Transcriptome Analysis of Retinoic Acid-Inducible Gene I Overexpression Reveals the Potential Genes for Autophagy-Related Negative Regulation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells, Virus, Plasmid, Reagents, and Antibodies
2.2. Cell Seeding, Transfection, RNA Extraction, and cDNA Synthesis
2.3. RNA Sequencing and Data Analysis
2.4. Functional Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Signaling Pathways Analysis
2.5. Fluorescence Analysis
2.6. Dual-Luciferase Assay
2.7. Real-Time qPCR (qPCR)
2.8. Western Blotting
2.9. Autophagy-Related Treatment
2.10. Statistical Analysis
3. Results
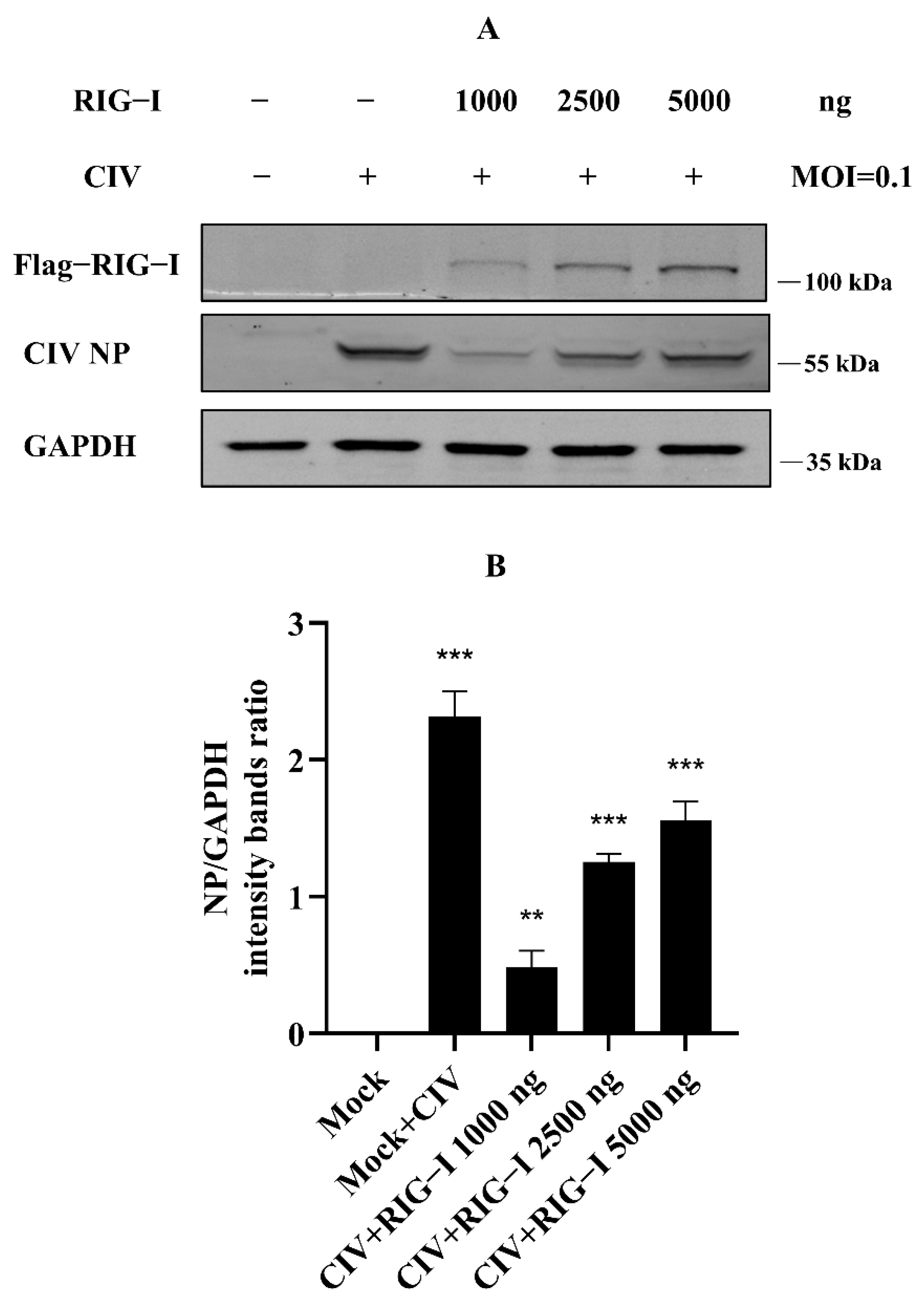
3.1. Excessive Expression of Canine RIG-I Triggered Unknown Negative Regulations
3.2. Cellular Autophagy Is Essential to the Regulation of Excessive RIG-I
3.3. Transcriptome Libraries Construction and Differentially Expressed Genes Analysis
3.4. Functional Enrichment of the Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Signaling Pathways
3.5. qPCR Validation of DEGs Related to Ubiquitination and Autophagy
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Fan, X.; Jin, T. Structures of RIG-I-Like receptors and insights into viral RNA sensing. Adv. Exp. Med. Biol. 2019, 1172, 157–188. [Google Scholar] [PubMed]
- Brisse, M.; Ly, H. Comparative structure and function analysis of the RIG-I-Like receptors: RIG-I and MDA5. Front. Immunol. 2019, 10, 1586. [Google Scholar] [CrossRef] [PubMed]
- Ferrage, F.; Dutta, K.; Nistal-Villán, E.; Patel, J.R.; Sánchez-Aparicio, M.T.; De Ioannes, P.; Buku, A.; Aseguinolaza, G.G.; García-Sastre, A.; Aggarwal, A.K. Structure and dynamics of the second CARD of human RIG-I provide mechanistic insights into regulation of RIG-I activation. Structure 2012, 20, 2048–2061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yong, H.Y.; Zheng, J.; Ho, V.C.Y.; Nguyen, M.T.; Fink, K.; Griffin, P.R.; Luo, D. Structure-Guided design of immunomodulatory RNAs specifically targeting the cytoplasmic viral RNA sensor RIG-I. Febs. Lett. 2019, 593, 3003–3014. [Google Scholar] [CrossRef] [Green Version]
- Lin, H.; Cao, X. Nuclear innate sensors for nucleic acids in immunity and inflammation. Immunol. Rev. 2020, 297, 162–173. [Google Scholar] [CrossRef]
- Chan, C.P.; Jin, D.Y. Cytoplasmic RNA sensors and their interplay with RNA-binding partners in innate antiviral response: Theme and variations. RNA 2022, 28, 449–477. [Google Scholar] [CrossRef]
- Feng, H.; Liu, H.; Kong, R.; Wang, L.; Wang, Y.; Hu, W.; Guo, Q. Expression profiles of carp IRF-3/-7 correlate with the up-regulation of RIG-I/MAVS/TRAF3/TBK1, four pivotal molecules in RIG-I signaling pathway. Fish Shellfish Immun. 2011, 30, 1159–1169. [Google Scholar] [CrossRef]
