High-Fat Diet Alters the Retinal Pigment Epithelium and Choroidal Transcriptome in the Absence of Gut Microbiota
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Diets
2.2. Sterility Monitoring
2.3. RNA Extraction
2.4. RNA Sequencing
2.5. Statistical Analysis
3. Results
3.1. High-Fat Diet Is Associated with Changes in the Rpe/Choroid Transcriptome
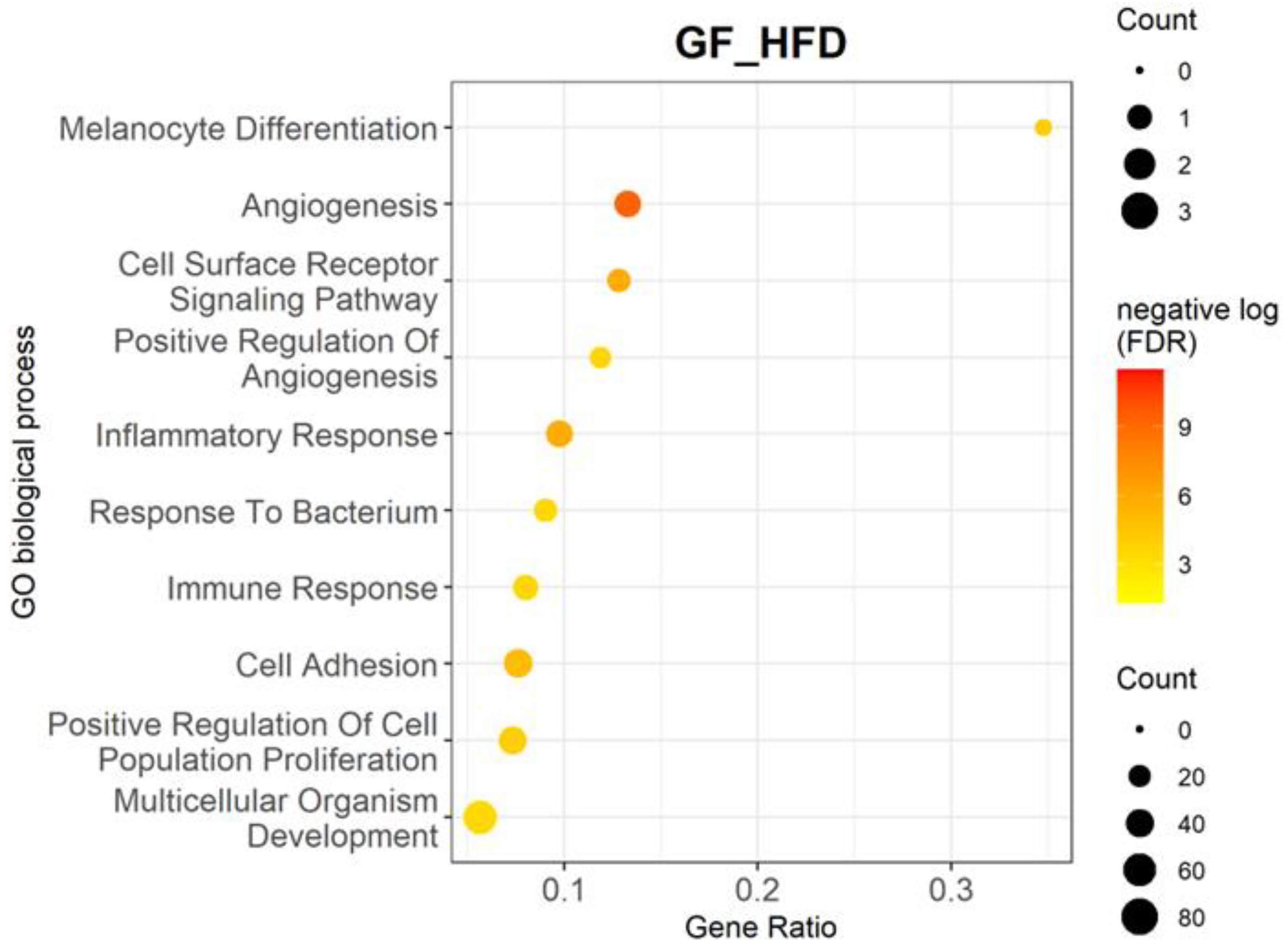
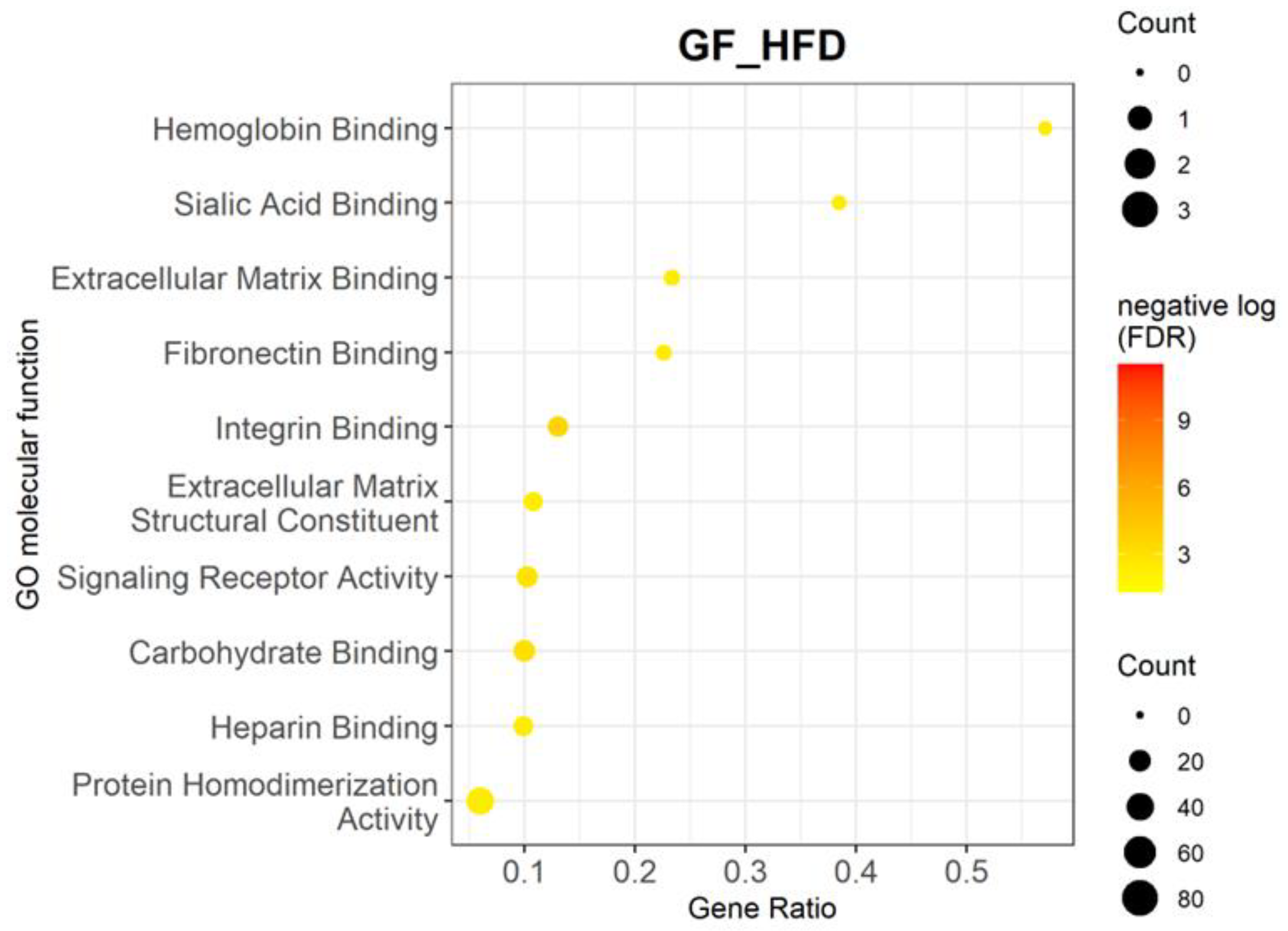
3.2. High-Fat Diet Upregulates Multiple Biological Processes and Genes Related to Inflammation and Angiogenesis
4. Discussion
4.1. High-Fat Diet Affects Gene Expression in Angiogenic Pathways in Germ-Free Mice
4.2. High-Fat Diet Alters Gene Expression Involved in Inflammatory and Immune Response Pathways in Germ-Free Mice
4.3. High-Fat Diet Affects Gene Expression Involved in the Complement System
4.4. Additional Genes and Pathways Are Differentially Represented by High-Fat Diet in Germ-Free Mice
5. Conclusions and Limitations
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wong, W.L.; Su, X.; Li, X.; Cheung, C.M.G.; Klein, R.; Cheng, C.-Y.; Wong, T.Y. Global Prevalence of Age-Related Macular Degeneration and Disease Burden Projection for 2020 and 2040: A Systematic Review and Meta-Analysis. Lancet Glob. Health 2014, 2, e106–e116. [Google Scholar] [CrossRef]
- Rinninella, E.; Mele, M.C.; Merendino, N.; Cintoni, M.; Anselmi, G.; Caporossi, A.; Gasbarrini, A.; Minnella, A.M. The Role of Diet, Micronutrients and the Gut Microbiota in Age-Related Macular Degeneration: New Perspectives from the Gut–Retina Axis. Nutrients 2018, 10, 1677. [Google Scholar] [CrossRef] [PubMed]
- Weikel, K.A.; Chiu, C.-J.; Taylor, A. Nutritional Modulation of Age-Related Macular Degeneration. Mol. Asp. Med. 2012, 33, 318–375. [Google Scholar] [CrossRef] [PubMed]
- Chapman, N.A.; Jacobs, R.J.; Braakhuis, A.J. Role of Diet and Food Intake in Age-Related Macular Degeneration: A Systematic Review. Clin. Exp. Ophthalmol. 2019, 47, 106–127. [Google Scholar] [CrossRef] [PubMed]
- Cundiff, D.K.; Nigg, C.R. Diet and Diabetic Retinopathy: Insights From the Diabetes Control and Complications Trial (DCCT). MedGenMed 2005, 7, 3. [Google Scholar]
- Parekh, N.; Voland, R.P.; Moeller, S.M.; Blodi, B.A.; Ritenbaugh, C.; Chappell, R.J.; Wallace, R.B.; Mares, J.A. CAREDS Research Study Group. Association between Dietary Fat Intake and Age-Related Macular Degeneration in the Carotenoids in Age-Related Eye Disease Study (CAREDS): An Ancillary Study of the Women’s Health Initiative. Arch. Ophthalmol. 2009, 127, 1483–1493. [Google Scholar] [CrossRef]
