Circadian Volume Changes in Hippocampal Glia Studied by Label-Free Interferometric Imaging
Abstract
1. Introduction
2. Results
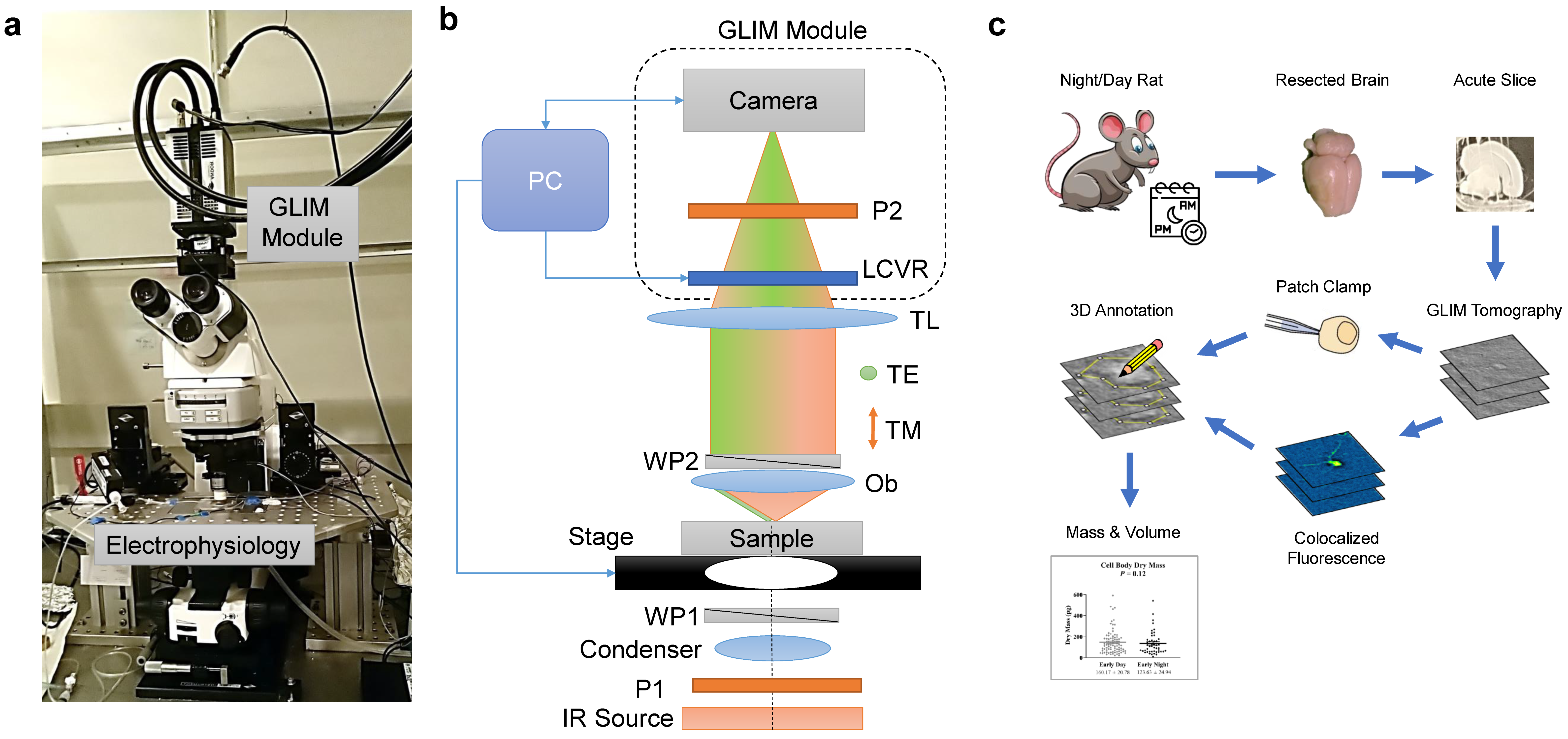
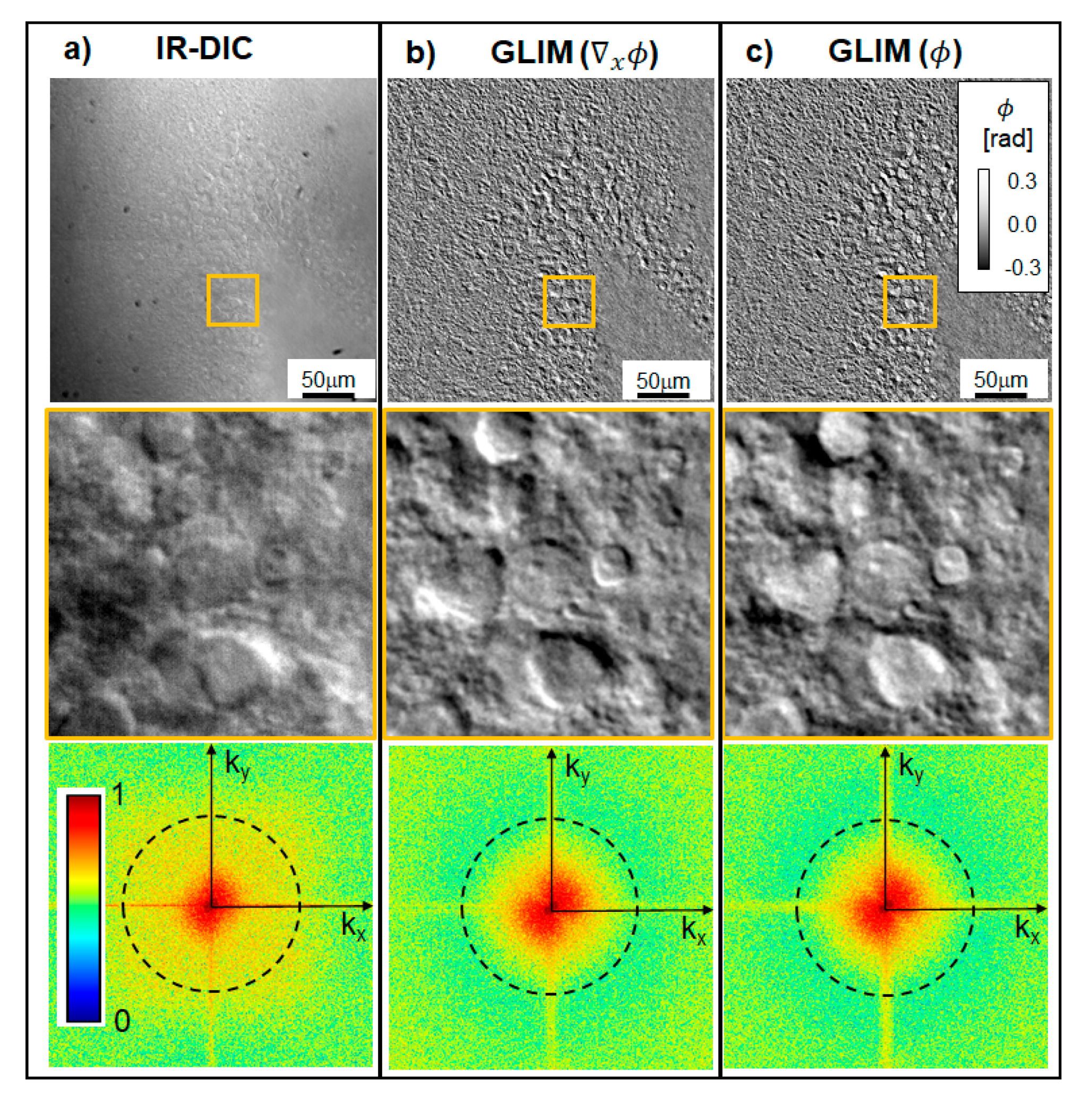
2.1. Optical System
2.2. Cell Dry Mass
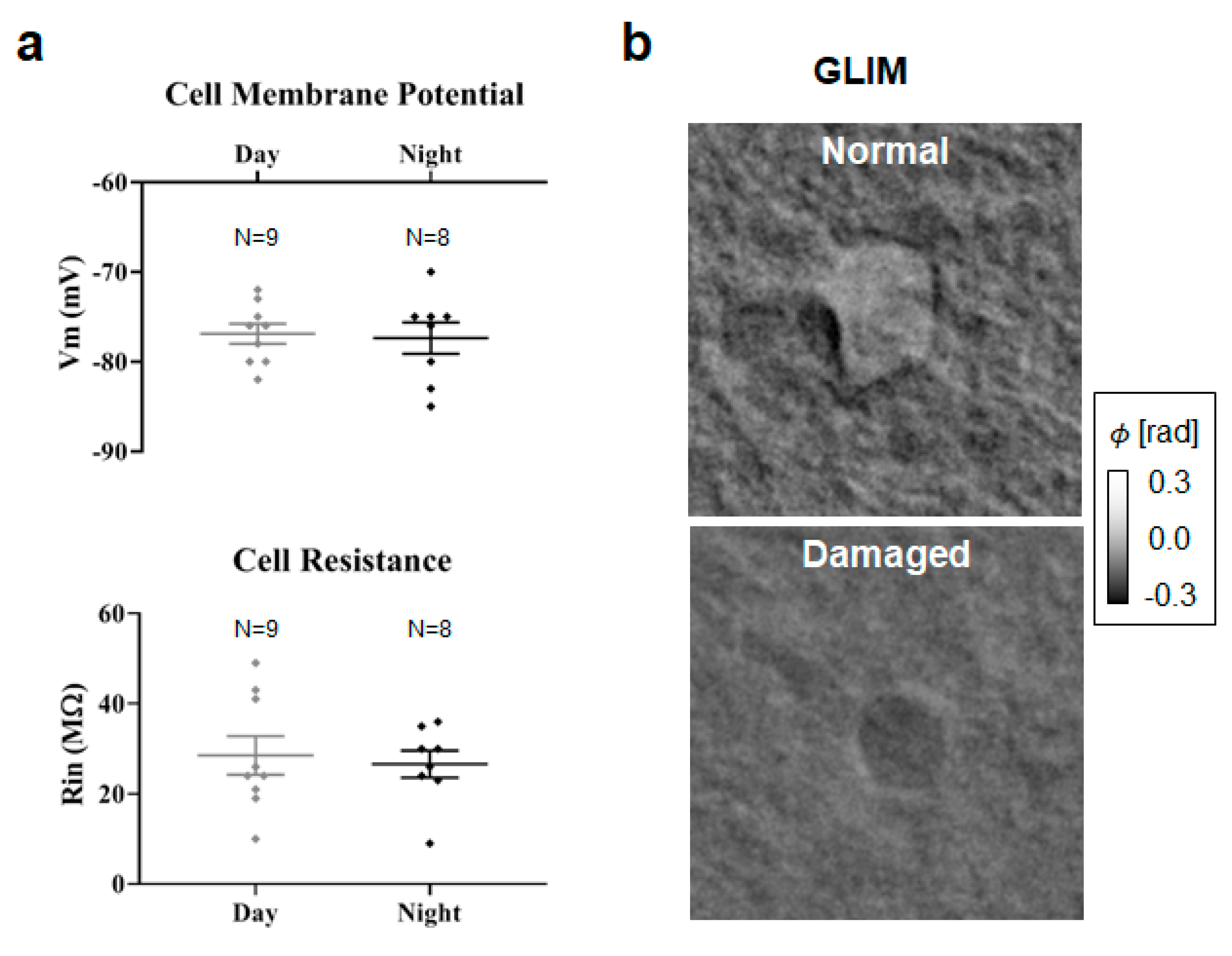
2.3. Cell Volume and Mass Changes during the Circadian Cycle
3. Discussion
4. Materials and Methods
4.1. Animals
4.2. Brain Slice Preparation and Patch-Clamp Recording
4.3. Colocalized Fluorescence and GLIM Imaging
4.4. Analysis of GLIM Data
4.5. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hammond, C. Cellular and Molecular Neurobiology, 2nd ed.; Academic Press: Cambridge, MA, USA, 2001. [Google Scholar]
- Pinar, C.; Fontaine, C.J.; Triviño-Paredes, J.; Lottenberg, C.P.; Gil-Mohapel, J.; Christie, B.R. Revisiting the flip side: Long-term depression of synaptic efficacy in the hippocampus. Neurosci. Biobehav. Rev. 2017, 80, 394–413. [Google Scholar] [CrossRef] [PubMed]
- Amaral, D.G.; Scharfman, H.E.; Lavenex, P. The dentate gyrus: Fundamental neuroanatomical organization (dentate gyrus for dummies). Prog. Brain Res. 2007, 163, 3–22. [Google Scholar] [CrossRef] [PubMed]