- Lazear, H.M.; Lancaster, A.; Wilkins, C.; Suthar, M.S.; Huang, A.; Vick, S.C.; Clepper, L.; Thackray, L.; Brassil, M.M.; Virgin, H.W.; et al. IRF-3, IRF-5, and IRF-7 coordinately regulate the type I IFN response in myeloid dendritic cells downstream of MAVS signaling. PLoS Pathog. 2013, 9, e1003118. [Google Scholar] [CrossRef]
- Zhao, K.; Du, J.; Peng, Y.; Li, P.; Wang, S.; Wang, Y.; Hou, J.; Kang, J.; Zheng, W.; Hua, S.; et al. LINE1 contributes to autoimmunity through both RIG-I- and MDA5-mediated RNA sensing pathways. J. Autoimmun. 2018, 90, 105–115. [Google Scholar] [CrossRef]
- Jena, K.K.; Mehto, S.; Nath, P.; Chauhan, N.R.; Sahu, R.; Dhar, K.; Das, S.K.; Kolapalli, S.P.; Murmu, K.C.; Jain, A.; et al. Autoimmunity gene IRGM suppresses cGAS-STING and RIG-I-MAVS signaling to control interferon response. Embo. Rep. 2020, 21, e50051. [Google Scholar] [CrossRef]
- Turianova, L.; Lachova, V.; Svetlikova, D.; Kostrabova, A.; Betakova, T. Comparison of cytokine profiles induced by nonlethal and lethal doses of influenza A virus in mice. Exp. Ther. Med. 2019, 18, 4397–4405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rudnicka, A.; Yamauchi, Y. Ubiquitin in influenza virus entry and innate immunity. Viruses 2016, 8, 293. [Google Scholar] [CrossRef] [Green Version]
- Wies, E.; Wang, M.K.; Maharaj, N.P.; Chen, K.; Zhou, S.; Finberg, R.W.; Gack, M.U. Dephosphorylation of the RNA sensors RIG-I and MDA5 by the phosphatase PP1 is essential for innate immune signaling. Immunity 2013, 38, 437–449. [Google Scholar] [CrossRef] [Green Version]
- Wu, N.; Nguyen, X.; Wang, L.; Appourchaux, R.; Zhang, C.; Panthu, B.; Gruffat, H.; Journo, C.; Alais, S.; Qin, J.; et al. The interferon stimulated gene 20 protein (ISG20) is an innate defense antiviral factor that discriminates self versus non-self translation. PLoS Pathog. 2019, 15, e1008093. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Duan, T.; Feng, Y.; Liu, Q.; Lin, M.; Cui, J.; Wang, R.F. LRRC25 inhibits type I IFN signaling by targeting ISG15-associated RIG-I for autophagic degradation. EMBO J. 2018, 37, 351–366. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Chen, Y.; Tooze, S.A. Autophagy pathway: Cellular and molecular mechanisms. Autophagy 2018, 14, 207–215. [Google Scholar] [CrossRef] [Green Version]
- Tal, M.C.; Iwasaki, A. Autophagic control of RLR signaling. Autophagy 2009, 5, 749–750. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Ye, S.; Yao, C.; Wang, J.; Mao, J.; Xu, L.; Liu, Y.; Fu, C.; Lu, G.; Li, S. Antiviral activity of canine RIG-I against canine influenza virus and interactions between canine RIG-I and CIV. Viruses 2021, 13, 2048. [Google Scholar] [CrossRef]