- Chiu, C.-J.; Chang, M.-L.; Zhang, F.F.; Li, T.; Gensler, G.; Schleicher, M.; Taylor, A. The Relationship of Major American Dietary Patterns to Age-Related Macular Degeneration. Am. J. Ophthalmol. 2014, 158, 118–127.e1. [Google Scholar] [CrossRef]
- Cho, E.; Hung, S.; Willett, W.C.; Spiegelman, D.; Rimm, E.B.; Seddon, J.M.; Colditz, G.A.; Hankinson, S.E. Prospective Study of Dietary Fat and the Risk of Age-Related Macular Degeneration. Am. J. Clin. Nutr. 2001, 73, 209–218. [Google Scholar] [CrossRef]
- Seddon, J.M.; Cote, J.; Rosner, B. Progression of Age-Related Macular Degeneration. Arch. Ophthalmol. 2003, 121, 1728–1737. [Google Scholar] [CrossRef]
- Wong-Riley, M. Energy Metabolism of the Visual System. Eye Brain 2010, 2, 99–116. [Google Scholar] [CrossRef]
- Berry, K.A.Z.; Gordon, W.C.; Murphy, R.C.; Bazan, N.G. Spatial Organization of Lipids in the Human Retina and Optic Nerve by MALDI Imaging Mass Spectrometry. J. Lipid Res. 2014, 55, 504–515. [Google Scholar] [CrossRef] [PubMed]
- Duda, M.; Kawula, K.; Pawlak, A.; Sarna, T.; Wisniewska-Becker, A. EPR Studies on the Properties of Model Photoreceptor Membranes Made of Natural and Synthetic Lipids. Cell Biochem. Biophys. 2017, 75, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.; Chen, C.T.; Cagnone, G.; Heckel, E.; Sun, Y.; Cakir, B.; Tomita, Y.; Huang, S.; Li, Q.; Britton, W.; et al. Dyslipidemia in Retinal Metabolic Disorders. EMBO Mol. Med. 2019, 11, e10473. [Google Scholar] [CrossRef] [PubMed]
- Bhutto, I.; Lutty, G. Understanding Age-Related Macular Degeneration (AMD): Relationships between the Photoreceptor/Retinal Pigment Epithelium/Bruch’s Membrane/Choriocapillaris Complex. Mol. Asp. Med. 2012, 33, 295–317. [Google Scholar] [CrossRef] [PubMed]
- Clarkson-Townsend, D.A.; Douglass, A.J.; Singh, A.; Allen, R.S.; Uwaifo, I.N.; Pardue, M.T. Impacts of High Fat Diet on Ocular Outcomes in Rodent Models of Visual Disease. Exp. Eye Res. 2021, 204, 108440. [Google Scholar] [CrossRef]
- Miceli, M.V.; Newsome, D.A.; Tate, D.J.; Sarphie, T.G. Pathologic Changes in the Retinal Pigment Epithelium and Bruch’s Membrane of Fat-Fed Atherogenic Mice. Curr. Eye Res. 2000, 20, 8–16. [Google Scholar] [CrossRef]
- Lee, J.-J.; Wang, P.-W.; Yang, I.-H.; Huang, H.-M.; Chang, C.-S.; Wu, C.-L.; Chuang, J.-H. High-Fat Diet Induces Toll-like Receptor 4-Dependent Macrophage/Microglial Cell Activation and Retinal Impairment. Investig. Ophthalmol. Vis. Sci. 2015, 56, 3041–3050. [Google Scholar] [CrossRef]