- Chaudhury, D.; Wang, L.M.; Colwell, C.S. Circadian regulation of hippocampal long-term potentiation. J. Biol. Rhythms 2005, 20, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Harris, K.M.; Teyler, T.J. Age differences in a circadian influence on hippocamapl LTP. Brain Res. 1983, 261, 69–73. [Google Scholar] [CrossRef]
- Bowden, J.B.; Abraham, W.C.; Harris, K.M. Differential effects of strain, circadian cycle, and stimulation pattern on LTP and concurrent LTD in the dentate gyrus of freely moving rats. Hippocampus 2012, 22, 1363–1370. [Google Scholar] [CrossRef]
- Schreurs, A.; Sabanov, V.; Balschun, D. Distinct properties of long-term potentiation in the dentate gyrus along the dorsoventral axis: Influence of age and inhibition. Sci. Rep. 2017, 7, 5157. [Google Scholar] [CrossRef]
- Tamai, S.; Sanada, K.; Fukada, Y. Time-of-day-dependent enhancement of adult neurogenesis in the hippocampus. PLoS ONE 2008, 3, e3835. [Google Scholar] [CrossRef]
- Heinemann, U.; Beck, H.; Dreier, J.; Ficker, E.; Stabel, J.; Zhang, C.L. The dentate gyrus as a regulated gate for the propagation of epileptiform activity. Epilepsy Res. 1992, 7, 273–280. [Google Scholar]
