Quantitative Succinyl-Proteome Profiling of Turnip (Brassica rapa var. rapa) in Response to Cadmium Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Cd Treatment
2.2. Cd Concentration Determination
2.3. Protein Extraction, Qualification, and Quantification
2.4. WB Analysis
2.5. Succinyl-Proteome Profiling
2.5.1. Trypsin Digestion
2.5.2. Succinylation Modification Enrichment
2.5.3. LC–MS/MS Analysis
2.5.4. Database Searching
2.5.5. Motif Analysis
2.5.6. Protein Function Annotation
2.5.7. Protein Interaction Network Analysis
2.6. Identification of Differentially Succinylated Sites (DSSs)
- Firstly, after the signal intensity value (I) of the peptide in different samples was changed by centralization, the relative quantification value (R) of the peptide in different samples was obtained. The calculation formula was as follows: where i and j represent the sample and the peptide, respectively:Rij = Iij/Mean (Ij)
- In order to eliminate the systematic error of sample loading in MS determination, the relative quantitative value of the peptide needed to be corrected by median normalization (NR). The calculation formula was as follows:NRij = Rij/Median (Ri)
- The relative quantitative value of the protein was expressed by the median of the relative quantitative value of the corresponding specific peptide to protein. The calculation formula was as follows: where k and j represent the protein and the corresponding specific peptide of the protein:Rik = Median (NRij, j ∈ k)
2.7. Analysis of Protein Sequence Characteristics
2.8. Enzyme Activity Assay
2.9. Hydrogen Peroxide (H2O2) Concentration Detection
2.10. Statistical Analysis
3. Results
3.1. Protein Acylation Levels in Turnip Shoots under Cd Stress
3.2. Global Lysine Succinylation Modification in Turnip Shoots
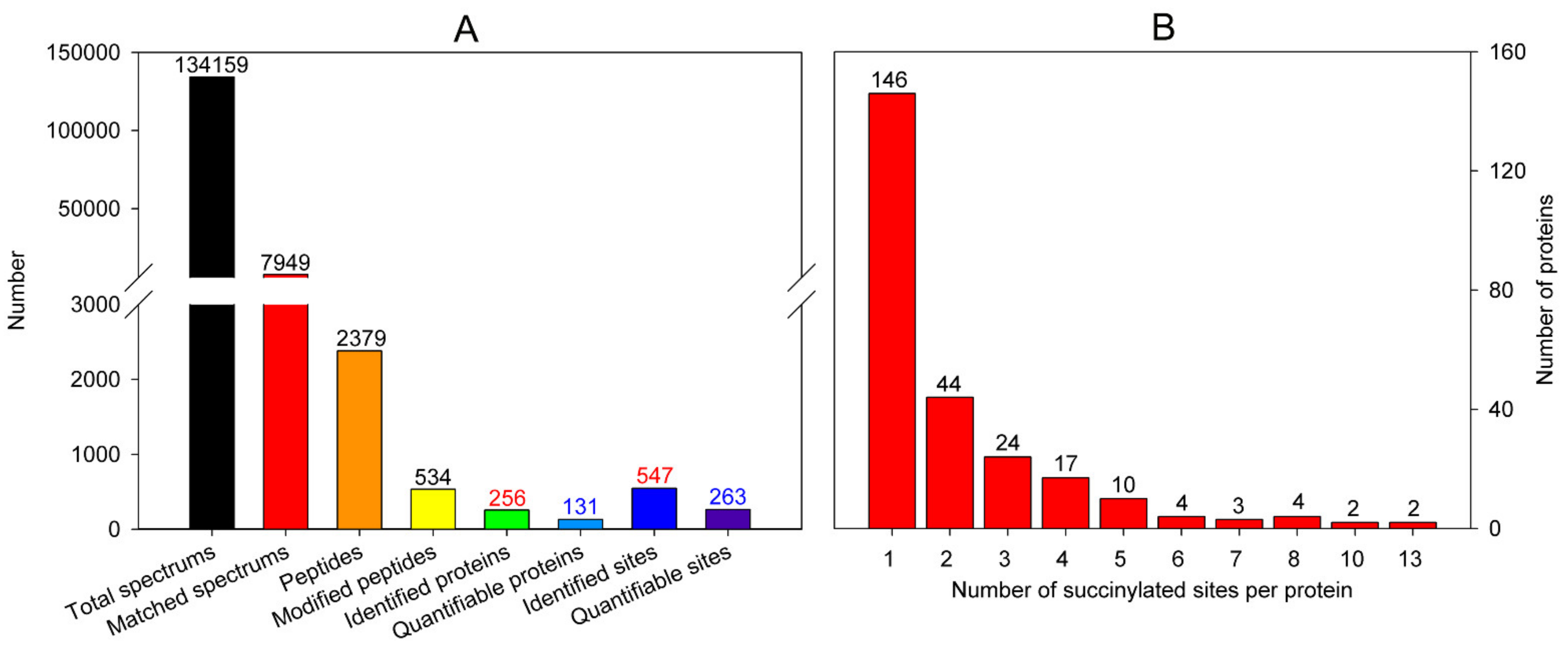
3.2.1. Identification of Lysine Succinylation in Turnip Shoots