- Albouery, M.; Buteau, B.; Grégoire, S.; Martine, L.; Gambert, S.; Bron, A.M.; Acar, N.; Chassaing, B.; Bringer, M.-A. Impact of a High-Fat Diet on the Fatty Acid Composition of the Retina. Exp. Eye Res. 2020, 196, 108059. [Google Scholar] [CrossRef]
- Skondra, D.; She, H.; Zambarakji, H.J.; Connolly, E.; Michaud, N.; Chan, P.; Kim, I.K.; Gragoudas, E.S.; Miller, J.W.; Hafezi-Moghadam, A. Effects of ApoE Deficiency, Aging and High Fat Diet on Laser-Induced Choroidal Neovascularization and Bruch’s Membrane-RPE Interface Morphology. Investig. Ophthalmol. Vis. Sci. 2007, 48, 1768. [Google Scholar]
- Biswas, L.; Ibrahim, K.S.; Li, X.; Zhou, X.; Zeng, Z.; Craft, J.; Shu, X. Effect of a TSPO Ligand on Retinal Pigment Epithelial Cholesterol Homeostasis in High-Fat Fed Mice, Implication for Age-Related Macular Degeneration. Exp. Eye Res. 2021, 208, 108625. [Google Scholar] [CrossRef]
- Cousins, S.W.; Espinosa-Heidmann, D.G.; Alexandridou, A.; Sall, J.; Dubovy, S.; Csaky, K. The Role of Aging, High Fat Diet and Blue Light Exposure in an Experimental Mouse Model for Basal Laminar Deposit Formation. Exp. Eye Res. 2002, 75, 543–553. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Chu, Y.; Mowery, J.; Konkel, B.; Galli, S.; Theos, A.C.; Golestaneh, N. Pgc-1α Repression and High-Fat Diet Induce Age-Related Macular Degeneration-like Phenotypes in Mice. Dis Model. Mech. 2018, 11, dmm032698. [Google Scholar] [CrossRef] [PubMed]
- Dithmar, S.; Sharara, N.A.; Curcio, C.A.; Le, N.-A.; Zhang, Y.; Brown, S.; Grossniklaus, H.E. Murine High-Fat Diet and Laser Photochemical Model of Basal Deposits in Bruch Membrane. Arch. Ophthalmol. 2001, 119, 1643–1649. [Google Scholar] [CrossRef]
- Jandhyala, S.M.; Talukdar, R.; Subramanyam, C.; Vuyyuru, H.; Sasikala, M.; Reddy, D.N. Role of the Normal Gut Microbiota. World J. Gastroenterol. 2015, 21, 8787–8803. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, B.O.; Bäckhed, F. Signals from the Gut Microbiota to Distant Organs in Physiology and Disease. Nat. Med. 2016, 22, 1079–1089. [Google Scholar] [CrossRef]
- Fujimura, K.E.; Lynch, S.V. Microbiota in Allergy and Asthma and the Emerging Relationship with the Gut Microbiome. Cell Host Microbe 2015, 17, 592–602. [Google Scholar] [CrossRef]
- Mohajeri, M.H.; La Fata, G.; Steinert, R.E.; Weber, P. Relationship between the Gut Microbiome and Brain Function. Nutr. Rev. 2018, 76, 481–496. [Google Scholar] [CrossRef]
