Kank1 Is Essential for Myogenic Differentiation by Regulating Actin Remodeling and Cell Proliferation in C2C12 Progenitor Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Differentiation
2.2. Transfection of Oligonucleotides
2.3. RNA Extraction and Quantitative Real-Time PCR (qRT-PCR)
2.4. Preparation of the Cytoplasmic and Nuclear Fractions
2.5. Immunoblot Analysis
2.6. Immunofluorescence Analysis
2.7. Cell Proliferation Assays
2.8. Cell Viability Analysis
2.9. Flow Cytometry
2.10. Statistical Analysis
3. Results
3.1. Kank1 Expression Was Altered during Myoblast Differentiation
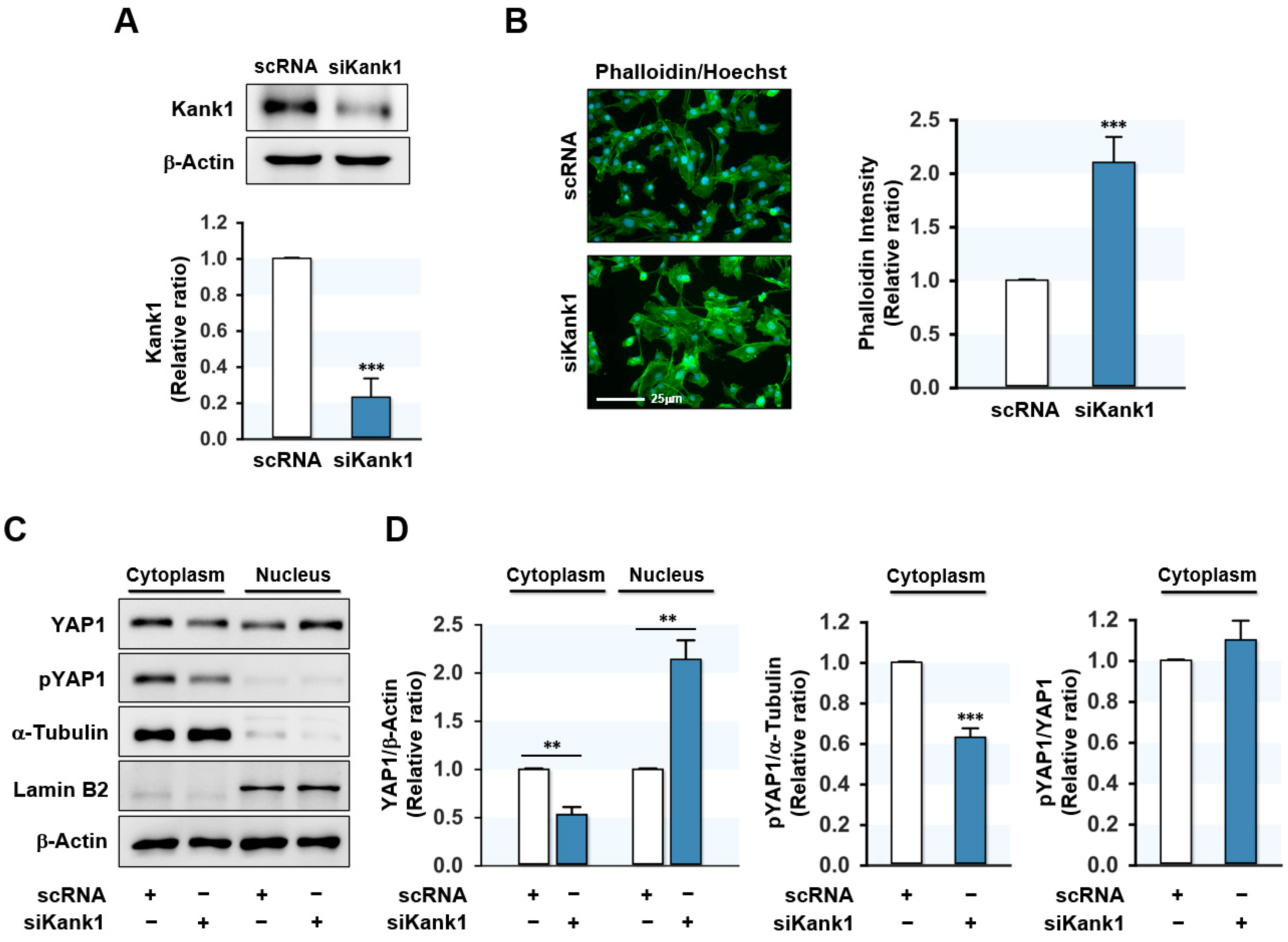
3.2. Depletion of Kank1 Promoted the Nuclear Translocation of YAP1
3.3. Kank1 Knockdown Induced Myoblast Proliferation
3.4. Kank1 Knockdown Suppressed Myogenic Differentiation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mukund, K.; Subramaniam, S. Skeletal muscle: A review of molecular structure and function, in health and disease. Wiley Interdiscip. Rev. Syst. Biol. Med. 2020, 12, e1462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chal, J.; Pourquie, O. Making muscle: Skeletal myogenesis in vivo and in vitro. Development 2017, 144, 2104–2122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sartori, R.; Romanello, V.; Sandri, M. Mechanisms of muscle atrophy and hypertrophy: Implications in health and disease. Nat. Commun. 2021, 12, 330. [Google Scholar] [CrossRef]
- Watt, K.I.; Goodman, C.A.; Hornberger, T.A.; Gregorevic, P. The Hippo Signaling Pathway in the Regulation of Skeletal Muscle Mass and Function. Exerc. Sport Sci. Rev. 2018, 46, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.; Rikeit, P.; Knaus, P.; Coirault, C. YAP-Mediated Mechanotransduction in Skeletal Muscle. Front. Physiol. 2016, 7, 41. [Google Scholar] [CrossRef]
- Guerin, C.M.; Kramer, S.G. Cytoskeletal remodeling during myotube assembly and guidance: Coordinating the actin and microtubule networks. Commun. Integr. Biol. 2009, 2, 452–457. [Google Scholar] [CrossRef] [Green Version]
- Pollard, T.D.; Borisy, G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell 2003, 112, 453–465. [Google Scholar] [CrossRef] [Green Version]
- Heng, Y.W.; Koh, C.G. Actin cytoskeleton dynamics and the cell division cycle. Int. J. Biochem. Cell Biol. 2010, 42, 1622–1633. [Google Scholar] [CrossRef]
- Bezanilla, M.; Gladfelter, A.S.; Kovar, D.R.; Lee, W.-L. Cytoskeletal dynamics: A view from the membrane. J. Cell Biol. 2015, 209, 329–337. [Google Scholar] [CrossRef] [Green Version]
- Dupont, S.; Morsut, L.; Aragona, M.; Enzo, E.; Giulitti, S.; Cordenonsi, M.; Zanconato, F.; Le Digabel, J.; Forcato, M.; Bicciato, S.; et al. Role of YAP/TAZ in mechanotransduction. Nature 2011, 474, 179–183. [Google Scholar] [CrossRef]