- Lin, L.; Liu, Q.; Berube, N.; Detmer, S.; Zhou, Y. 5'-Triphosphate-short interfering RNA: Potent inhibition of influenza a virus infection by gene silencing and RIG-I activation. J. Virol. 2012, 86, 10359–10369. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Park, H.S.; Pyo, H.M.; Liu, Q.; Zhou, Y. Influenza a virus panhandle structure is directly involved in RIG-I activation and interferon induction. J. Virol. 2015, 89, 6067–6079. [Google Scholar] [CrossRef] [Green Version]
- Biondo, C.; Lentini, G.; Beninati, C.; Teti, G. The dual role of innate immunity during influenza. Biomed. J. 2019, 42, 8–18. [Google Scholar] [CrossRef] [PubMed]
- Urban, C.; Welsch, H.; Heine, K.; Wüst, S.; Haas, D.A.; Dächert, C.; Pandey, A.; Pichlmair, A.; Binder, M. Persistent innate immune stimulation results in IRF3-Mediated but caspase-independent cytostasis. Viruses 2020, 12, 635. [Google Scholar] [CrossRef] [PubMed]
- Seth, R.B.; Sun, L.; Ea, C.; Chen, Z.J. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF3. Cell 2005, 122, 669–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, Z.; Fang, L.; Jing, H.; Zeng, S.; Wang, D.; Liu, L.; Zhang, H.; Luo, R.; Chen, H.; Xiao, S. Porcine epidemic diarrhea virus nucleocapsid protein antagonizes beta interferon production by sequestering the interaction between IRF3 and TBK1. J. Virol. 2014, 88, 8936–8945. [Google Scholar] [CrossRef] [Green Version]
- Rong, E.; Wang, X.; Chen, H.; Yang, C.; Hu, J.; Liu, W.; Wang, Z.; Chen, X.; Zheng, H.; Pu, J.; et al. Molecular mechanisms for the adaptive switching between the OAS/RNase L and OASL/RIG-I pathways in birds and mammals. Front. Immunol. 2018, 9, 1398. [Google Scholar] [CrossRef]
- Yap, G.L.R.; Sachaphibulkij, K.; Foo, S.L.; Cui, J.; Fairhurst, A.; Lim, L.H.K. Annexin-A1 promotes RIG-I-dependent signaling and apoptosis via regulation of the IRF3–IFNAR–STAT1–IFIT1 pathway in A549 lung epithelial cells. Cell Death Dis. 2020, 11, 463. [Google Scholar] [CrossRef]
- Seitz, C.; Frensing, T.; Hoper, D.; Kochs, G.; Reichl, U. High yields of influenza A virus in Madin-Darby canine kidney cells are promoted by an insufficient interferon-induced antiviral state. J. Gen. Virol. 2010, 91, 1754–1763. [Google Scholar] [CrossRef]
- Nakamura, T.; Asano, A.; Okano, S.; Ko, J.H.; Kon, Y.; Watanabe, T.; Agui, T. Intracellular localization and antiviral property of canine Mx proteins. J. Interferon Cytokine Res. 2005, 25, 169–173. [Google Scholar] [CrossRef]
- Bu, L.; Wang, H.; Hou, P.; Guo, S.; He, M.; Xiao, J.; Li, P.; Zhong, Y.; Jia, P.; Cao, Y.; et al. The Ubiquitin E3 Ligase Parkin Inhibits Innate Antiviral Immunity Through K48-Linked Polyubiquitination of RIG-I and MDA5. Front. Immunol. 2020, 11, 1926. [Google Scholar] [CrossRef]