3.2.2. Site Properties of the Succinylated Peptides
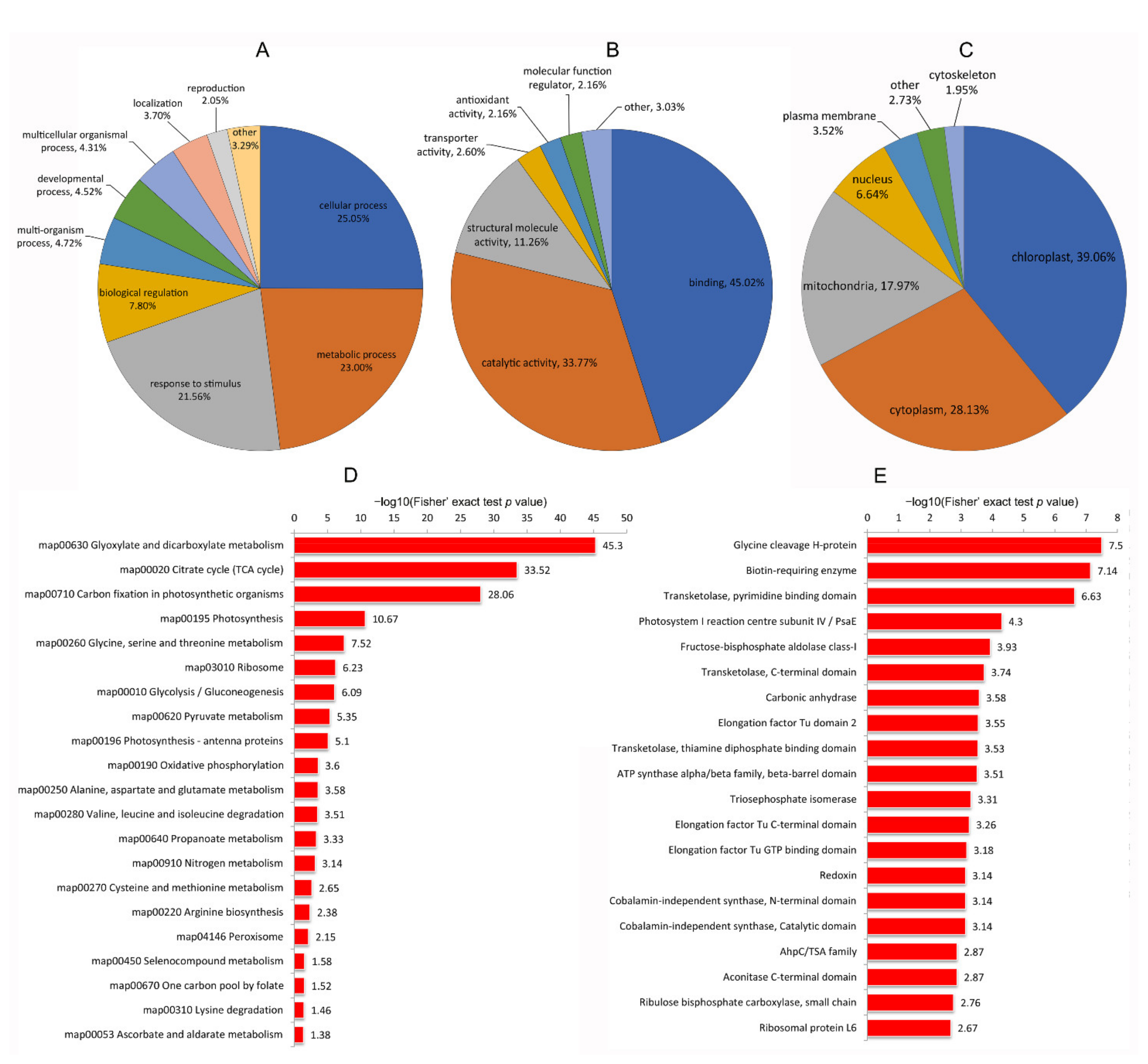
3.2.3. Functional Classification and Subcellular Location Analysis
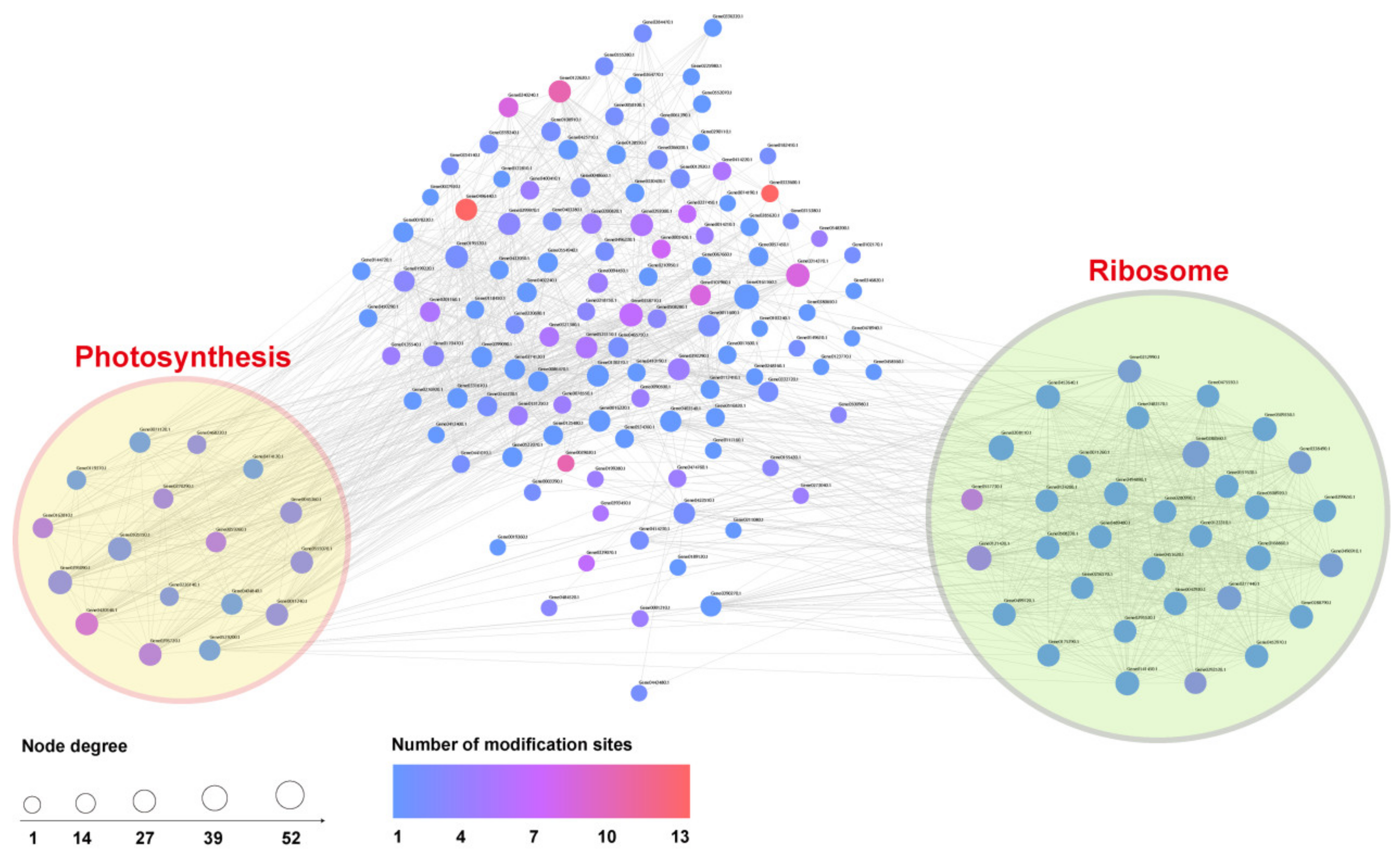
3.2.4. PPI Network Construction
3.3. Differentially Succinylated Sites (DSSs) in Turnip Shoots in Response to Cd Stress
3.3.1. Identification of DSSs
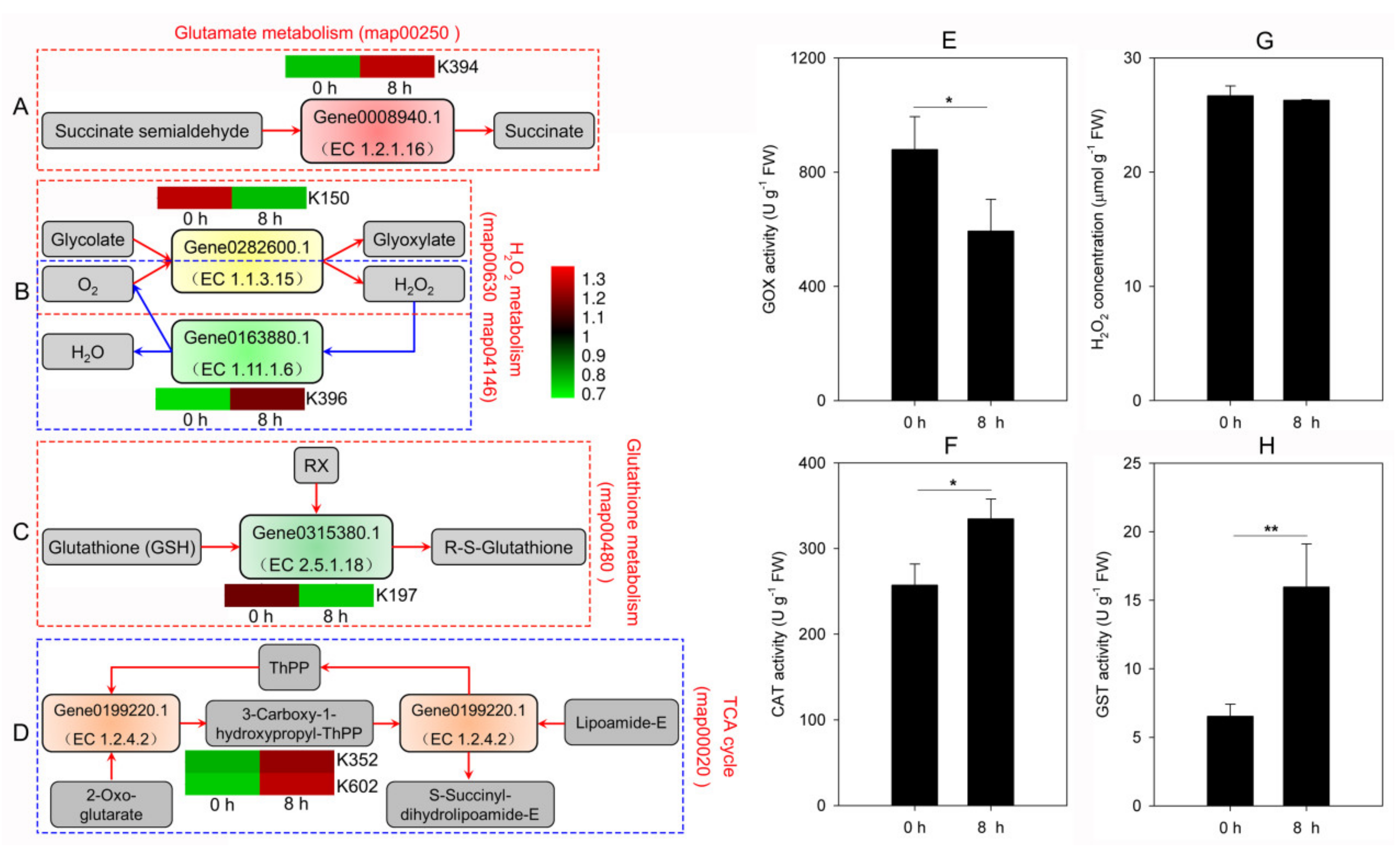
3.3.2. Variations in H2O2 Metabolism under Cd Stress
3.3.3. Variations in GSH Metabolism under Cd Stress
4. Discussion
4.1. Changes in Protein Acylation Levels in Turnip Shoots under Cd Stress
4.2. Global Analysis of Lysine Succinylation in Turnip Shoots
4.3. DSSs in Turnip Shoots in Response to Cd Stress
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sebastian, A.; Prasad, M.N.V. Cadmium minimization in rice. A review. Agron. Sustain. Dev. 2014, 34, 155–173. [Google Scholar] [CrossRef]
- Hussain, B.; Umer, M.J.; Li, J.M.; Ma, Y.B.; Abbas, Y.; Ashraf, M.N.; Tahir, N.; Ullah, A.; Gogoi, N.; Farooq, M. Strategies for reducing cadmium accumulation in rice grains. J. Clean. Prod. 2021, 286, 125557. [Google Scholar] [CrossRef]
- Zulfiqar, U.; Jiang, W.; Xiukang, W.; Hussain, S.; Ahmad, M.; Maqsood, M.F.; Ali, N.; Ishfaq, M.; Kaleem, M.; Haider, F.U.; et al. Cadmium phytotoxicity, tolerance, and advanced remediation approaches in agricultural soils; A comprehensive review. Front. Plant Sci. 2022, 13, 773815. [Google Scholar] [CrossRef] [PubMed]
- Zulfiqar, U.; Ayub, A.; Hussain, S.; Waraich, E.A.; El-Esawi, M.A.; Ishfaq, M.; Ahmad, M.; Ali, N.; Maqsood, M.F. Cadmium toxicity in plants: Recent progress on morpho-physiological effects and remediation strategies. J. Soil Sci. Plant Nutr. 2022, 22, 212–269. [Google Scholar] [CrossRef]
- Chen, Q.; Lu, X.; Guo, X.; Pan, Y.; Yu, B.; Tang, Z.; Guo, Q. Differential responses to Cd stress induced by exogenous application of Cu, Zn or Ca in the medicinal plant Catharanthus roseus. Ecotoxicol. Environ. Saf. 2018, 157, 266–275. [Google Scholar] [CrossRef]
