, I. Clinicopathologic study of dextran sulfate sodium experimental acute colitis. JPN J. Pharmacol. 1998, 76, 297. [Google Scholar] [CrossRef]
- Erben, U.; Loddenkemper, C.; Doerfel, K.; Spieckermann, S.; Haller, D.; Heimesaat, M.M.; Zeitz, M.; Siegmund, B.; Kühl, A.A. A guide to histomorphological evaluation of intestinal inflammation in mouse models. Int. J. Clin. Exp. Pathol. 2014, 7, 4557–4576. [Google Scholar]
- Sandelin, A.; Alkema, W.; Engstrom, P.; Wasserman, W.W.; Lenhard, B. JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004, 32, 91D–94D. [Google Scholar] [CrossRef]
- De Iudicibus, S.; Franca, R.; Martelossi, S.; Ventura, A.; Decorti, G. Molecular mechanism of glucocorticoid resistance in inflammatory bowel disease. World J. Gastroenterol. 2011, 17, 1095–1108. [Google Scholar] [CrossRef] [PubMed]
- Díaz-Jiménez, D.; Núñez, L.; De La Fuente, M.; Dubois-Camacho, K.; Sepúlveda, H.; Montecino, M.; Torres-Riquelme, A.; García-González, P.; Chnaiderman, J.; Vossenkamper, A.; et al. A functional IL1RL1 variant regulates corticosteroid-induced sST2 expression in ulcerative colitis. Sci. Rep. Sci. Rep. 2017, 7, 10180. [Google Scholar] [CrossRef]
- Muzzi, C.; Watanabe, N.; Twomey, E.; Meers, G.K.; Reichardt, H.M.; Bohnenberger, H.; Reichardt, S.D. The glucocorticoid receptor in intestinal epithelial cells alleviates colitis and associated colorectal cancer in mice. Cell. Mol. Gastroenterol. Hepatol. 2021, 11, 1505–1518. [Google Scholar] [CrossRef] [PubMed]
- Makita, S.; Takatori, H.; Nakajima, H. Post-Transcriptional Regulation of Immune Responses and Inflammatory Diseases by RNA-Binding ZFP36 Family Proteins. Front. Immunol. 2021, 12, 2636. [Google Scholar] [CrossRef] [PubMed]
- Fornes, O.; Castro-Mondragon, J.A.; Khan, A.; van der Lee, R.; Zhang, X.; Richmond, P.A.; Modi, B.P.; Correard, S.; Gheorghe, M.; Baranašić, D.; et al. JASPAR 2020: Update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020, 48, D87–D92. [Google Scholar] [CrossRef]
- Travis, S.P.L. Review article: The management of mild to severe acute ulcerative colitis. Aliment. Pharmacol. Ther. Suppl. 2004, 20, 88–92. [Google Scholar] [CrossRef]
- Wang, H.; Lee, J.M.; Zhou, Y.; Moore, D.D. Targeting the Nuclear Receptor LRH-1 to Treat Inflammatory Bowel Disease in Mice. Gastroenterology 2017, 152, S90. [Google Scholar] [CrossRef]
- Corzo, C.A.; Mari, Y.; Chang, M.R.; Khan, T.; Kuruvilla, D.; Nuhant, P.; Kumar, N.; West, G.M.; Duckett, D.R.; Roush, W.R.; et al. Antiproliferation Activity of a Small Molecule Repressor of Liver Receptor Homolog 1. Mol. Pharmacol. 2015, 87, 296–304. [Google Scholar] [CrossRef]
- Borgius, L.J.; Steffensen, K.R.; Gustafsson, J.-Å.; Treuter, E. Glucocorticoid Signaling Is Perturbed by the Atypical Orphan Receptor and Corepressor SHP. J. Biol. Chem. 2002, 277, 49761–49766. [Google Scholar] [CrossRef]
- Mays, S.G.; Okafor, C.D.; Tuntland, M.L.; Whitby, R.J.; Dharmarajan, V.; Stec, J.; Griffin, P.R.; Ortlund, E.A. Structure and Dynamics of the Liver Receptor Homolog 1–PGC1 α Complex. Mol. Pharmacol. 2017, 92, 1–11. [Google Scholar] [CrossRef]
- Huang, J.; Jia, R.; Brunner, T. Local synthesis of immunosuppressive glucocorticoids in the intestinal epithelium regulates anti-viral immune responses. Cell. Immunol. 2018, 334, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Dubois-Camacho, K.; Diaz-Jimenez, D.; De la Fuente, M.; Quera, R.; Simian, D.; Martínez, M.; Landskron, G.; Olivares-Morales, M.; Cidlowski, J.A.; Xu, X.; et al. Inhibition of miR-378a-3p by Inflammation Enhances IL-33 Levels: A Novel Mechanism of Alarmin Modulation in Ulcerative Colitis. Front. Immunol. 2019, 10, 2449. [Google Scholar] [CrossRef] [PubMed]