- Lothman, E.W.; Stringer, J.L.; Bertram, E.H. The dentate gyrus as a control point for seizures in the hippocampus and beyond. Epilepsy Res. 1992, 7, 301–313. [Google Scholar]
- Oberheim, N.A.; Tian, G.-F.; Han, X.; Peng, W.; Takano, T.; Ransom, B.R.; Nedergaard, M. Loss of astrocytic domain organization in the epileptic brain. J. Neurosci. 2008, 28, 3264–3276. [Google Scholar] [CrossRef]
- Binder, D.K.; Carson, M.J. Glial cells as primary therapeutic targets for epilepsy. Neurochem. Int. 2013, 63, 635–637. [Google Scholar] [CrossRef] [PubMed]
- Devinsky, O.; Vezzani, A.; Najjar, S.; De Lanerolle, N.C.; Rogawski, M.A. Glia and epilepsy: Excitability and inflammation. Trends Neurosci. 2013, 36, 174–184. [Google Scholar] [CrossRef] [PubMed]
- Volterra, A.; Steinhauser, C. Glial modulation of synaptic transmission in the hippocampus. Glia 2004, 47, 249–257. [Google Scholar] [CrossRef] [PubMed]
- Perea, G.; Navarrete, M.; Araque, A. Tripartite synapses: Astrocytes process and control synaptic information. Trends Neurosci. 2009, 32, 421–431. [Google Scholar] [CrossRef] [PubMed]