- Davis, M.E.; Gack, M.U. Ubiquitination in the antiviral immune response. Virology 2015, 479–480, 52–65. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.; Choi, J.; Yoon, I.; Lee, J.K.; Inn, K. Positive regulatory role of c-Src-mediated TRIM25 tyrosine phosphorylation on RIG-I ubiquitination and RIG-I-mediated antiviral signaling pathway. Cell. Immunol. 2018, 332, 94–100. [Google Scholar] [CrossRef] [PubMed]
- Peisley, A.; Wu, B.; Yao, H.; Walz, T.; Hur, S. RIG-I forms Signaling-Competent filaments in an ATP-Dependent, Ubiquitin-Independent manner. Mol. Cell 2013, 51, 573–583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, C.; Liu, G.; Gack, M.U. Viral evasion of RIG-I-Like receptor-mediated immunity through dysregulation of ubiquitination and ISGylation. Viruses 2021, 13, 182. [Google Scholar] [CrossRef] [PubMed]
- Onomoto, K.; Onoguchi, K.; Yoneyama, M. Regulation of RIG-I-like receptor-mediated signaling: Interaction between host and viral factors. Cell. Mol. Immunol. 2021, 18, 539–555. [Google Scholar] [CrossRef]
- Jounai, N.; Takeshita, F.; Kobiyama, K.; Sawano, A.; Miyawaki, A.; Xin, K.Q.; Ishii, K.J.; Kawai, T.; Akira, S.; Suzuki, K.; et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc. Natl. Acad. Sci. USA 2007, 104, 14050–14055. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.; Ban, J.; Lee, N.; Yi, C.; Choi, J.; Kim, H.; Lee, J.K.; Seong, J.; Cho, N.; Jung, J.U.; et al. Activation of RIG-I-Mediated antiviral signaling triggers autophagy through the MAVS-TRAF6-Beclin-1 signaling axis. Front. Immunol. 2018, 9, 2096. [Google Scholar] [CrossRef] [Green Version]
- Tal, M.C.; Sasai, M.; Lee, H.K.; Yordy, B.; Shadel, G.S.; Iwasaki, A. Absence of autophagy results in reactive oxygen species-dependent amplification of RLR signaling. Proc. Natl. Acad. Sci. USA 2009, 106, 2770–2775. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Jin, S.; Liu, Q.; Zhang, Y.; Ma, L.; Zhao, Z.; Yang, S.; Li, Y.P.; Cui, J. Selective autophagy controls the stability of transcription factor IRF3 to balance type I interferon production and immune suppression. Autophagy 2021, 17, 1379–1392. [Google Scholar] [CrossRef]
- Jin, S.; Tian, S.; Chen, Y.; Zhang, C.; Xie, W.; Xia, X.; Cui, J.; Wang, R.F. USP19 modulates autophagy and antiviral immune responses by deubiquitinating Beclin-1. EMBO J. 2016, 35, 866–880. [Google Scholar] [CrossRef] [Green Version]
- Kuma, A.; Matsui, M.; Mizushima, N. LC3, an autophagosome marker, can be incorporated into protein aggregates independent of autophagy: Caution in the interpretation of LC3 localization. Autophagy 2007, 3, 323–328. [Google Scholar] [CrossRef] [Green Version]
- Yoshii, S.R.; Mizushima, N. Monitoring and measuring autophagy. Int. J. Mol. Sci. 2017, 18, 1865. [Google Scholar] [CrossRef] [PubMed]