- Czarnewski, P.; Parigi, S.M.; Sorini, C.; Diaz, O.E.; Das, S.; Gagliani, N.; Villablanca, E.J. Conserved transcriptomic profile between mouse and human colitis allows unsupervised patient stratification. Nat. Commun. 2019, 10, 2892. [Google Scholar] [CrossRef]
- Schoonjans, K.; Dubuquoy, L.; Mebis, J.; Fayard, E.; Wendling, O.; Haby, C.; Geboes, K.; Auwerx, J. Liver receptor homolog 1 contributes to intestinal tumor formation through effects on cell cycle and inflammation. Proc. Natl. Acad. Sci. USA 2005, 102, 2058–2062. [Google Scholar] [CrossRef] [PubMed]
- Flood, L.; Innala, E.; Löfberg, R.; Wikström, A.-C. Patients with ulcerative colitis responding to steroid treatment up-regulate glucocorticoid receptor levels in colorectal mucosa. J. Crohn’s Colitis 2008, 2, 123–130. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Honda, M.; Orii, F.; Ayabe, T.; Imai, S.; Ashida, T.; Obara, T.; Kohgo, Y. Expression of glucocorticoid receptor β in lymphocytes of patients with glucocorticoid-resistant ulcerative colitis. Gastroenterology 2000, 118, 859–866. [Google Scholar] [CrossRef]
- Webster, J.C.; Oakley, R.H.; Jewell, C.M.; Cidlowski, J.A. Proinflammatory cytokines regulate human glucocorticoid receptor gene expression and lead to the accumulation of the dominant negative beta isoform: A mechanism for the generation of glucocorticoid resistance. Proc. Natl. Acad. Sci. USA 2001, 98, 6865–6870. [Google Scholar] [CrossRef]
- González, S.; Aguilera, S.; Alliende, C.; Urzúa, U.; Quest, A.F.G.; Herrera, L.; Molina, C.; Hermoso, M.; Ewert, P.; Brito, M.; et al. Alterations in type I hemidesmosome components suggestive of epigenetic control in the salivary glands of patients with Sjögren’s syndrome. Arthritis Rheum. 2011, 63, 1106–1115. [Google Scholar] [CrossRef]
- Vogel, C.; De Sousa Abreu, R.; Ko, D.; Le, S.Y.; Shapiro, B.A.; Burns, S.C.; Sandhu, D.; Boutz, D.R.; Marcotte, E.M.; Penalva, L.O. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol. Syst. Biol. 2010, 6, 400. [Google Scholar] [CrossRef]
- Lai, T.C.; Hu, M.C. Regulation of liver receptor homologue-1 by DDB2 E3 ligase activity is critical for hepatic glucose metabolism. Sci. Rep. 2019, 9, 5304. [Google Scholar] [CrossRef]
- Yang, C.; Zhou, W.; Jeon, M.; Demydenko, D.; Harada, Y.; Zhou, H.; Liu, Y.-C. Negative Regulation of the E3 Ubiquitin Ligase Itch via Fyn-Mediated Tyrosine Phosphorylation. Mol. Cell 2006, 21, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Labuda, T.; Xia, Y.; Gallagher, E.; Fang, D.; Liu, Y.-C.; Karin, M. Jun turnover is controlled through JNK-dependent phosphorylation of the E3 ligase Itch. Science 2004, 306, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Quax, R.A.; van Laar, J.A.; van Heerebeek, R.; Greiner, K.; Ben-Chetrit, E.; Stanford, M.; Wallace, G.R.; Fortune, F.; Ghabra, M.; Soylu, M.; et al. Glucocorticoid sensitivity in Behçet’s disease. Endocr. Connect. 2012, 1, 103–111. [Google Scholar] [CrossRef] [PubMed]
| Demographic/Clinical Data | Healthy Controls n = 10 | Ulcerative Colitis Patients | ||
|---|---|---|---|---|
| Responders n = 13 | Steroid-Refractory n = 4 | Steroid-Dependent n = 4 | ||