- Halassa, M.M.; Fellin, T.; Haydon, P.G. The tripartite synapse: Roles for gliotransmission in health and disease. Trends Mol. Med. 2007, 13, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Smith, N.A.; Xu, Q.; Fujita, T.; Baba, A.; Matsuda, T.; Takano, T.; Bekar, L.; Nedergaard, M. Astrocytes modulate neural network activity by Ca2+-dependent uptake of extracellular K+. Sci. Signal. 2012, 5, ra26. [Google Scholar] [CrossRef] [PubMed]
- Ransom, B.; Behar, T.; Nedergaard, M. New roles for astrocytes (stars at last). Trends Neurosci. 2003, 26, 520–522. [Google Scholar] [CrossRef]
- Rouach, N.; Koulakoff, A.; Abudara, V.; Willecke, K.; Giaume, C. Astroglial metabolic networks sustain hippocampal synaptic transmission. Science 2008, 322, 1551–1555. [Google Scholar] [CrossRef]
- Iliff, J.J.; Wang, M.; Liao, Y.; Plogg, B.A.; Peng, W.; Gundersen, G.A.; Benveniste, H.; Vates, G.E.; Deane, R.; Goldman, S.A.; et al. A paravascular pathway facilitates CSF flow through the brain parenchyma and the clearance of interstitial solutes, including amyloid beta. Sci. Transl. Med. 2012, 4, 147ra111. [Google Scholar] [CrossRef]
- Veitinger, S. The Patch-Clamp Technique; Leica Microsystems: Wetzlar, Germany, 2011; Available online: https://www.leica-microsystems.com/science-lab/the-patch-clamp-technique/ (accessed on 25 June 2022).