- Aragona, M.; Panciera, T.; Manfrin, A.; Giulitti, S.; Michielin, F.; Elvassore, N.; Dupont, S.; Piccolo, S. A mechanical checkpoint controls multicellular growth through YAP/TAZ regulation by actin-processing factors. Cell 2013, 154, 1047–1059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, N.U.; Liang, V.R.; Wang, H.V. Actin-associated protein palladin is required for migration behavior and differentiation potential of C2C12 myoblast cells. Biochem. Biophys. Res. Commun. 2014, 452, 728–733. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Hou, L.; Zhang, Y.; Jiang, F.; Zhu, Y.; Li, Q.X.; Hu, C.Y.; Wang, C. PFN2a Suppresses C2C12 Myogenic Development by Inhibiting Proliferation and Promoting Apoptosis via the p53 Pathway. Cells 2019, 8, 959. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, M.T.; Min, K.H.; Kim, D.; Park, S.Y.; Lee, W. CFL2 is an essential mediator for myogenic differentiation in C2C12 myoblasts. Biochem. Biophys. Res. Commun. 2020, 533, 710–716. [Google Scholar] [CrossRef]
- Horton, E.R.; Byron, A.; Askari, J.A.; Ng, D.H.J.; Millon-Fremillon, A.; Robertson, J.; Koper, E.J.; Paul, N.R.; Warwood, S.; Knight, D.; et al. Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat. Cell Biol. 2015, 17, 1577–1587. [Google Scholar] [CrossRef] [Green Version]
- Paradzik, M.; Humphries, J.D.; Stojanovic, N.; Nestic, D.; Majhen, D.; Dekanic, A.; Samarzija, I.; Sedda, D.; Weber, I.; Humphries, M.J.; et al. KANK2 Links alphaVbeta5 Focal Adhesions to Microtubules and Regulates Sensitivity to Microtubule Poisons and Cell Migration. Front. Cell Dev. Biol. 2020, 8, 125. [Google Scholar] [CrossRef]
- Bouchet, B.P.; Gough, R.E.; Ammon, Y.C.; van de Willige, D.; Post, H.; Jacquemet, G.; Altelaar, A.M.; Heck, A.J.; Goult, B.T.; Akhmanova, A. Talin-KANK1 interaction controls the recruitment of cortical microtubule stabilizing complexes to focal adhesions. eLife 2016, 5, e18124. [Google Scholar] [CrossRef]
- Stubb, A.; Guzman, C.; Narva, E.; Aaron, J.; Chew, T.L.; Saari, M.; Miihkinen, M.; Jacquemet, G.; Ivaska, J. Superresolution architecture of cornerstone focal adhesions in human pluripotent stem cells. Nat. Commun. 2019, 10, 4756. [Google Scholar] [CrossRef] [Green Version]
- Chen, N.P.; Sun, Z.; Fassler, R. The Kank family proteins in adhesion dynamics. Curr. Opin. Cell Biol. 2018, 54, 130–136. [Google Scholar] [CrossRef]
- Kakinuma, N.; Zhu, Y.; Wang, Y.; Roy, B.C.; Kiyama, R. Kank proteins: Structure, functions and diseases. Cell Mol. Life Sci. 2009, 66, 2651–2659. [Google Scholar] [CrossRef]
- Roy, B.C.; Aoyagi, T.; Sarkar, S.; Nomura, K.; Kanda, H.; Iwaya, K.; Tachibana, M.; Kiyama, R. Pathological characterization of Kank in renal cell carcinoma. Exp. Mol. Pathol. 2005, 78, 41–48. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, J.I.; Roy, B.C.; Ogaeri, T.; Kakinuma, N.; Kiyama, R. Depletion of tumor suppressor Kank1 induces centrosomal amplification via hyperactivation of RhoA. Exp. Cell Res. 2017, 353, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, S.; Roy, B.C.; Hatano, N.; Aoyagi, T.; Gohji, K.; Kiyama, R. A novel ankyrin repeat-containing gene (Kank) located at 9p24 is a growth suppressor of renal cell carcinoma. J. Biol. Chem. 2002, 277, 36585–36591. [Google Scholar] [CrossRef] [Green Version]