| Gender (n; %) Male Female | 5 (50) 5 (50) | 6 (46) 7 (54) | 2 (50) 2 (50) | 1 (25) 3 (75) |
| Age in years (median; range) | 53 (41–66) | 32 (20–63) | 29 (22–38) | 38 (24–41) |
| Body mass index (median; range) | 26 (20.7–29.8) | 24.1 (16.9–29.8) | 22 (14.7–32.1) | 21 (18–26.3) |
| Smoking habit (n; %) | 1 (10) | 1 (8) | 1 (25) | 1 (25) |
| Family history of IBD (n; %) | 0 (0) | 2 (15) | 0 (0) | 1 (25) |
| Years of disease (median; range) | - | 1 (0–8) | 1 (1–2) | 8 (3–20) |
| Extraintestinal manifestations (n; %) | - | 5 (38) | 0 (0) | 3 (75) |
| Montreal classification (n; %) E1: Extensive colitis E2: Left-sided colitis E3: Proctitis | - | 11 (85) 2 (15) 0 (0) | 3 (75) 1 (25) 0 (0) | 3 (75) 1 (25) 0 (0) |
| IBD current treatment (n; %) 5—Aminosalicylates Azathioprine Methotrexate 6-Mercaptopurine | - | 9 (69) 2 (15) 1 (8) 1 (8) | 4 (100) 0 (0) 0 (0) 0 (0) | 2 (50) 2 (50) 0 (0) 0 (0) |
| Clinical Mayo score (median; range) | - | 4 (1–8) | 4 (2–7) | 5 (3–5) |
| Endoscopic Mayo score 2 (Moderate activity) 3 (Severe activity) | - | 9 (69) 4 (31) | 2 (50) 2 (50) | 3 (75) 1 (25) |
| Fecal calprotectin (median; range) | - | 939 (254–2410) | 600 (258–1320) | 640 (600–1200) |
| Evaluation | GRflox (n = 10) | GRiKO (n = 8) |
|---|---|---|
| Rectal bleeding (% of mice at day 7) | 40 | 59 |
| Rectal bleeding severity (% of mice at day 7) | 5% severe, 20% mild | 18% severe, 36% mild |
| Colon length (cm) | 6.7 ± 0.2 | 5.9 ± 0.1 |
| Body weight loss (% of initial weight) *** | 91 ± 1 | 86 ± 1 |
| Disease activity index (DAI) *** | 1.8 ± 0.2 | 3.15 ± 0.15 |
| Pathological Scores: | ||
| Inflammation (p = 0.06) | 2.3 ± 0.4 | 3.4 ± 0.3 |
| Erosion * | 2.1 ± 0.6 | 3.8 ± 0.1 |
| Atrophy | 2.9 ± 0.5 | 3.7 ± 0.2 |
| Fibrosis | 2.5 ± 0.4 | 2.9 ± 0.1 |
| Edema * | 1.3 ± 0.5 | 3 ± 0.7 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Landskron, G.; Dubois-Camacho, K.; Orellana-Serradell, O.; De la Fuente, M.; Parada-Venegas, D.; Bitrán, M.; Diaz-Jimenez, D.; Tang, S.; Cidlowski, J.A.; Li, X.; et al. Regulation of the Intestinal Extra-Adrenal Steroidogenic Pathway Component LRH-1 by Glucocorticoids in Ulcerative Colitis. Cells 2022, 11, 1905. https://doi.org/10.3390/cells11121905
Landskron G, Dubois-Camacho K, Orellana-Serradell O, De la Fuente M, Parada-Venegas D, Bitrán M, Diaz-Jimenez D, Tang S, Cidlowski JA, Li X, et al. Regulation of the Intestinal Extra-Adrenal Steroidogenic Pathway Component LRH-1 by Glucocorticoids in Ulcerative Colitis. Cells. 2022; 11(12):1905. https://doi.org/10.3390/cells11121905
Chicago/Turabian StyleLandskron, Glauben, Karen Dubois-Camacho, Octavio Orellana-Serradell, Marjorie De la Fuente, Daniela Parada-Venegas, Mirit Bitrán, David Diaz-Jimenez, Shuang Tang, John A. Cidlowski, Xiaoling Li, and et al. 2022. "Regulation of the Intestinal Extra-Adrenal Steroidogenic Pathway Component LRH-1 by Glucocorticoids in Ulcerative Colitis" Cells 11, no. 12: 1905. https://doi.org/10.3390/cells11121905
APA StyleLandskron, G., Dubois-Camacho, K., Orellana-Serradell, O., De la Fuente, M., Parada-Venegas, D., Bitrán, M., Diaz-Jimenez, D., Tang, S., Cidlowski, J. A., Li, X., Molina, H., Gonzalez, C. M., Simian, D., Lubascher, J., Pola, V., Montecino, M., Blokzijl, T., Faber, K. N., González, M.-J., ... Hermoso, M. A. (2022). Regulation of the Intestinal Extra-Adrenal Steroidogenic Pathway Component LRH-1 by Glucocorticoids in Ulcerative Colitis. Cells, 11(12), 1905. https://doi.org/10.3390/cells11121905