- Popescu, G. Quantitative Phase Imaging of Cells and Tissues; McGraw-Hill: New York, NY, USA, 2011. [Google Scholar]
- Chen, X.; Kandel, M.E.; Popescu, G. Spatial light interference microscopy: Principle and applications to biomedicine. Adv. Opt. Photon. 2021, 13, 353–425. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Sridharan, S.; Macias, V.; Kajdacsy-Balla, A.; Melamed, J.; Do, M.N.; Popescu, G. Automatic Gleason grading of prostate cancer using quantitative phase imaging and machine learning. J. Biomed. Opt. 2017, 22, 036015. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.; Depeursinge, C.; Popescu, G. Quantitative phase imaging in biomedicine. Nat. Photon. 2018, 12, 578–589. [Google Scholar] [CrossRef]
- Merola, F.; Memmolo, P.; Miccio, L.; Mugnano, M.; Ferraro, P. Phase contrast tomography at lab on chip scale by digital holography. Methods 2018, 136, 108–115. [Google Scholar] [CrossRef] [PubMed]
- Rivenson, Y.; Zhang, Y.; Günaydın, H.; Teng, D.; Ozcan, A. Phase recovery and holographic image reconstruction using deep learning in neural networks. Light Sci. Appl. 2018, 7, 17141. [Google Scholar] [CrossRef]
- Jo, Y.; Cho, H.; Lee, S.Y.; Choi, G.; Kim, G.; Min, H.-S.; Park, Y. Quantitative phase imaging and artificial intelligence: A review. IEEE J. Sel. Top. Quantum Electron. 2019, 25, 6800914. [Google Scholar] [CrossRef]
- Uttam, S.; Pham, H.V.; LaFace, J.; Leibowitz, B.; Yu, J.; Brand, R.E.; Hartman, D.J.; Liu, Y. Early prediction of cancer progression by depth-resolved nanoscale maps of nuclear architecture from unstained tissue specimens. Cancer Res. 2015, 75, 4718–4727. [Google Scholar] [CrossRef]
- Wang, Z.; Millet, L.J.; Gillette, M.U.; Popescu, G. Jones phase microscopy of transparent and anisotropic samples. Opt. Lett. 2008, 33, 1270–1272. [Google Scholar] [CrossRef]
- Lue, N.; Choi, W.; Popescu, G.; Badizadegan, K.; Dasari, R.R.; Feld, M.S. Synthetic aperture tomographic phase microscopy for 3D imaging of live cells in translational motion. Opt. Express 2008, 16, 16240–16246. [Google Scholar] [CrossRef]
- Kandel, M.E.; Hu, C.; Kouzehgarani, G.N.; Min, E.; Sullivan, K.M.; Kong, H.; Li, J.M.; Robson, D.N.; Gillette, M.U.; Best-Popescu, C.; et al. Epi-illumination gradient light interference microscopy for imaging opaque structures. Nat. Commun. 2019, 10, 4691. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Kandel, M.E.; Rubessa, M.; Wheeler, M.B.; Popescu, G. Gradient light interference microscopy for 3D imaging of unlabeled specimens. Nat. Commun. 2017, 8, 210. [Google Scholar] [CrossRef]
- Kim, T.; Zhou, R.; Goddard, L.L.; Popescu, G. Solving inverse scattering problems in biological samples by quantitative phase imaging. Laser Photon. Rev. 2016, 10, 13–39. [Google Scholar] [CrossRef]
- Mir, M.; Kim, T.; Majumder, A.A.S.; Xiang, M.; Wang, R.; Liu, S.C.; Gillette, M.U.; Stice, S.L.; Popescu, G. Label-free characterization of emerging human neuronal networks. Sci. Rep. 2014, 4, 4434. [Google Scholar] [CrossRef]
- Wang, Z.; Chun, I.S.; Li, X.; Ong, Z.-Y.; Pop, E.; Millet, L.; Gillette, M.; Popescu, G. Topography and refractometry of nanostructures using spatial light interference microscopy. Opt. Lett. 2010, 35, 208–210. [Google Scholar] [CrossRef]
- Mir, M.; Wang, Z.; Shen, Z.; Bednarz, M.; Bashir, R.; Golding, I.; Prasanth, S.G.; Popescu, G. Optical measurement of cycle-dependent cell growth. Proc. Nat. Acad. Sci. USA 2011, 108, 13124–13129. [Google Scholar] [CrossRef]
- Challa, S.; Chan, F.K.-M. Going up in flames: Necrotic cell injury and inflammatory diseases. Cell. Mol. Life Sci. 2010, 67, 3241–3253. [Google Scholar] [CrossRef] [PubMed]
- Milo, R. What is the Density of Cells? John Wiley and Sons: Hoboken, NJ, USA, 2013; Available online: http://book.bionumbers.org/what-is-the-density-of-cells (accessed on 25 June 2022).
