Programmed Cell Death-1/Programmed Cell Death-1 Ligand as Prognostic Markers of Coronavirus Disease 2019 Severity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Groups
2.2. Ethical Issues
2.3. Sampling and Data Collection Methodology
2.4. Immunophenotyping
2.5. Statistics
3. Results
3.1. Clinical Characteristics of Patients with COVID-19
| Study Group | Non-ICU (n = 17) | ICU (n = 35) | p Value | Total |
|---|---|---|---|---|
| Mortality, n (%) | ||||
| Survived | 16 (94.1) | 22 (62.9) | 0.017 | 38 (73.1) |
| Died | 1 (5.9) | 13 (37.1) | 14 (26.9) | |
| Sex, n (%) | ||||
| Male | 24 (68.6) | 13 (76.5) | n.s. | 37 (71.2) |
| Female | 11 (31.4) | 4 (23.5) | 15 (28.9) | |
| Age, median (IQR) | 74 (63–92) | 69 (59–73) | n.s. | 69 (59.5–74) |
| Days of hospitalization, median (IQR) | 17 (15–29) | 45 (31–80) | <0.0001 | 32 (17.5–61.5) |
| Anti-COVID-19 treatment during in-hospital stay, n (%) | ||||
| Remdesivir | 11 (31.4) | 2 (15.4) | n.s. | 19 (36.5) |
| Tocilizumab | 14 (40.0) | 5 (29.4) | 13 (25.0) | |
| Convalescent plasma | 7 (20.0) | 2 (11.8) | 9 (17.3) | |
| Selected baseline laboratory parameters, median (IQR) | ||||
| WBC [cells/μL] | 7.36 (4.93–14.9) | 11.49 (7.19–14.43) | 0.004 | 9.82 (6.11–13.43) |
| NEU [cells/μL] | 5.8 (3.3–13) | 9.4 (5.9–12.7) | 0.004 | 7.7 (4.25–11.4) |
| CRP [mg/L] | 49.39 (26.42–241.94) | 107.09 (41.12–195.92) | 0.05 | 80.78 (29.77–158.995) |
| IL-6 [pg/mL] | 36.5 (22.6–454) | 122 (40.3–335) | 0.037 | 83.4 (28.65–206.5) |
| LDH [U/L] | 308 (246–528) | 567 (321–690) | 0.0002 | 418.5 (304.5–602) |
| D-dimer [µg/L] | 583 (323–60102) | 1547 (856–2790) | 0.031 | 1333 (496.5–2309) |
| Creatinine levels [mg/dL] | 0.94 (0.88–11.85) | 0.85 (0.61–1.12) | n.s. | 0.9 (0.74–1.17) |
3.2. Immunophenotype Differences between the Analysed Groups and Controls
| Characteristics | Patients with COVID-19 Hospitalized in the ICU | Patients with COVID-19 not Hospitalized in the ICU | Healthy Controls | p Values | Entire COVID-19 Group | |||
|---|---|---|---|---|---|---|---|---|
| Median (IQR) | Median (IQR) | Median (IQR) | ICU Versus Controls | Non-ICU Versus Controls | ICU Versus Non-ICU | Median (IQR) | ||
| Frequencies of individual cells (%) | CD3−CD16+CD56+ NK cells | 12.85 (6.63–15.96) | 12.22 (10.16–13.74) | 14.35 (12.97–16.96) | 0.027 | 0.001 | 0.91 | 12.54 (8.45–15.53) |
| CD3+CD16+CD56+ NK-like cells | 2.14 (0.50–6.87) | 4.09 (1.03–5.59) | 3.34 (2.51–3.57) | 0.54 | 0.56 | 0.77 | 2.46 (0.73–6.74) | |
| CD3+ T cells | 69.70 (64.77–74.43) | 71.36 (65.6–73.21) | 67.57 (64.86–69.81) | 0.08 | 0.09 | 0.91 | 69.76 (65.09–74.14) | |