- Rowan, S.; Taylor, A. The Role of Microbiota in Retinal Disease. Adv. Exp. Med. Biol. 2018, 1074, 429–435. [Google Scholar] [CrossRef]
- Zysset-Burri, D.C.; Keller, I.; Berger, L.E.; Largiadèr, C.R.; Wittwer, M.; Wolf, S.; Zinkernagel, M.S. Associations of the Intestinal Microbiome with the Complement System in Neovascular Age-Related Macular Degeneration. NPJ Genom. Med. 2020, 5, 34. [Google Scholar] [CrossRef]
- Zinkernagel, M.S.; Zysset-Burri, D.C.; Keller, I.; Berger, L.E.; Leichtle, A.B.; Largiadèr, C.R.; Fiedler, G.M.; Wolf, S. Association of the Intestinal Microbiome with the Development of Neovascular Age-Related Macular Degeneration. Sci. Rep. 2017, 7, 40826. [Google Scholar] [CrossRef]
- Andriessen, E.M.; Wilson, A.M.; Mawambo, G.; Dejda, A.; Miloudi, K.; Sennlaub, F.; Sapieha, P. Gut Microbiota Influences Pathological Angiogenesis in Obesity-driven Choroidal Neovascularization. EMBO Mol. Med. 2016, 8, 1366–1379. [Google Scholar] [CrossRef] [PubMed]
- Murphy, E.A.; Velazquez, K.T.; Herbert, K.M. Influence of High-Fat-Diet on Gut Microbiota: A Driving Force for Chronic Disease Risk. Curr. Opin. Clin. Nutr. Metab. Care 2015, 18, 515–520. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Sonne, S.B.; Feng, Q.; Chen, N.; Xia, Z.; Li, X.; Fang, Z.; Zhang, D.; Fjære, E.; Midtbø, L.K.; et al. High-Fat Feeding Rather than Obesity Drives Taxonomical and Functional Changes in the Gut Microbiota in Mice. Microbiome 2017, 5, 43. [Google Scholar] [CrossRef] [PubMed]
- Dao, D.; Xie, B.; Nadeem, U.; Xiao, J.; Movahedan, A.; D’Souza, M.; Leone, V.; Hariprasad, S.M.; Chang, E.B.; Sulakhe, D.; et al. High-Fat Diet Alters the Retinal Transcriptome in the Absence of Gut Microbiota. Cells 2021, 10, 2119. [Google Scholar] [CrossRef]
- National Research Council (US) Committee for the Update of the Guide for the Care and Use of Laboratory Animals. The National Academies Collection: Reports funded by National Institutes of Health. In Guide for the Care and Use of Laboratory Animals, 8th ed.; National Academies Press (US): Washington, DC, USA, 2011; ISBN 978-0-309-15400-0. [Google Scholar]