- Oskarsson, A.; Widell, A.; Olsson, I.M.; Grawe, K.P. Cadmium in food chain and health effects in sensitive population groups. Biometals 2004, 17, 531–534. [Google Scholar] [CrossRef]
- Yan, A.; Wang, Y.; Tan, S.N.; Yusof, M.L.M.; Ghosh, S.; Chen, Z. Phytoremediation: A promising approach for revegetation of heavy metal-polluted land. Front. Plant Sci. 2020, 11, 359. [Google Scholar] [CrossRef]
- Zeremski, T.; Randelovic, D.; Jakovljevic, K.; Marjanovic Jeromela, A.; Milic, S. Brassica Species in phytoextractions: Real potentials and challenges. Plants 2021, 10, 2340. [Google Scholar] [CrossRef]
- Cai, X.; Chang, L.C.; Zhang, T.T.; Chen, H.X.; Zhang, L.; Lin, R.M.; Liang, J.L.; Wu, J.; Freeling, M.; Wang, X.W. Impacts of allopolyploidization and structural variation on intraspecific diversification in Brassica rapa. Genome Biol. 2021, 22, 166. [Google Scholar] [CrossRef]
- Zhang, Z.; Guo, J.; Cai, X.; Li, Y.; Xi, X.; Lin, R.; Liang, J.; Wang, X.; Wu, J. Improved reference genome annotation of Brassica rapa by pacific biosciences RNA sequencing. Front. Plant Sci. 2022, 13, 841618. [Google Scholar] [CrossRef]
- Chen, Y.T.; Mao, W.W.; Liu, T.; Feng, Q.Q.; Li, L.; Li, B.B. Genome editing as a versatile tool to improve horticultural crop qualities. Hortic. Plant J. 2020, 6, 372–384. [Google Scholar] [CrossRef]
- Li, X.; Zhang, X.M.; Yang, Y.; Li, B.Q.; Wu, Y.S.; Sun, H.; Yang, Y.P. Cadmium accumulation characteristics in turnip landraces from China and assessment of their phytoremediation potential for contaminated soils. Front. Plant Sci. 2016, 7, 1862. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhang, X.; Wu, Y.; Li, B.; Yang, Y. Physiological and biochemical analysis of mechanisms underlying cadmium tolerance and accumulation in turnip. Plant Divers. 2018, 40, 19–27. [Google Scholar] [CrossRef]