- Cassé, F.; Richetin, K.; Toni, N. Astrocytes’ contribution to adult neurogenesis in physiology and Alzheimer’s disease. Front. Cell. Neurosci. 2018, 12, 432. [Google Scholar] [CrossRef] [PubMed]
- Lavialle, M.; Begue, A.; Papillon, C.; Vilaplana, J. Modifications of retinal afferent activity induce changes in astroglial plasticity in the hamster circadian clock. Glia 2001, 34, 88–100. [Google Scholar] [CrossRef] [PubMed]
- Lavialle, M.; Aumann, G.; Anlauf, E.; Pröls, F.; Arpin, M.; Derouiche, A. Structural plasticity of perisynaptic astrocyte processes involves ezrin and metabotropic glutamate receptors. Proc. Natl. Acad. Sci. USA 2011, 108, 12915–12919. [Google Scholar] [CrossRef] [PubMed]
- Bellesi, M.; de Vivo, L.; Tononi, G.; Cirelli, C. Effects of sleep and wake on astrocytes: Clues from molecular and ultrastructural studies. BMC Biol. 2015, 13, 66. [Google Scholar] [CrossRef]
- Theodosis, D.T. Oxytocin-secreting neurons: A physiological model of morphological neuronal and glial plasticity in the adult hypothalamus. Front. Neuroendocr. 2002, 23, 101–135. [Google Scholar] [CrossRef]
- Theodosis, D.T.; Poulain, D.A.; Oliet, S.H. Activity-dependent structural and functional plasticity of astrocyte-neuron interactions. Physiol. Rev. 2008, 88, 983–1008. [Google Scholar] [CrossRef] [PubMed]
- Hawrylak, N.; Fleming, J.C.; Salm, A.K. Dehydration and rehydration selectively and reversibly alter glial fibrillary acidic protein immunoreactivity in the rat supraoptic nucleus and subjacent glial limitans. Glia 1998, 22, 260–271. [Google Scholar] [CrossRef]
- Xie, L.; Kang, H.; Xu, Q.; Chen, M.J.; Liao, Y.; Thiyagarajan, M.; O’Donnell, J.; Christensen, D.J.; Nicholson, C.; Iliff, J.J.; et al. Sleep drives metabolite clearance from the adult brain. Science 2013, 342, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Wilson, C.S.; Mongin, A.A. Cell volume control in healthy brain and neuropathologies. Curr. Top. Membr. 2018, 81, 385–455. [Google Scholar] [CrossRef]
- Walch, E.; Murphy, T.R.; Cuvelier, N.; Aldoghmi, M.; Morozova, C.; Donohue, J.; Young, G.; Samant, A.; Garcia, S.; Alvarez, C.; et al. Astrocyte-selective volume increase in elevated extracellular potassium conditions is mediated by the Na(+)/K(+) ATPase and occurs independently of aquaporin 4. ASN Neuro 2020, 12, 1759091420967152. [Google Scholar] [CrossRef]
- Risher, W.C.; Andrew, R.D.; Kirov, S.A. Real-time passive volume responses of astrocytes to acute osmotic and ischemic stress in cortical slices and in vivo revealed by two-photon microscopy. Glia 2009, 57, 207–221. [Google Scholar] [CrossRef]
- Lux, H.D.; Heinemann, U.; Dietzel, I. Ionic changes and alterations in the size of the extracellular space during epileptic activity. Adv. Neurol. 1986, 44, 619–639. [Google Scholar]
- Florence, C.M.; Baillie, L.D.; Mulligan, S.J. Dynamic volume changes in astrocytes are an intrinsic phenomenon mediated by bicarbonate ion flux. PLoS ONE 2012, 7, e51124. [Google Scholar] [CrossRef]
- Amzica, F.; Neckelmann, D. Membrane capacitance of cortical neurons and glia during sleep oscillations and spike-wave seizures. J. Neurophysiol. 1999, 82, 2731–2746. [Google Scholar] [CrossRef]
- Naseri Kouzehgarani, G.; Bothwell, M.Y.; Gillette, M.U. Circadian rhythm of redox state regulates membrane excitability in hippocampal CA1 neurons. Eur. J. Neurosci. 2020, 51, 34–46. [Google Scholar] [CrossRef]