| CD19+ B cells | 11.73 (8.90–15.34) | 11.63 (8.59–12.98) | 11.21 (9.71–12.28) | 0.44 | 0.83 | 0.51 | 11.68 (8.75–14.87) | |
| CD3+/CD4+ T cells | 43.06 (37.02–48.81) | 44.41 (41.09–48.3) | 43.91 (42.42–45.60) | 0.46 | 0.80 | 0.70 | 43.26 (37.71–48.65) | |
| CD3+/CD8+ T cells | 26.04 (21.91–29.98) | 29.22 (25.4–33.85) | 34.81 (31.39–36.66) | <0.001 | 0.0004 | 0.35 | 26.99 (22.58–32.69) | |
| Ratio of CD3+/CD4+ T cells to CD3+/CD8+ T cells | 1.68 (1.33–1.97) | 1.565 (1.13–1.90) | 1.28 (1.19–1.42) | <0.001 | 0.06 | 0.66 | 1.58 (1.18–1.96) | |
| CD4+PD-1+ T cells | 29.08 (20.52–38.34) | 14.14 (11.58–15.53) | 5.53 (4.14–6.77) | <0.001 | <0.001 | <0.001 | 21.16 (15.47–32.76) | |
| CD4+PD-L1+ T cells | 22.49 (14.76–27.61) | 8.69 (6.20–11.30) | 1.92 (1.49–2.57) | <0.001 | <0.001 | <0.001 | 19.19 (10.03–25.30) | |
| CD8+PD-1+ T cells | 12.72 (6.76–16.76) | 10.00 (7.80–10.91) | 3.77 (2.38–4.63) | <0.001 | <0.001 | 0.08 | 11.01 (6.90–14.62) | |
| CD8+PD-L1+ T cells | 18.38 (15.50–19.33) | 3.12 (2.57–3.66) | 0.42 (0.35–0.52) | <0.001 | <0.001 | <0.001 | 15.65 (3.67–18.87) | |
| CD19+PD-1+ B cells | 7.14 (3.97–12.1) | 2.46 (1.96–4.81) | 1.82 (0.80–2.44) | <0.001 | 0.004 | <0.001 | 5.37 (2.53–9.63) | |
| CD19+PD-L1+ B cells | 5.05 (3.00–11.19) | 3.51 (2.62–4.18) | 0.20 (0.14–0.27) | <0.001 | <0.001 | 0.024 | 4.05 (2.82–8.32) | |
| Time | Parameters | Patients with COVID-19 Hospitalized in the ICU | Patients with COVID-19 Hospitalized in Non-ICU Departments | p Values |
|---|---|---|---|---|
| Median (IQR) | Median (IQR) | |||
| Frequencies of individual cells (%)—day 7 | CD3−CD16+CD56+ NK cells | 12.63 (7.56–16.04) | 11.23 (8.22–13.74) | 0.85 |
| CD3+CD16+CD56+ NK-like cells | 1.66 (0.77–5.79) | 2.82 (1.4–5.21) | 0.52 | |
| CD3+ T cells | 71.73 (65.86–74.92) | 69.46 (66.17–73.99) | 0.66 | |
| CD19+ B cells | 11.22 (7.7–15.62) | 9.95 (8.26–11.9) | 0.54 | |
| CD3+/CD4+ T cells | 43.2 (35.95–45.69) | 44.94 (40.75–48.1) | 0.23 | |
| CD3+/CD8+ T cells | 26.64 (22.22–33.18) | 29.52 (25.44–31.54) | 0.39 | |
| Ratio of CD3+/CD4+ T cells to CD3+/CD8+ T cells | 1.62 (1.25–1.96) | 1.56 (1.21–2) | 0.99 | |
| CD4+PD-1+ T cells | 30.07 (21.13–36.61) | 14.1 (11.53–15.65) | <0.001 | |
| CD4+PD-L1+ T cells | 23.56 (14.71–29.29) | 7.82 (5.52–10.21) | <0.001 | |
| CD8+PD-1+ T cells | 11.89 (6.9–15.66) | 9.97 (7.8–10.53) | 0.14 | |
| CD8+PD-L1+ T cells | 17.37 (12.79–18.55) | 3.2 (2.31–3.62) | <0.001 | |
| CD19+PD-1+ B cells | 6.89 (4.75–12.09) | 2.45 (1.78–4.16) | <0.001 | |
| CD19+PD-L1+ B cells | 4.83 (2.29–9.96) | 3.48 (2.62–4.17) | 0.13 | |