- Li, X.; Chen, D.; Yang, Y.; Liu, Y.; Luo, L.; Chen, Q.; Yang, Y. Comparative transcriptomics analysis reveals differential Cd response processes in roots of two turnip landraces with different Cd accumulation capacities. Ecotoxicol. Environ. Saf. 2021, 220, 112392. [Google Scholar] [CrossRef]
- Li, X.; Chen, D.; Li, B.Q.; Yang, Y.; Yang, Y.P. Combined transcriptomic, proteomic and biochemical approaches to identify the cadmium hyper-tolerance mechanism of turnip seedling leaves. Environ. Sci. Pollut. R. 2021, 28, 22458–22473. [Google Scholar] [CrossRef]
- Ruan, B.J.; Dai, P.; Wang, W.; Sun, J.B.; Zhang, W.T.; Yan, Z.; Yang, J.H. Progress on post-translational modification of proteins. Chin. J. Cell Biol. 2014, 36, 1027–1037. [Google Scholar]
- Millar, A.H.; Heazlewood, J.L.; Giglione, C.; Holdsworth, M.J.; Bachmair, A.; Schulze, W.X. The scope, functions, and dynamics of posttranslational protein modifications. Annu. Rev. Plant Biol. 2019, 70, 119–151. [Google Scholar] [CrossRef]
- Buuh, Z.Y.; Lyu, Z.; Wang, R.E. Interrogating the roles of post-translational modifications of non-histone proteins. J. Med. Chem. 2018, 61, 3239–3252. [Google Scholar] [CrossRef]
- Zhou, H.; Finkemeier, I.; Guan, W.; Tossounian, M.A.; Wei, B.; Young, D.; Huang, J.; Messens, J.; Yang, X.; Zhu, J.; et al. Oxidative stress-triggered interactions between the succinyl- and acetyl-proteomes of rice leaves. Plant Cell Environ. 2018, 41, 1139–1153. [Google Scholar] [CrossRef]
- Weinert, B.T.; Scholz, C.; Wagner, S.A.; Iesmantavicius, V.; Su, D.; Daniel, J.A.; Choudhary, C. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013, 4, 842–851. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhang, X.M.; Li, B.Q.; Wu, Y.S.; Sun, H.; Yang, Y.P. Cadmium phytoremediation potential of turnip compared with three common high Cd-accumulating plants. Environ. Sci. Pollut. R. 2017, 24, 21660–21670. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Li, P.; Li, P.; Chen, C. First comprehensive analysis of lysine succinylation in paper mulberry (Broussonetia papyrifera). BMC Genom. 2021, 22, 255. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Xiong, Y.; Sun, W.; Wang, G.L.; Liu, W. Global proteomic analysis reveals widespread lysine succinylation in rice seedlings. Int. J. Mol. Sci. 2019, 20, 5911. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Xu, Y.X.; Shen, C.J.; Ma, J.Q.; Chen, W.; Mao, J.; Zhou, Y.Y.; Chen, L. Quantitative succinyl-proteome profiling of Camellia sinensis cv. ‘Anji Baicha’ during periodic albinism. Sci. Rep. 2017, 7, 1873. [Google Scholar] [CrossRef] [Green Version]
- Zhen, S.; Deng, X.; Wang, J.; Zhu, G.; Cao, H.; Yuan, L.; Yan, Y. First Comprehensive proteome analyses of lysine acetylation and succinylation in seedling leaves of Brachypodium distachyon L. Sci. Rep. 2016, 6, 31576. [Google Scholar] [CrossRef] [Green Version]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic. Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING database in 2011: Functional interaction networks of proteins, globally integrated and scored. Nucleic. Acids Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Zhu, H.; Ai, H.; Hu, Z.; Du, D.; Sun, J.; Chen, K.; Chen, L. Comparative transcriptome combined with metabolome analyses revealed key factors involved in nitric oxide (NO)-regulated cadmium stress adaptation in tall fescue. BMC Genom. 2020, 21, 601. [Google Scholar] [CrossRef]
- Cao, Y.; Fan, G.; Wang, Z.; Gu, Z. Phytoplasma-induced changes in the acetylome and succinylome of Paulownia tomentosa provide evidence for involvement of acetylated proteins in witches’ broom disease. Mol. Cell Proteom. 2019, 18, 1210–1226. [Google Scholar] [CrossRef]