- Tadijan, A.; Samarzija, I.; Humphries, J.D.; Humphries, M.J.; Ambriovic-Ristov, A. KANK family proteins in cancer. Int. J. Biochem. Cell Biol. 2021, 131, 105903. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Wang, K.; Tong, X. In vivo and in vitro inhibition of human gastric cancer progress by upregulating Kank1 gene. Oncol. Rep. 2017, 38, 1663–1669. [Google Scholar] [CrossRef] [Green Version]
- Fan, H.; Tian, H.; Cheng, X.; Chen, Y.; Liang, S.; Zhang, Z.; Liao, Y.; Xu, P. Aberrant Kank1 expression regulates YAP to promote apoptosis and inhibit proliferation in OSCC. J. Cell Physiol. 2020, 235, 1850–1865. [Google Scholar] [CrossRef] [Green Version]
- Ren, T.J.; Liu, C.; Hou, J.F.; Shan, F.X. CircDDX17 reduces 5-fluorouracil resistance and hinders tumorigenesis in colorectal cancer by regulating miR-31-5p/KANK1 axis. Eur. Rev. Med. Pharmacol. Sci. 2020, 24, 1743–1754. [Google Scholar] [CrossRef]
- Kariri, Y.A.; Joseph, C.; Kurozumi, S.; Toss, M.S.; Alsaleem, M.; Raafat, S.; Mongan, N.P.; Aleskandarany, M.A.; Green, A.R.; Rakha, E.A. Prognostic significance of KN motif and ankyrin repeat domains 1 (KANK1) in invasive breast cancer. Breast Cancer Res. Treat. 2020, 179, 349–357. [Google Scholar] [CrossRef] [Green Version]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Dupont, S. Role of YAP/TAZ in cell-matrix adhesion-mediated signalling and mechanotransduction. Exp. Cell Res. 2016, 343, 42–53. [Google Scholar] [CrossRef]
- Roy, B.C.; Kakinuma, N.; Kiyama, R. Kank attenuates actin remodeling by preventing interaction between IRSp53 and Rac1. J. Cell Biol. 2009, 184, 253–267. [Google Scholar] [CrossRef] [Green Version]
- Kakinuma, N.; Roy, B.C.; Zhu, Y.; Wang, Y.; Kiyama, R. Kank regulates RhoA-dependent formation of actin stress fibers and cell migration via 14-3-3 in PI3K-Akt signaling. J. Cell Biol. 2008, 181, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Dumont, N.A.; Bentzinger, C.F.; Sincennes, M.-C.; Rudnicki, M.A. Satellite cells and skeletal muscle regeneration. Compr. Physiol. 2015, 5, 1027–1059. [Google Scholar] [PubMed]
- Wang, J.; Jia, J.; Zhou, L. Long non-coding RNA CASC2 enhances cisplatin sensitivity in oral squamous cell cancer cells by the miR-31-5p/KANK1 axis. Neoplasma 2020, 67, 1279–1292. [Google Scholar] [CrossRef] [PubMed]
- Cui, Z.B.; Shen, Y.J.; Chen, K.H.; Mittal, S.K.; Yang, J.Y.; Zhang, G.J. KANK1 inhibits cell growth by inducing apoptosis though regulating CXXC5 in human malignant peripheral nerve sheath tumors. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef]
- Pu, J.; Shen, J.; Zhong, Z.; Yanling, M.; Gao, J. KANK1 regulates paclitaxel resistance in lung adenocarcinoma A549 cells. Artif. Cells Nanomed. Biotechnol. 2020, 48, 639–647. [Google Scholar] [CrossRef] [Green Version]
- Rafiq, N.B.M.; Nishimura, Y.; Plotnikov, S.V.; Thiagarajan, V.; Zhang, Z.; Shi, S.; Natarajan, M.; Viasnoff, V.; Kanchanawong, P.; Jones, G.E. A mechano-signalling network linking microtubules, myosin IIA filaments and integrin-based adhesions. Nat. Mater. 2019, 18, 638–649. [Google Scholar] [CrossRef] [Green Version]