| Frequencies of individual cells (%)—day 14 | CD3−CD16+CD56+ NK cells | 12.03 (7.78–15.6) | 12.92 (10.5–14.61) | 0.47 |
| CD3+CD16+CD56+ NK-like cells | 1.8 (0.78–5.86) | 1.75 (1.34–2.94) | 0.95 | |
| CD3+ T cells | 71.88 (68.94–76.18) | 71.4 (67.81–73.67) | 0.20 | |
| CD19+ B cells | 11.43 (9.09–14.84) | 10.67 (9.47–14.27) | 0.85 | |
| CD3+/CD4+ T cells | 40.15 (33.08–47.31) | 44.15 (42.41–47.59) | 0.20 | |
| CD3+/CD8+ T cells | 27.19 (24.72–34.33) | 29.06 (25.47–33.2) | 0.66 | |
| Ratio of CD3+/CD4+ T cells to CD3+/CD8+ T cells | 1.43 (1.09–1.83) | 1.56 (1.1–1.8) | 0.77 | |
| CD4+PD-1+ T cells | 26.55 (20.52–36.17) | 14.27 (11.7–15.49) | <0.001 | |
| CD4+PD-L1+ T cells | 21.95 (15.02–28.28) | 9.3 (6.29–10.29) | <0.001 | |
| CD8+PD-1+ T cells | 13.57 (7.65–18.22) | 9.53 (7.99–10.79) | 0.08 | |
| CD8+PD-L1+ T cells | 17.69 (15.94–19.22) | 3.31 (2.31–4.06) | <0.001 | |
| CD19+PD-1+ B cells | 7.78 (4.31–12.46) | 2.48 (1.56–4.95) | <0.001 | |
| CD19+PD-L1+ B cells | 5.98 (3.06–10.08) | 3.54 (2.42–4.56) | <0.001 |
| Characteristic | Prognostic Value | AUC | 95% CI | Youden Index | p Value |
|---|---|---|---|---|---|
| Frequency of CD4+PD-1+ T cells [%] Day 0 | 17.52 | 0.99 | 0.96–1.0 | 0.91 | <0.0001 |
| Frequency of CD4+PD-1+ T cells [%] Day 7 | 17.43 | 0.995 | 0.98–1.0 | 0.94 | <0.0001 |
| Frequency of CD4+PD-1+ T cells [%] Day 14 | 17.35 | 0.97 | 0.92–1.0 | 0.88 | <0.0001 |
| Frequency of CD4+PD-L1+ T cells [%] Day 0 | 12.17 | 0.89 | 0.79–0.99 | 0.71 | <0.0001 |
| Frequency of CD4+PD-L1+ T cells [%] Day 7 | 11.51 | 0.91 | 0.82–0.99 | 0.77 | <0.0001 |
| Frequency of CD4+PD-L1+ T cells [%] Day 14 | 10.82 | 0.89 | 0.80–0.99 | 0.74 | <0.0001 |
| Frequency of CD8+PD-1+ T cells [%] Day 0 | 14.06 | 0.65 | 0.51–0.80 | 0.46 | 0.042 |
| Frequency of CD8+PD-1+ T cells [%] Day 7 | 13.91 | 0.71 | 0.57–0.85 | 0.47 | 0.004 |
| Frequency of CD8+PD-1+ T cells [%] Day 14 | 13.87 | 0.80 | 0.68–0.92 | 0.71 | <0.0001 |
| Frequency of CD8+PD-L1+ T cells [%] Day 0 | 5.76 | 1.00 | 1.0 | 1.0 | <0.0001 |
| Frequency of CD8+PD-L1+ T cells [%] Day 7 | 5.48 | 0.97 | 0.92–1.0 | 0.97 | <0.0001 |
| Frequency of CD8+PD-L1+ T cells [%] Day 14 | 6.86 | 0.998 | 0.99–1.0 | 0.97 | <0.0001 |
| Frequency of CD19+PD-1+ B cells [%] Day 0 | 5.96 | 0.86 | 0.76–0.96 | 0.69 | <0.0001 |
| Frequency of CD19+PD-1+ B cells [%] Day 7 | 6.08 | 0.88 | 0.78–0.97 | 0.74 | <0.0001 |
| Frequency of CD19+PD-1+ B cells [%] Day 14 | 5.96 | 0.89 | 0.81–0.98 | 0.82 | <0.0001 |
| Frequency of CD19+PD-L1+ B cells [%] Day 0 | 7.66 | 0.69 | 0.55–0.84 | 0.40 | 0.0068 |
| Frequency of CD19+PD-L1+ B cells [%] Day 7 | 7.1 | 0.75 | 0.62–0.88 | 0.54 | 0.0002 |