- Rosen, R.; Becher, D.; Buttner, K.; Biran, D.; Hecker, M.; Ron, E.Z. Probing the active site of homoserine trans-succinylase. FEBS Lett. 2004, 577, 386–392. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.C.; Ma, Z.K.; Li, C.D.; Ren, P.R.; Yao, L.R.; Li, B.C.; Meng, Y.X.; Ma, X.L.; Si, E.J.; Yang, K.; et al. Dynamic responses of barley root succinyl-proteome to short-term phosphate starvation and recovery. Front. Plant Sci. 2021, 12, 649147. [Google Scholar] [CrossRef]
- Chen, J.; Li, F.B.; Liu, Y.; Shen, W.F.; Du, X.; He, L.H.; Meng, Z.Q.; Ma, X.L.; Wang, Y.Q. Systematic identification of mitochondrial lysine succinylome in silkworm (Bombyx mori) midgut during the larval gluttonous stage. J. Proteom. 2018, 174, 61–70. [Google Scholar] [CrossRef]
- Wang, J.Y.; Li, L.; Chai, R.Y.; Zhang, Z.; Qiu, H.P.; Mao, X.Q.; Hao, Z.N.; Wang, Y.L.; Sun, G.C. Succinyl-proteome profiling of Pyricularia oryzae, a devastating phytopathogenic fungus that causes rice blast disease. Sci. Rep. 2019, 9, 3490. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Chen, X.; Li, J.; Zhou, X.; Liu, Y.; Zhong, L.; Tang, Y.; Zheng, H.; Liu, J.; Zhan, R.; et al. Global analysis of lysine succinylation in patchouli plant leaves. Hortic. Res. 2019, 6, 133. [Google Scholar] [CrossRef] [Green Version]
- Broz, A.K.; Tovar-Mendez, A.; Mooney, B.P.; Johnston, M.L.; Miernyk, J.A.; Randall, D.D. A novel regulatory mechanism based upon a dynamic core structure for the mitochondrial pyruvate dehydrogenase complex? Mitochondrion 2014, 19, 144–153. [Google Scholar] [CrossRef]
- Fang, X.; Xin, Y.; Sheng, Z.; Liu, H.; Jiang, A.; Wang, F.; Yang, J.; Xi, X.; Zha, Q.; Zhang, L.; et al. Systematic identification and analysis of lysine succinylation in strawberry stigmata. J. Agric. Food Chem. 2018, 66, 13310–13320. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, G.; Song, L.; Mu, P.; Wang, S.; Liang, W.; Lin, Q. Global analysis of protein lysine succinylation profiles in common wheat. BMC Genom. 