- Rodley, P.; Hatano, N.; Nishikawa, N.; Roy, B.; Sarkar, S.; Kiyama, R. A differential genomic cloning for cancer study: An outline and applications. Recent Res. Dev. Mol. Biol. 2003, 1, 13–27. [Google Scholar]
- Mendez, M.G.; Janmey, P.A. Transcription factor regulation by mechanical stress. Int. J. Biochem. Cell Biol. 2012, 44, 728–732. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, M.T.; Won, Y.H.; Kwon, T.W.; Lee, W. Twinfilin-1 is an essential regulator of myogenic differentiation through the modulation of YAP in C2C12 myoblasts. Biochem. Biophys. Res. Commun. 2022, 599, 17–23. [Google Scholar] [CrossRef]
- Pan, D. The hippo signaling pathway in development and cancer. Dev. Cell 2010, 19, 491–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seo, J.; Kim, J. Regulation of Hippo signaling by actin remodeling. BMB Rep. 2018, 51, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Sansores-Garcia, L.; Bossuyt, W.; Wada, K.; Yonemura, S.; Tao, C.; Sasaki, H.; Halder, G. Modulating F-actin organization induces organ growth by affecting the Hippo pathway. EMBO J. 2011, 30, 2325–2335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, F.X.; Zhao, B.; Guan, K.L. Hippo Pathway in Organ Size Control, Tissue Homeostasis, and Cancer. Cell 2015, 163, 811–828. [Google Scholar] [CrossRef] [Green Version]
- Bernstein, B.W.; Bamburg, J.R. ADF/cofilin: A functional node in cell biology. Trends Cell Biol. 2010, 20, 187–195. [Google Scholar] [CrossRef] [Green Version]
- Watt, K.I.; Turner, B.J.; Hagg, A.; Zhang, X.; Davey, J.R.; Qian, H.; Beyer, C.; Winbanks, C.E.; Harvey, K.F.; Gregorevic, P. The Hippo pathway effector YAP is a critical regulator of skeletal muscle fibre size. Nat. Commun. 2015, 6, 1–13. [Google Scholar] [CrossRef]
- Goodman, C.A.; Dietz, J.M.; Jacobs, B.L.; McNally, R.M.; You, J.S.; Hornberger, T.A. Yes-Associated Protein is up-regulated by mechanical overload and is sufficient to induce skeletal muscle hypertrophy. FEBS Lett. 2015, 589, 1491–1497. [Google Scholar] [CrossRef] [Green Version]
- Judson, R.N.; Gray, S.R.; Walker, C.; Carroll, A.M.; Itzstein, C.; Lionikas, A.; Zammit, P.S.; De Bari, C.; Wackerhage, H. Constitutive expression of Yes-associated protein (Yap) in adult skeletal muscle fibres induces muscle atrophy and myopathy. PLoS ONE 2013, 8, e59622. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, M.T.; Lee, W. Kank1 Is Essential for Myogenic Differentiation by Regulating Actin Remodeling and Cell Proliferation in C2C12 Progenitor Cells. Cells 2022, 11, 2030. https://doi.org/10.3390/cells11132030
Nguyen MT, Lee W. Kank1 Is Essential for Myogenic Differentiation by Regulating Actin Remodeling and Cell Proliferation in C2C12 Progenitor Cells. Cells. 2022; 11(13):2030. https://doi.org/10.3390/cells11132030
Chicago/Turabian StyleNguyen, Mai Thi, and Wan Lee. 2022. "Kank1 Is Essential for Myogenic Differentiation by Regulating Actin Remodeling and Cell Proliferation in C2C12 Progenitor Cells" Cells 11, no. 13: 2030. https://doi.org/10.3390/cells11132030