- Rasmussen, R.; Nedergaard, M.; Petersen, N.C. Sulforhodamine 101, a widely used astrocyte marker, can induce cortical seizure-like activity at concentrations commonly used. Sci. Rep. 2016, 6, 30433. [Google Scholar] [CrossRef] [PubMed]
- Kafitz, K.W.; Meier, S.D.; Stephan, J.; Rose, C.R. Developmental profile and properties of sulforhodamine 101—Labeled glial cells in acute brain slices of rat hippocampus. J. Neurosci. Methods 2008, 169, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Meier, S.D.; Kafitz, K.W.; Rose, C.R. Developmental profile and mechanisms of GABA-induced calcium signaling in hippocampal astrocytes. Glia 2008, 56, 1127–1137. [Google Scholar] [CrossRef] [PubMed]
- Schnell, C.; Shahmoradi, A.; Wichert, S.P.; Mayerl, S.; Hagos, Y.; Heuer, H.; Rossner, M.J.; Hülsmann, S. The multispecific thyroid hormone transporter OATP1C1 mediates cell-specific sulforhodamine 101-labeling of hippocampal astrocytes. Brain Struct. Funct. 2015, 220, 193–203. [Google Scholar] [CrossRef][Green Version]
- Hagos, L.; Hulsmann, S. Unspecific labelling of oligodendrocytes by sulforhodamine 101 depends on astrocytic uptake via the thyroid hormone transporter OATP1C1 (SLCO1C1). Neurosci. Lett. 2016, 631, 13–18. [Google Scholar] [CrossRef]
- Hulsmann, S.; Hagos, L.; Heuer, H.; Schnell, C. Limitations of Sulforhodamine 101 for Brain Imaging. Front. Cell. Neurosci. 2017, 11, 44. [Google Scholar] [CrossRef]
- Cahoy, J.D.; Emery, B.; Kaushal, A.; Foo, L.C.; Zamanian, J.L.; Christopherson, K.S.; Xing, Y.; Lubischer, J.L.; Krieg, P.A.; Krupenko, S.A.; et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J. Neurosci. 2008, 28, 264–278. [Google Scholar] [CrossRef]
- Kandel, M.E.; He, Y.R.; Lee, Y.J.; Chen, T.H.-Y.; Sullivan, K.M.; Aydin, O.; Saif, M.T.A.; Kong, H.; Sobh, N.; Popescu, G. Phase imaging with computational specificity (PICS) for measuring dry mass changes in sub-cellular compartments. Nat. Commun. 2020, 11, 6256. [Google Scholar] [CrossRef]
- Kandel, M.E. Convert Delaunay Trinagulation to Volume; Stack Overflow: New York, NY, USA, 2018; Available online: https://stackoverflow.com/questions/52662007/convert-delaunay-trinagulation-to-volume (accessed on 25 June 2022).



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Naseri Kouzehgarani, G.; Kandel, M.E.; Sakakura, M.; Dupaty, J.S.; Popescu, G.; Gillette, M.U. Circadian Volume Changes in Hippocampal Glia Studied by Label-Free Interferometric Imaging. Cells 2022, 11, 2073. https://doi.org/10.3390/cells11132073
Naseri Kouzehgarani G, Kandel ME, Sakakura M, Dupaty JS, Popescu G, Gillette MU. Circadian Volume Changes in Hippocampal Glia Studied by Label-Free Interferometric Imaging. Cells. 2022; 11(13):2073. https://doi.org/10.3390/cells11132073
Chicago/Turabian StyleNaseri Kouzehgarani, Ghazal, Mikhail E. Kandel, Masayoshi Sakakura, Joshua S. Dupaty, Gabriel Popescu, and Martha U. Gillette. 2022. "Circadian Volume Changes in Hippocampal Glia Studied by Label-Free Interferometric Imaging" Cells 11, no. 13: 2073. https://doi.org/10.3390/cells11132073
APA StyleNaseri Kouzehgarani, G., Kandel, M. E., Sakakura, M., Dupaty, J. S., Popescu, G., & Gillette, M. U. (2022). Circadian Volume Changes in Hippocampal Glia Studied by Label-Free Interferometric Imaging. Cells, 11(13), 2073. https://doi.org/10.3390/cells11132073