2017, 18, 309. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Jiao, K.; Guo, H.; Jiang, M.; Hao, J.; Wang, H.; Shen, C. Succinyl-proteome profiling of Dendrobium officinale, an important traditional Chinese orchid herb, revealed involvement of succinylation in the glycolysis pathway. BMC Genom. 2017, 18, 598. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Z.F.; Xu, H.B.; Wang, J.Y.; Lin, Q.; Ruan, Z.; Liu, F.B.; Jin, W.; Huang, H.H.; Chen, X. SIRT5 desuccinylates and activates SOD1 to eliminate ROS. Biochem. Biophys. Res. Commun. 2013, 441, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Nazir, F.; Fariduddin, Q.; Khan, T.A. Hydrogen peroxide as a signalling molecule in plants and its crosstalk with other plant growth regulators under heavy metal stress. Chemosphere 2020, 252, 126486. [Google Scholar] [CrossRef]
- Foyer, C.H.; Noctor, G. Redox sensing and signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol. Plant. 2003, 119, 355–364. [Google Scholar] [CrossRef] [Green Version]
- Mhamdi, A.; Noctor, G.; Baker, A. Plant catalases: Peroxisomal redox guardians. Arch. Biochem. Biophys. 2012, 525, 181–194. [Google Scholar] [CrossRef]
- Kieffer, P.; Planchon, S.; Oufir, M.; Ziebel, J.; Dommes, J.; Hoffmann, L.; Hausman, J.F.; Renaut, J. Combining proteomics and metabolite analyses to unravel cadmium stress-response in poplar leaves. J. Proteome Res. 2009, 8, 400–417. [Google Scholar] [CrossRef]

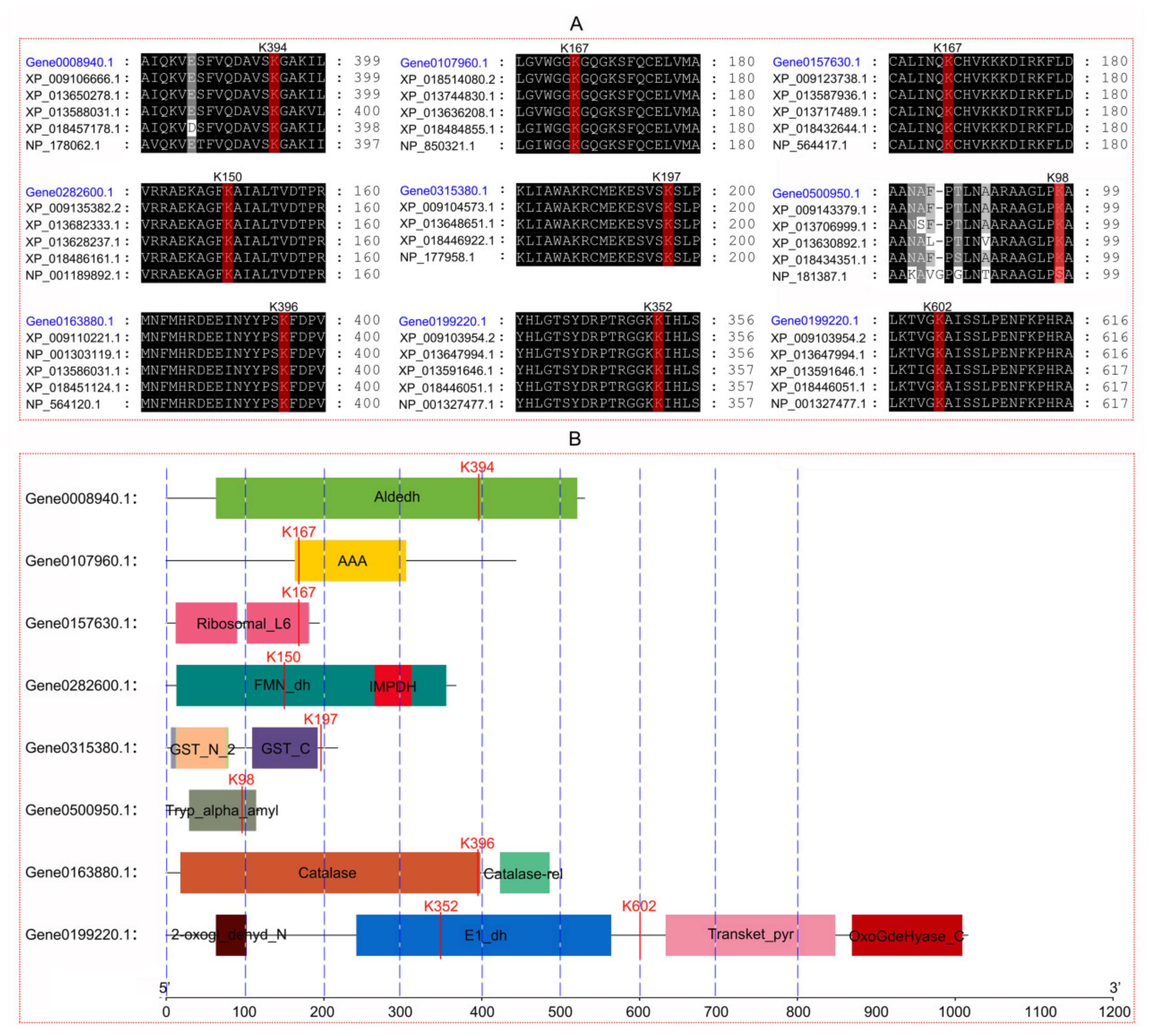
| Protein Accession | Protein Description | Subcellular Localization | Protein Size (aa) | Succinylated Site (s) | Differentially Succinylated Site | Fold Change (8 h/0 h) |
|---|---|---|---|---|---|---|
| Gene0008940.1 | Succinate-semialdehyde dehydrogenase | mitochondria | 530 | K394/K442 | K394 | 1.66 |