- Bao, W.; Gu, Y.; Ta, L.; Wang, K.; Xu, Z. Induction of autophagy by the MG132 proteasome inhibitor is associated with endoplasmic reticulum stress in MCF7 cells. Mol. Med. Rep. 2016, 13, 796–804. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.; Xu, W.; Chen, W.; Zhou, Q. Interplay of autophagy inducer rapamycin and proteasome inhibitor MG132 in reduction of foam cell formation and inflammatory cytokine expression. Cell Transpl. 2018, 27, 1235–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harhouri, K.; Navarro, C.; Depetris, D.; Mattei, M.G.; Nissan, X.; Cau, P.; De Sandre-Giovannoli, A.; Levy, N. MG132-induced progerin clearance is mediated by autophagy activation and splicing regulation. EMBO Mol. Med. 2017, 9, 1294–1313. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.H.; Hou, Y.; Meng, F.K.; Wang, X.F.; Luo, Y.N.; Ge, P.F. Proteasome inhibitor MG-132 induces C6 glioma cell apoptosis via oxidative stress. Acta Pharmacol. Sin. 2011, 32, 619–625. [Google Scholar] [CrossRef]
- Spalinger, M.R.; Kasper, S.; Gottier, C.; Lang, S.; Atrott, K.; Vavricka, S.R.; Scharl, S.; Gutte, P.M.; Grütter, M.G.; Beer, H.; et al. NLRP3 tyrosine phosphorylation is controlled by protein tyrosine phosphatase PTPN22. J. Clin. Investig. 2016, 126, 1783–1800. [Google Scholar] [CrossRef] [Green Version]
- Cubas, R.; Khan, Z.; Gong, Q.; Moskalenko, M.; Xiong, H.; Ou, Q.; Pai, C.; Rodriguez, R.; Cheung, J.; Chan, A.C. Autoimmunity linked protein phosphatase PTPN22 as a target for cancer immunotherapy. J. Immunother. Cancer 2020, 8, e1439. [Google Scholar] [CrossRef]
- Spalinger, M.R.; Lang, S.; Gottier, C.; Dai, X.; Rawlings, D.J.; Chan, A.C.; Rogler, G.; Scharl, M. PTPN22 regulates NLRP3-mediated IL1B secretion in an autophagy-dependent manner. Autophagy 2017, 13, 1590–1601. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Ma, C.; Long, F.; Yang, D.; Liu, X.; Hu, Y.; Wu, C.; Wang, B.; Wang, M.; Chen, Y.; et al. Parkin impairs antiviral immunity by suppressing the mitochondrial reactive oxygen Species-Nlrp3 axis and antiviral inflammation. iScience 2019, 16, 468–484. [Google Scholar] [CrossRef] [Green Version]
- Xin, D.; Gu, H.; Liu, E.; Sun, Q. Parkin negatively regulates the antiviral signaling pathway by targeting TRAF3 for degradation. J. Biol. Chem. 2018, 293, 11996–12010. [Google Scholar] [CrossRef] [Green Version]