- Theriault, B.; Wang, Y.; Chen, L.; Vest, A.; Bartman, C.; Alegre, M.-L. Long-Term Maintenance of Sterility After Skin Transplantation in Germ-Free Mice. Transplant. Direct 2015, 1, e28. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. Limma Powers Differential Expression Analyses for RNA-Sequencing and Microarray Studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Smyth, G.K. Limma: Linear Models for Microarray Data. In Bioinformatics and Computational Biology Solutions Using R and Bioconductor; Gentleman, R., Carey, V.J., Huber, W., Irizarry, R.A., Dudoit, S., Eds.; Statistics for Biology and Health; Springer: New York, NY, USA, 2005; pp. 397–420. ISBN 978-0-387-29362-2. [Google Scholar]
- Frankish, A.; Diekhans, M.; Ferreira, A.-M.; Johnson, R.; Jungreis, I.; Loveland, J.; Mudge, J.M.; Sisu, C.; Wright, J.; Armstrong, J.; et al. GENCODE Reference Annotation for the Human and Mouse Genomes. Nucleic Acids Res. 2019, 47, D766–D773. [Google Scholar] [CrossRef]
- Chen, Y.; Lun, A.T.L.; Smyth, G.K. From Reads to Genes to Pathways: Differential Expression Analysis of RNA-Seq Experiments Using Rsubread and the EdgeR Quasi-Likelihood Pipeline. F1000Research 2016, 5, 1438. [Google Scholar]
- Robinson, M.D.; McCarthy, D.J.; Smyth, K.G. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Sulakhe, D.; Balasubramanian, S.; Xie, B.; Feng, B.; Taylor, A.; Wang, S.; Berrocal, E.; Dave, U.; Xu, J.; Börnigen, D.; et al. Lynx: A Database and Knowledge Extraction Engine for Integrative Medicine. Nucleic Acids Res. 2014, 42, D1007–D1012. [Google Scholar] [CrossRef] [PubMed]
- Nickla, D.L.; Wallman, J. The multifunctional choroid. Prog. Retin. Eye Res. 2010, 29, 144–168. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, R.A.; Mousavi, M. Overview of Risk Factors for Age-Related Macular Degeneration (AMD). J. Stem Cells 2015, 10, 171–191. [Google Scholar]
- Dow, C.; Mancini, F.; Rajaobelina, K.; Boutron-Ruault, M.-C.; Balkau, B.; Bonnet, F.; Fagherazzi, G. Diet and Risk of Diabetic Retinopathy: A Systematic Review. Eur. J. Epidemiol. 2018, 33, 141–156. [Google Scholar] [CrossRef]
- Rowan, S. Involvement of a Gut–Retina Axis in Protection against Dietary Glycemia-Induced Age-Related Macular Degeneration. Proc. Natl. Acad. Sci. USA 2017, 114, 4472–4481. [Google Scholar] [CrossRef]
- Bretillon, L.; Thuret, G.; Grégoire, S.; Acar, N.; Joffre, C.; Bron, A.M.; Gain, P.; Creuzot-Garcher, C.P. Lipid and Fatty Acid Profile of the Retina, Retinal Pigment Epithelium/Choroid, and the Lacrimal Gland, and Associations with Adipose Tissue Fatty Acids in Human Subjects. Exp. Eye Res. 2008, 87, 521–528. [Google Scholar] [CrossRef]
- Shao, J.; Choudhary, M.M.; Schachat, A.P. Neovascular Age-Related Macular Degeneration. Retin. Pharmacother. 2016, 55, 125–136. [Google Scholar] [CrossRef]