| Gene0107960.1 | ATPase_AAA_core domain-containing protein | chloroplast | 443 | K147/K167/K 171/K218/K221/K302/K359/K368 | K167 | 0.57 |
| Gene0157630.1 | 60S ribosomal protein L9 | cytoplasm | 194 | K167 | K167 | 0.66 |
| Gene0282600.1 | Glycolate oxidase | cytoplasm | 367 | K132/K135/K150 | K150 | 0.61 |
| Gene0315380.1 | Glutathione S-transferase | chloroplast | 217 | K52/K197 | K197 | 0.64 |
| Gene0500950.1 | Non-specific lipid-transfer protein | extracellular | 118 | K98/K107/K110 | K98 | 2.01 |
| Gene0163880.1 | Catalase 3 | peroxisome | 492 | K396/K481 | K396 | 1.62 |
| Gene0199220.1 | 2-oxoglutarate dehydrogenase, E1 subunit | mitochondria | 1016 | K352/K518/K602 | K352 | 1.70 |
| Gene0199220.1 | 2-oxoglutarate dehydrogenase, E1 subunit | mitochondria | 1016 | K352/K518/K602 | K602 | 1.55 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, X.; Yang, D.; Yang, Y.; Jin, G.; Yin, X.; Zheng, Y.; Xu, J.; Yang, Y. Quantitative Succinyl-Proteome Profiling of Turnip (Brassica rapa var. rapa) in Response to Cadmium Stress. Cells 2022, 11, 1947. https://doi.org/10.3390/cells11121947
Li X, Yang D, Yang Y, Jin G, Yin X, Zheng Y, Xu J, Yang Y. Quantitative Succinyl-Proteome Profiling of Turnip (Brassica rapa var. rapa) in Response to Cadmium Stress. Cells. 2022; 11(12):1947. https://doi.org/10.3390/cells11121947
Chicago/Turabian StyleLi, Xiong, Danni Yang, Yunqiang Yang, Guihua Jin, Xin Yin, Yan Zheng, Jianchu Xu, and Yongping Yang. 2022. "Quantitative Succinyl-Proteome Profiling of Turnip (Brassica rapa var. rapa) in Response to Cadmium Stress" Cells 11, no. 12: 1947. https://doi.org/10.3390/cells11121947
APA StyleLi, X., Yang, D., Yang, Y., Jin, G., Yin, X., Zheng, Y., Xu, J., & Yang, Y. (2022). Quantitative Succinyl-Proteome Profiling of Turnip (Brassica rapa var. rapa) in Response to Cadmium Stress. Cells, 11(12), 1947. https://doi.org/10.3390/cells11121947