- Hu, H.; Brittain, G.C.; Chang, J.; Puebla-Osorio, N.; Jin, J.; Zal, A.; Xiao, Y.; Cheng, X.; Chang, M.; Fu, Y.; et al. OTUD7B controls non-canonical NF-κB activation through deubiquitination of TRAF3. Nature 2013, 494, 371–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, W.; Tian, S.; Yang, J.; Cai, S.; Jin, S.; Zhou, T.; Wu, Y.; Chen, Z.; Ji, Y.; Cui, J. OTUD7B deubiquitinates SQSTM1/p62 and promotes IRF3 degradation to regulate antiviral immunity. Autophagy 2022, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Gal, J.; Bang, Y.; Choi, H.J. SIRT2 interferes with autophagy-mediated degradation of protein aggregates in neuronal cells under proteasome inhibition. Neurochem. Int. 2012, 61, 992–1000. [Google Scholar] [CrossRef] [PubMed]
- Roychowdhury, S.; Gandhirajan, A.; Kibler, C.; Wang, X.; Vachharajani, V. Sirtuin 2 Dysregulates Autophagy in High-Fat-Exposed Immune-Tolerant Macrophages. Cells 2021, 10, 731. [Google Scholar] [CrossRef]
- Inoue, T.; Nakayama, Y.; Li, Y.; Matsumori, H.; Takahashi, H.; Kojima, H.; Wanibuchi, H.; Katoh, M.; Oshimura, M. SIRT2 knockdown increases basal autophagy and prevents postslippage death by abnormally prolonging the mitotic arrest that is induced by microtubule inhibitors. FEBS J. 2014, 281, 2623–2637. [Google Scholar] [CrossRef]











| Protein-Coding Gene Name | NGS (log2 Fold Change) | Regulation Style | qRT-PCR (2−∆∆Ct Fold Change) |
|---|---|---|---|
| PTPN22 | 4.97 | Up | 2.31 * |
| UBA7 | 2.48 | Up | 2.97 * |
| RASD2 | 2.46 | Up | 1.26 |
| PRKN | 1.47 | Up | 2.32 * |
| SMAD7 | 1.31 | Up | 1.41 * |
| TRIM72 | 1.18 | Up | 1.67 * |
| OTUD7B | 1.11 | Up | 2.01 * |
| SIRT2 | −2.83 | Down | −2.56 * |
| BFAR | −3.75 | Down | −2.75 * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, S.; Tan, C.; Yang, X.; Wang, J.; Li, Q.; Xu, L.; Wang, Z.; Mao, J.; Wang, J.; Cheng, K.; et al. Transcriptome Analysis of Retinoic Acid-Inducible Gene I Overexpression Reveals the Potential Genes for Autophagy-Related Negative Regulation. Cells 2022, 11, 2009. https://doi.org/10.3390/cells11132009
Ye S, Tan C, Yang X, Wang J, Li Q, Xu L, Wang Z, Mao J, Wang J, Cheng K, et al. Transcriptome Analysis of Retinoic Acid-Inducible Gene I Overexpression Reveals the Potential Genes for Autophagy-Related Negative Regulation. Cells. 2022; 11(13):2009. https://doi.org/10.3390/cells11132009
Chicago/Turabian StyleYe, Shaotang, Chen Tan, Xiaoyun Yang, Ji Wang, Qi Li, Liang Xu, Zhen Wang, Jianwei Mao, Jingyu Wang, Kui Cheng, and et al. 2022. "Transcriptome Analysis of Retinoic Acid-Inducible Gene I Overexpression Reveals the Potential Genes for Autophagy-Related Negative Regulation" Cells 11, no. 13: 2009. https://doi.org/10.3390/cells11132009
APA StyleYe, S., Tan, C., Yang, X., Wang, J., Li, Q., Xu, L., Wang, Z., Mao, J., Wang, J., Cheng, K., Chen, A., Zhou, P., & Li, S. (2022). Transcriptome Analysis of Retinoic Acid-Inducible Gene I Overexpression Reveals the Potential Genes for Autophagy-Related Negative Regulation. Cells, 11(13), 2009. https://doi.org/10.3390/cells11132009