- Campochiaro, P.A. Molecular Pathogenesis of Retinal and Choroidal Vascular Diseases. Prog. Retin. Eye Res. 2015, 49, 67–81. [Google Scholar] [CrossRef]
- Khanna, S.; Komati, R.; Eichenbaum, D.A.; Hariprasad, I.; Ciulla, T.A.; Hariprasad, S.M. Current and Upcoming Anti-VEGF Therapies and Dosing Strategies for the Treatment of Neovascular AMD: A Comparative Review. BMJ Open Ophthalmol. 2019, 4, e000398. [Google Scholar] [CrossRef]
- Saharinen, P.; Eklund, L.; Alitalo, K. Therapeutic Targeting of the Angiopoietin–TIE Pathway. Nat. Rev. Drug Discov. 2017, 16, 635–661. [Google Scholar] [CrossRef] [PubMed]
- Fagiani, E.; Christofori, G. Angiopoietins in Angiogenesis. Cancer Lett. 2013, 328, 18–26. [Google Scholar] [CrossRef] [PubMed]
- Thurston, G. Complementary Actions of VEGF and Angiopoietin-1 on Blood Vessel Growth and Leakage. J. Anat. 2002, 200, 575–580. [Google Scholar] [CrossRef]
- Brindle, N.P.J.; Saharinen, P.; Alitalo, K. Signaling and Functions of Angiopoietin-1 in Vascular Protection. Circ. Res. 2006, 98, 1014–1023. [Google Scholar] [CrossRef] [PubMed]
- Richardson, M.R.; Robbins, E.P.; Vemula, S.; Critser, P.J.; Whittington, C.; Voytik-Harbin, S.L.; Yoder, M.C. Angiopoietin-like Protein 2 Regulates Endothelial Colony Forming Cell Vasculogenesis. Angiogenesis 2014, 17, 675–683. [Google Scholar] [CrossRef] [PubMed]
- Kadomatsu, T.; Endo, M.; Miyata, K.; Oike, Y. Diverse Roles of ANGPTL2 in Physiology and Pathophysiology. Trends Endocrinol. Metab. 2014, 25, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Lux, A.; Llacer, H.; Heussen, F.M.A.; Joussen, A.M. Non-Responders to Bevacizumab (Avastin) Therapy of Choroidal Neovascular Lesions. Br. J. Ophthalmol. 2007, 91, 1318–1322. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, P.J.; Brown, D.M.; Heier, J.S.; Boyer, D.S.; Kaiser, P.K.; Chung, C.Y.; Kim, R.Y.; MARINA Study Group. Ranibizumab for Neovascular Age-Related Macular Degeneration. N. Engl. J. Med. 2006, 355, 1419–1431. [Google Scholar] [CrossRef]
- Yang, S.; Zhao, J.; Sun, X. Resistance to Anti-VEGF Therapy in Neovascular Age-Related Macular Degeneration: A Comprehensive Review. Drug Des. Dev. Ther. 2016, 10, 1857–1867. [Google Scholar] [CrossRef]
- Regeneron Pharmaceuticals. An Open-Label, Dose-Escalation Study of the Safety and Tolerability of Intravitreal (IVT) REGN910-3 and IVT REGN910 in Patients with Either Neovascular AMD or DME. 2016. Available online: https://clinicaltrials.gov (accessed on 6 May 2022).