| Frequency of CD19+PD-L1+ B cells [%] Day 14 | 6.39 | 0.79 | 0.66–0.91 | 0.62 | <0.0001 |
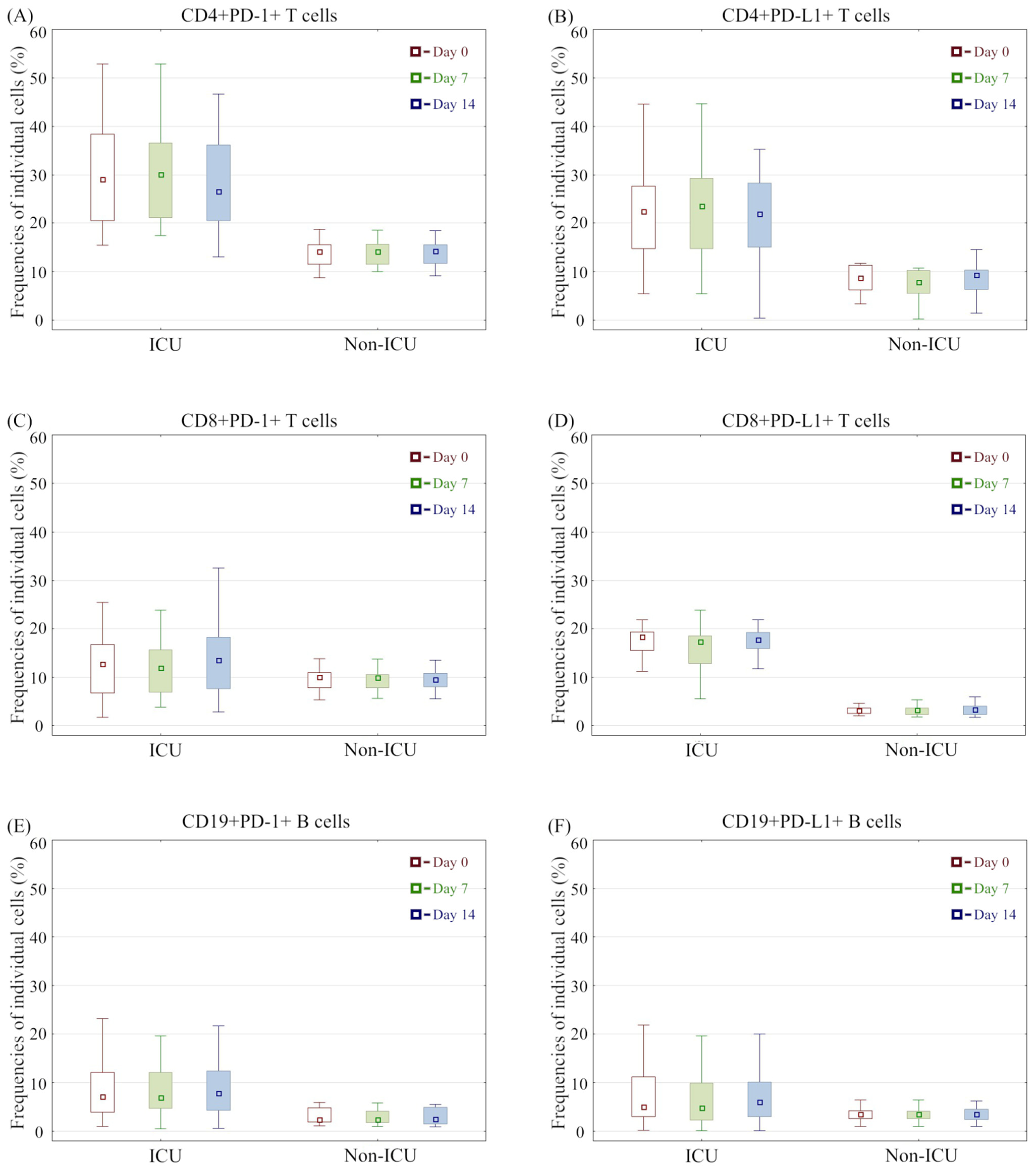
3.3. Immunophenotype Differences between COVID-19 Groups and Association with Mortality
| Characteristic | Patients with Fatal COVID-19 | Surviving Patients with COVID-19 | p Values | |
|---|---|---|---|---|
| Time | Parameters | Median (Range) | Median (Range) | |
| Frequencies of individual cells (%)—day 0 | CD3−CD16+CD56+ NK cells | 14.97 (12.03–16.94) | 10.38 (6.88–14.9) | 0.035 |
| CD3+CD16+CD56+ NK-like cells | 6.47 (1.03–8.41) | 2.08 (0.57–5.59) | 0.06 | |
| CD3+ T cells | 68.14 (63.76–70.58) | 70.85 (65.41–74.71) | 0.16 | |
| CD19+ B cells | 14.11 (11.73–15.34) | 10.74 (7.69–13.81) | 0.042 | |
| CD3+/CD4+ T cells | 41.14 (35.42–47.73) | 43.8 (39.08–50.53) | 0.12 | |
| CD3+/CD8+ T cells | 24.68 (20.74–29.17) | 28.07 (24.54–34.42) | 0.23 | |
| Ratio of CD3+/CD4+ T cells to CD3+/CD8+ T cells | 1.71 (1.43–1.96) | 1.58 (1.13–1.96) | 0.98 | |
| CD4+PD-1+ T cells | 27.15 (20.52–38.47) | 18.16 (14.86–29.42) | 0.008 | |
| CD4+PD-L1+ T cells | 23.27 (19.18–27.61) | 13.12 (8.69–23.8) | 0.03 | |
| CD8+PD-1+ T cells | 12.1 (6.29–14.99) | 10.69 (7.8–14.25) | 0.88 | |
| CD8+PD-L1+ T cells | 16.36 (14.13–19.08) | 10.55 (3.22–18.81) | 0.15 | |
| CD19+PD-1+ B cells | 6.65 (5.26–11.18) | 4.87 (2.16–9.04) | 0.07 | |
| CD19+PD-L1+ B cells | 8.03 (1.88–12.04) | 4 (2.91–5.88) | 0.46 | |
| Frequencies of individual cells (%)—day 7 | CD3−CD16+CD56+ NK cells | 13.11 (11.73–15.9) | 10.59 (7.47–15.86) | 0.07 |
| CD3+CD16+CD56+ NK-like cells | 5.07 (1.08–7.16) | 1.65 (0.94–5.18) | 0.20 | |
| CD3+ T cells | 71.64 (68.18–74.55) | 69.6 (65.86–74.71) | 0.68 | |
| CD19+ B cells | 11.82 (9.66–15.99) | 10.47 (7.86–12.81) | 0.20 | |
| CD3+/CD4+ T cells | 44.69 (37.18–48.92) | 43.11 (36.2–47.31) | 0.43 | |
| CD3+/CD8+ T cells | 24.42 (20.72–28.86) | 28.74 (25.31–33.04) | 0.12 | |
| Ratio of CD3+/CD4+ T cells to CD3+/CD8+ T cells | 1.78 (1.45–2.15) | 1.56 (1.11–1.92) | 0.29 | |
| CD4+PD-1+ T cells | 25.77 (20.15–30.24) | 18.83 (14.75–35.55) | 0.22 | |
| CD4+PD-L1+ T cells | 20.25 (16.51–25.32) | 14.47 (7.82–28.3) | 0.25 | |
| CD8+PD-1+ T cells | 11.99 (6.91–15.66) | 10.33 (7–13.72) | 0.36 | |
| CD8+PD-L1+ T cells | 15.72 (11.86–18.37) | 7.85 (3.2–17.68) | 0.07 | |
| CD19+PD-1+ B cells | 7.32 (5.52–12.96) | 4.69 (2.18–7.03) | 0.027 | |
| CD19+PD-L1+ B cells | 8.02 (2.46–11.91) | 3.78 (2.29–5.07) | 0.24 | |
| Frequencies of individual cells (%)—day 14 | CD3−CD16+CD56+ NK cells | 14.89 (12.15–17.14) | 10.66 (7.59–13.98) | 0.003 |