- Patel, S.S.; Sahni, J.; Sadikhov, S.; Pauly-Evers, M.; Szczesny, P.; Weikert, R. Anti-VEGF/Anti-Angiopoietin-2 Bispecific Antibody RG7716 in Diabetic Macular Edema: Complete 36-Week Results from the Phase 2, Multicenter, Randomized, Active Treatment-Controlled BOULEVARD Clinical Trial. Investig. Ophthalmol. Vis. Sci. 2018, 59, 1959. [Google Scholar]
- Kumar, A.; Li, X. PDGF-C and PDGF-D in Ocular Diseases. Mol. Asp. Med. 2018, 62, 33–43. [Google Scholar] [CrossRef] [PubMed]
- Folestad, E.; Kunath, A.; Wågsäter, D. PDGF-C and PDGF-D Signaling in Vascular Diseases and Animal Models. Mol. Asp. Med. 2018, 62, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Sun, X.; Huang, Z.; Zhou, T.; Zhu, X.; Liu, Y.; Wang, J.; Cheng, B.; Li, M.; He, C.; et al. Imatinib Ameliorated Retinal Neovascularization by Suppressing PDGFR-α and PDGFR-β. CPB 2018, 48, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, G.J.; Ciulla, T.A.; Ciardella, A.P.; Devin, F.; Dugel, P.U.; Eandi, C.M.; Masonson, H.; Monés, J.; Pearlman, J.A.; Quaranta-El Maftouhi, M.; et al. Dual Antagonism of PDGF and VEGF in Neovascular Age-Related Macular Degeneration: A Phase IIb, Multicenter, Randomized Controlled Trial. Ophthalmology 2017, 124, 224–234. [Google Scholar] [CrossRef]
- Falk, M.K.; Singh, A.; Faber, C.; Nissen, M.H.; Hviid, T.; Sørensen, T.L. Dysregulation of CXCR3 Expression on Peripheral Blood Leukocytes in Patients With Neovascular Age-Related Macular Degeneration. Investig. Ophthalmol. Vis. Sci. 2014, 55, 4050–4056. [Google Scholar] [CrossRef]
- Palenski, T.L.; Gurel, Z.; Sorenson, C.M.; Hankenson, K.D.; Sheibani, N. Cyp1B1 Expression Promotes Angiogenesis by Suppressing NF-ΚB Activity. Am. J. Physiol. Cell Physiol. 2013, 305, C1170–C1184. [Google Scholar] [CrossRef]
- Tang, Y.; Scheef, E.A.; Wang, S.; Sorenson, C.M.; Marcus, C.B.; Jefcoate, C.R.; Sheibani, N. CYP1B1 Expression Promotes the Proangiogenic Phenotype of Endothelium through Decreased Intracellular Oxidative Stress and Thrombospondin-2 Expression. Blood 2009, 113, 744–754. [Google Scholar] [CrossRef]
- Kauppinen, A.; Paterno, J.J.; Blasiak, J.; Salminen, A.; Kaarniranta, K. Inflammation and Its Role in Age-Related Macular Degeneration. Cell. Mol. Life Sci. 2016, 73, 1765–1786. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, V.M.; Chan, C.-C. The Role of Anti-Inflammatory Agents in Age-Related Macular Degeneration (AMD) Treatment. Eye 2011, 25, 127–139. [Google Scholar] [CrossRef]
- Hageman, G.S.; Luthert, P.J.; Victor Chong, N.H.; Johnson, L.V.; Anderson, D.H.; Mullins, R.F. An Integrated Hypothesis That Considers Drusen as Biomarkers of Immune-Mediated Processes at the RPE-Bruch’s Membrane Interface in Aging and Age-Related Macular Degeneration. Prog. Retin. Eye Res. 2001, 20, 705–732. [Google Scholar] [CrossRef]
- Hong, T.; Tan, A.G.; Mitchell, P.; Wang, J.J. A Review and Meta-Analysis of the Association between C-Reactive Protein and Age-Related Macular Degeneration. Surv. Ophthalmol. 2011, 56, 184–194. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.; Myers, C.E.; Cruickshanks, K.J.; Gangnon, R.E.; Danforth, L.G.; Sivakumaran, T.A.; Iyengar, S.K.; Tsai, M.Y.; Klein, B.E.K. Markers of Inflammation, Oxidative Stress, and Endothelial Dysfunction and the 20-Year Cumulative Incidence of Early Age-Related Macular Degeneration: The Beaver Dam Eye Study. JAMA Ophthalmol. 2014, 132, 446–455. [Google Scholar] [CrossRef] [PubMed]