| CD3+CD16+CD56+ NK-like cells | 2.95 (0.78–8.19) | 1.74 (0.96–4.31) | 0.37 | |
| CD3+ T cells | 68.85 (66.88–71.8) | 73.24 (70.39–75.38) | 0.005 | |
| CD19+ B cells | 12.79 (10.56–15.8) | 10.85 (9.09–14.27) | 0.42 | |
| CD3+/CD4+ T cells | 39.49 (30.54–47.7) | 43.73 (37.93–47.31) | 0.40 | |
| CD3+/CD8+ T cells | 27.41 (23.11–34.33) | 28.11 (25.38–33.2) | 0.67 | |
| Ratio of CD3+/CD4+ T cells to CD3+/CD8+ T cells | 1.29 (0.97–1.9) | 1.52 (1.12–1.8) | 0.84 | |
| CD4+PD-1+ T cells | 25.55 (18.49–34.38) | 17.65 (14.27–27.07) | 0.02 | |
| CD4+PD-L1+ T cells | 21.32 (15.28–25.28) | 14.52 (9.3–23.39) | 0.11 | |
| CD8+PD-1+ T cells | 11.22 (6.47–15.65) | 10.25 (7.76–14.72) | 0.89 | |
| CD8+PD-L1+ T cells | 17.69 (16.56–19.85) | 7.26 (3.67–17.87) | 0.007 | |
| CD19+PD-1+ B cells | 7.76 (4.31–11.57) | 4.97 (2.35–8.52) | 0.06 | |
| CD19+PD-L1+ B cells | 7.92 (4.05–12.71) | 3.64 (2.78–5.85) | 0.039 | |
| Characteristic | Prognostic Value | AUC | 95% CI | Youden Index | p Value |
|---|---|---|---|---|---|
| Frequency of CD3−CD16+CD56+ NK cells [%] Day 0 | 9.49 | 0.71 | 0.53–0.88 | 0.51 | 0.022 |
| Frequency of CD3−CD16+CD56+ NK cells [%] Day 14 | 11.98 | 0.84 | 0.70–0.97 | 0.64 | <0.0001 |
| Frequency of CD3+CD16+CD56+ NKT-like cells [%] Day 0 | 2.29 | 0.73 | 0.54–0.91 | 0.45 | 0.017 |
| Frequency of CD3+CD16+CD56+ NKT-like cells [%] Day 7 | 4.51 | 0.70 | 0.51–0.89 | 0.43 | 0.035 |
| Frequency of CD3+ T cells [%] Day 14 | 70.06 | 0.90 | 0.79–1.0 | 0.72 | <0.0001 |
| Frequency of CD3+/CD4+ T cells [%] Day 0 | 48.49 | 0.70 | 0.52–0.88 | 0.41 | 0.026 |
| Frequency of CD4+PD-1+ T cells [%] Day 0 | 30.1 | 0.76 | 0.6–0.92 | 0.48 | 0.0015 |
| Frequency of CD4+PD-1+ T cells [%] Day 14 | 34.3 | 0.82 | 0.67–0.96 | 0.65 | <0.0001 |
| Frequency of CD4+PD-L1+ T cells [%] Day 14 | 20.9 | 0.72 | 0.55–0.90 | 0.38 | 0.011 |
| Frequency of CD8+PD-1+ T cells [%] Day 14 | 19.6 | 0.84 | 0.71–0.98 | 0.65 | <0.0001 |
| Frequency of CD8+PD-L1+ T cells [%] Day 14 | 20.1 | 0.91 | 0.79–1.0 | 0.83 | <0.0001 |
| Frequency of CD19+PD-1+ B cells [%] Day 7 | 12.96 | 0.87 | 0.74–1.0 | 0.74 | <0.0001 |
| Frequency of CD19+PD-1+ B cells [%] Day 14 | 11.1 | 0.89 | 0.78–1.0 | 0.67 | <0.0001 |
| Frequency of CD19+PD-L1+ B cells [%] Day 7 | 10.2 | 0.93 | 0.84–1.0 | 0.86 | <0.0001 |
| Frequency of CD19+PD-L1+ B cells [%] Day 14 | 15.1 | 0.97 | 0.92–1.0 | 0.90 | <0.0001 |

4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Weekly Epidemiological Update on COVID-19—8 February 2022. Available online: https://www.who.int/publications/m/item/weekly-epidemiological-update-on-covid-19 (accessed on 8 February 2022).
- Brodin, P. Immune determinants of COVID-19 disease presentation and severity. Nat. Med. 2021, 27, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Melenotte, C.; Silvin, A.; Goubet, A.-G.; Lahmar, I.; Dubuisson, A.; Zumla, A.; Raoult, D.; Merad, M.; Gachot, B.; Hénon, C.; et al. Immune responses during COVID-19 infection. Null 2020, 9, 1807836. [Google Scholar] [CrossRef]
- Ñamendys-Silva, S.A.; Alvarado-Ávila, P.E.; Domínguez-Cherit, G.; Rivero-Sigarroa, E.; Sánchez-Hurtado, L.A.; Gutiérrez-Villaseñor, A.; Romero-González, J.P. Outcomes of patients with COVID-19 in the intensive care unit in Mexico: A multicenter observational study. Heart Lung J. Cardiopulm. Acute Care 2021, 50, 28–32. [Google Scholar] [CrossRef] [PubMed]
- Pijls, B.G.; Jolani, S.; Atherley, A.; Derckx, R.T.; Dijkstra, J.I.R.; Franssen, G.H.L.; Hendriks, S.; Richters, A.; Venemans-Jellema, A.; Zalpuri, S.; et al. Demographic risk factors for COVID-19 infection, severity, ICU admission and death: A meta-analysis of 59 studies. BMJ Open 2021, 11, e044640. [Google Scholar] [CrossRef] [PubMed]
- Cai, H.; Liu, G.; Zhong, J.; Zheng, K.; Xiao, H.; Li, C.; Song, X.; Li, Y.; Xu, C.; Wu, H.; et al. Immune Checkpoints in Viral Infections. Viruses 2020, 12, 1051. [Google Scholar] [CrossRef] [PubMed]
- Velu, V.; Shetty, R.D.; Larsson, M.; Shankar, E.M. Role of PD-1 co-inhibitory pathway in HIV infection and potential therapeutic options. Retrovirology 2015, 12, 14. [Google Scholar] [CrossRef] [Green Version]
- Cho, H.; Kang, H.; Lee, H.H.; Kim, C.W. Programmed Cell Death 1 (PD-1) and Cytotoxic T Lymphocyte-Associated Antigen 4 (CTLA-4) in Viral Hepatitis. Int. J. Mol. Sci. 2017, 18, 1517. [Google Scholar] [CrossRef]
- Jubel, J.M.; Barbati, Z.R.; Burger, C.; Wirtz, D.C.; Schildberg, F.A. The Role of PD-1 in Acute and Chronic Infection. Front. Immunol. 2020, 11, 487. Available online: https://www.frontiersin.org/article/10.3389/fimmu.2020.00487 (accessed on 8 February 2022). [CrossRef] [PubMed] [Green Version]
- Murata, T. Human Herpesvirus and the Immune Checkpoint PD-1/PD-L1 Pathway: Disorders and Strategies for Survival. Microorganisms 2021, 9, 778. [Google Scholar] [CrossRef]
- Grywalska, E.; Smarz-Widelska, I.; Korona-Głowniak, I.; Mertowski, S.; Gosik, K.; Hymos, A.; Ludian, J.; Niedźwiedzka-Rystwej, P.; Roliński, J.; Załuska, W. PD-1 and PD-L1 Expression on Circulating Lymphocytes as a Marker of Epstein-Barr Virus Reactivation-Associated Proliferative Glomerulonephritis. Int. J. Mol. Sci. 2020, 21, 8001. [Google Scholar] [CrossRef]
- Jones, D.; Como, C.N.; Jing, L.; Blackmon, A.; Neff, C.P.; Krueger, O.; Bubak, A.N.; Palmer, B.E.; Koelle, D.M.; Nagel, M.A. Varicella zoster virus productively infects human peripheral blood mononuclear cells to modulate expression of immunoinhibitory proteins and blocking PD-L1 enhances virus-specific CD8+ T cell effector function. PLOS Pathog. 2019, 15, e1007650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farrukh, H.; El-Sayes, N.; Mossman, K. Mechanisms of PD-L1 Regulation in Malignant and Virus-Infected Cells. Int. J. Mol. Sci. 2021, 22, 4893. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, E.L.; Meier, P. Nonparametric Estimation from Incomplete Observations. J. Am. Stat. Assoc. 1958, 53, 457–481. [Google Scholar] [CrossRef]
- Azen, S.P.; Roy, S.; Pike, M.C.; Casagrande, J.; Mishell, D.R., Jr. A New Procedure for the Statistical Evaluation of Intrauterine Contraception. Am. J. Obstet. Gynecol. 1977, 128, 329–335. [Google Scholar] [CrossRef]
- Syn, N.L.; Teng, M.W.L.; Mok, T.S.K.; Soo, R.A. De-novo and acquired resistance to immune checkpoint targeting. Lancet Oncol. 2017, 18, e731–e741. [Google Scholar] [CrossRef]
- Alsaab, H.O.; Sau, S.; Alzhrani, R.; Tatiparti, K.; Bhise, K.; Kashaw, S.K.; Iyer, A.K. PD-1 and PD-L1 Checkpoint Signaling Inhibition for Cancer Immunotherapy: Mechanism, Combinations, and Clinical Outcome. Front. Pharmacol. 2017, 8, 561. Available online: https://www.frontiersin.org/article/10.3389/fphar.2017.00561 (accessed on 8 February 2022). [CrossRef]
- Wykes, M.N.; Lewin, S.R. Immune checkpoint blockade in infectious diseases. Nat. Rev. Immunol. 2018, 18, 91–104. [Google Scholar] [CrossRef]
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and Functional Exhaustion of T Cells in Patients with Coronavirus Disease 2019 (COVID-19). Front. Immunol. 2020, 11, 827. Available online: https://www.frontiersin.org/article/10.3389/fimmu.2020.00827 (accessed on 8 February 2022). [CrossRef]
- Chen, R.; Sang, L.; Jiang, M.; Yang, Z.; Jia, N.; Fu, W.; Xie, J.; Guan, W.; Liang, W.; Ni, Z.; et al. Longitudinal hematologic and immunologic variations associated with the progression of COVID-19 patients in China. J. Allergy Clin. Immunol. 2020, 146, 89–100. [Google Scholar] [CrossRef]
- Jiang, M.; Guo, Y.; Luo, Q.; Huang, Z.; Zhao, R.; Liu, S.; Le, A.; Li, J.; Wan, L. T-Cell Subset Counts in Peripheral Blood Can Be Used as Discriminatory Biomarkers for Diagnosis and Severity Prediction of Coronavirus Disease 2019. J. Infect. Dis. 2020, 222, 198–202. [Google Scholar] [CrossRef]
- Liu, R.; Wang, Y.; Li, J.; Han, H.; Xia, Z.; Liu, F.; Wu, K.; Yang, L.; Liu, X.; Zhu, C. Decreased T cell populations contribute to the increased severity of COVID-19. Clin. Chim. Acta 2020, 508, 110–114. [Google Scholar] [CrossRef]
- Song, J.-W.; Zhang, C.; Fan, X.; Meng, F.-P.; Xu, Z.; Xia, P.; Cao, W.-J.; Yang, T.; Dai, X.-P.; Wang, S.-Y.; et al. Immunological and inflammatory profiles in mild and severe cases of COVID-19. Nat. Commun. 2020, 11, 3410. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients with 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061–1069. [Google Scholar] [CrossRef] [PubMed]
- Kuri-Cervantes, L.; Pampena, M.B.; Meng, W.; Rosenfeld, A.M.; Ittner, C.A.G.; Weisman, A.R.; Agyekum, R.S.; Mathew, D.; Baxter, A.E.; Vella, L.A. Comprehensive mapping of immune perturbations associated with severe COVID-19. Sci. Immunol. 2020, 5, eabd7114. [Google Scholar] [CrossRef] [PubMed]
- Bobcakova, A.; Petriskova, J.; Vysehradsky, R.; Kocan, I.; Kapustova, L.; Barnova, M.; Diamant, Z.; Jesenak, M. Immune Profile in Patients with COVID-19: Lymphocytes Exhaustion Markers in Relationship to Clinical Outcome. Front. Cell. Infect. Microbiol. 2021, 11, 646688. Available online: https://www.frontiersin.org/article/10.3389/fcimb.2021.646688 (accessed on 8 February 2022). [CrossRef] [PubMed]
- Kong, Y.; Wang, Y.; Wu, X.; Han, J.; Li, G.; Hua, M.; Han, K.; Zhang, H.; Li, A.; Zeng, H. Storm of soluble immune checkpoints associated with disease severity of COVID-19. Signal Transduct. Target. Ther. 2020, 5, 192. [Google Scholar] [CrossRef]
- Demaria, O.; Carvelli, J.; Batista, L.; Thibult, M.-L.; Morel, A.; André, P.; Morel, Y.; Vély, F.; Vivier, E. Identification of druggable inhibitory immune checkpoints on Natural Killer cells in COVID-19. Cell. Mol. Immunol. 2020, 17, 995–997. [Google Scholar] [CrossRef]
- Ng, C.T.; Snell, L.M.; Brooks, D.G.; Oldstone, M.B.A. Networking at the level of host immunity: Immune cell interactions during persistent viral infections. Cell Host Microbe 2013, 13, 652–664. [Google Scholar] [CrossRef] [Green Version]
- Rha, M.-S.; Jeong, H.W.; Ko, J.-H.; Choi, S.J.; Seo, I.H.; Lee, J.S.; Sa, M.; Kim, A.R.; Joo, E.-J.; Ahn, J.Y.; et al. PD-1-Expressing SARS-CoV-2-Specific CD8(+) T Cells Are Not Exhausted, but Functional in Patients with COVID-19. Immunity 2021, 54, 44–52.e3. [Google Scholar] [CrossRef] [PubMed]
- Varchetta, S.; Mele, D.; Oliviero, B.; Mantovani, S.; Ludovisi, S.; Cerino, A.; Bruno, R.; Castelli, A.; Mosconi, M.; Vecchia, M.; et al. Unique immunological profile in patients with COVID-19. Cell. Mol. Immunol. 2021, 18, 604–612. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.-Y.; Zhang, M.; Yang, C.-X.; Zhang, N.; Wang, X.-C.; Yang, X.-P.; Dong, X.-Q.; Zheng, Y.-T. Elevated exhaustion levels and reduced functional diversity of T cells in peripheral blood may predict severe progression in COVID-19 patients. Cell. Mol. Immunol. 2020, 17, 541–543. [Google Scholar] [CrossRef] [PubMed]
- Schultheiß, C.; Paschold, L.; Simnica, D.; Mohme, M.; Willscher, E.; von Wenserski, L.; Scholz, R.; Wieters, I.; Dahlke, C.; Tolosa, E.; et al. Next-Generation Sequencing of T and B Cell Receptor Repertoires from COVID-19 Patients Showed Signatures Associated with Severity of Disease. Immunity 2020, 53, 442–455.e4. [Google Scholar] [CrossRef] [PubMed]
- Awadasseid, A.; Yin, Q.; Wu, Y.; Zhang, W. Potential protective role of the anti-PD-1 blockade against SARS-CoV-2 infection. Biomed. Pharmacother. 2021, 142, 111957. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Niedźwiedzka-Rystwej, P.; Majchrzak, A.; Aksak-Wąs, B.; Serwin, K.; Czajkowski, Z.; Grywalska, E.; Korona-Głowniak, I.; Roliński, J.; Parczewski, M. Programmed Cell Death-1/Programmed Cell Death-1 Ligand as Prognostic Markers of Coronavirus Disease 2019 Severity. Cells 2022, 11, 1978. https://doi.org/10.3390/cells11121978
Niedźwiedzka-Rystwej P, Majchrzak A, Aksak-Wąs B, Serwin K, Czajkowski Z, Grywalska E, Korona-Głowniak I, Roliński J, Parczewski M. Programmed Cell Death-1/Programmed Cell Death-1 Ligand as Prognostic Markers of Coronavirus Disease 2019 Severity. Cells. 2022; 11(12):1978. https://doi.org/10.3390/cells11121978
Chicago/Turabian StyleNiedźwiedzka-Rystwej, Paulina, Adam Majchrzak, Bogusz Aksak-Wąs, Karol Serwin, Zenon Czajkowski, Ewelina Grywalska, Izabela Korona-Głowniak, Jacek Roliński, and Miłosz Parczewski. 2022. "Programmed Cell Death-1/Programmed Cell Death-1 Ligand as Prognostic Markers of Coronavirus Disease 2019 Severity" Cells 11, no. 12: 1978. https://doi.org/10.3390/cells11121978
APA StyleNiedźwiedzka-Rystwej, P., Majchrzak, A., Aksak-Wąs, B., Serwin, K., Czajkowski, Z., Grywalska, E., Korona-Głowniak, I., Roliński, J., & Parczewski, M. (2022). Programmed Cell Death-1/Programmed Cell Death-1 Ligand as Prognostic Markers of Coronavirus Disease 2019 Severity. Cells, 11(12), 1978. https://doi.org/10.3390/cells11121978