- Nussenblatt, R.B.; Byrnes, G.; Sen, H.N.; Yeh, S.; Faia, L.; Meyerle, C.; Wroblewski, K.; Li, Z.; Liu, B.; Chew, E.; et al. A Randomized Pilot Study of Systemic Immunosuppression in the Treatment of Age-Related Macular Degeneration with Choroidal Neovascularization. RETINA 2010, 30, 1579. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Zeng, L.; Zheng, C.; Song, B.; Li, F.; Kong, X.; Xu, K. Inflammatory Links Between High Fat Diets and Diseases. Front. Immunol. 2018, 9, 2649. [Google Scholar] [CrossRef] [PubMed]
- Franceschi, C.; Garagnani, P.; Parini, P.; Giuliani, C.; Santoro, A. Inflammaging: A New Immune–Metabolic Viewpoint for Age-Related Diseases. Nat. Rev. Endocrinol. 2018, 14, 576–590. [Google Scholar] [CrossRef]
- Boulangé, C.L.; Neves, A.L.; Chilloux, J.; Nicholson, J.K.; Dumas, M.-E. Impact of the Gut Microbiota on Inflammation, Obesity, and Metabolic Disease. Genome Med. 2016, 8, 42. [Google Scholar] [CrossRef]
- Camous, X.; Pera, A.; Solana, R.; Larbi, A. NK Cells in Healthy Aging and Age-Associated Diseases. J. Biomed. Biotechnol. 2012, 2012, 195956. [Google Scholar] [CrossRef]
- Prolo, P.; Chiappelli, F.; Angeli, A.; Dovio, A.; Perotti, P.; Pautasso, M.; Sartori, M.L.; Saba, L.; Mussino, S.; Fraccalini, T.; et al. Physiologic Modulation of Natural Killer Cell Activity as an Index of Alzheimer’s Disease Progression. Bioinformation 2007, 1, 363–366. [Google Scholar] [CrossRef]
- Cui, Y.; Wan, Q. NKT Cells in Neurological Diseases. Front. Cell. Neurosci. 2019, 13, 245. [Google Scholar] [CrossRef]
- Baudouin, C.; Peyman, G.A.; Fredj-Reygrobellet, D.; Gordon, W.C.; Lapalus, P.; Gastaud, P.; Bazan, N.G. Immunohistological Study of Subretinal Membranes in Age-Related Macular Degeneration. Jpn. J. Ophthalmol. 1992, 36, 443–451. [Google Scholar]
- Hijioka, K.; Sonoda, K.-H.; Tsutsumi-Miyahara, C.; Fujimoto, T.; Oshima, Y.; Taniguchi, M.; Ishibashi, T. Investigation of the Role of CD1d-Restricted Invariant NKT Cells in Experimental Choroidal Neovascularization. Biochem. Biophys. Res. Commun. 2008, 374, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Goverdhan, S.V.; Khakoo, S.I.; Gaston, H.; Chen, X.; Lotery, A.J. Age-Related Macular Degeneration Is Associated with the HLA-Cw*0701 Genotype and the Natural Killer Cell Receptor AA Haplotype. Investig. Ophthalmol. Vis. Sci. 2008, 49, 5077–5082. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, A.; Yamada, R.; Kochi, Y.; Sawada, T.; Okada, Y.; Matsuda, K.; Kamatani, Y.; Mori, M.; Shimane, K.; Hirabayashi, Y.; et al. Functional SNPs in CD244 Increase the Risk of Rheumatoid Arthritis in a Japanese Population. Nat. Genet. 2008, 40, 1224–1229. [Google Scholar] [CrossRef]
- Lee, K.-M.; Forman, J.P.; McNerney, M.E.; Stepp, S.; Kuppireddi, S.; Guzior, D.; Latchman, Y.E.; Sayegh, M.H.; Yagita, H.; Park, C.-K.; et al. Requirement of Homotypic NK-Cell Interactions through 2B4(CD244)/CD48 in the Generation of NK Effector Functions. Blood 2006, 107, 3181–3188. [Google Scholar] [CrossRef]
- McArdel, S.L.; Terhorst, C.; Sharpe, A.H. Roles of CD48 in Regulating Immunity and Tolerance. Clin. Immunol. 2016, 164, 10–20. [Google Scholar] [CrossRef]
- Voskoboinik, I.; Whisstock, J.C.; Trapani, J.A. Perforin and Granzymes: Function, Dysfunction and Human Pathology. Nat. Rev. Immunol. 2015, 15, 388–400. [Google Scholar] [CrossRef] [PubMed]
- Vazirinejad, R.; Ahmadi, Z.; Arababadi, M.K.; Hassanshahi, G.; Kennedy, D. The Biological Functions, Structure and Sources of CXCL10 and Its Outstanding Part in the Pathophysiology of Multiple Sclerosis. NIM 2014, 21, 322–330. [Google Scholar] [CrossRef]
- Picard, C.; Fieschi, C.; Altare, F.; Al-Jumaah, S.; Al-Hajjar, S.; Feinberg, J.; Dupuis, S.; Soudais, C.; Al-Mohsen, I.Z.; Génin, E.; et al. Inherited Interleukin-12 Deficiency: IL12B Genotype and Clinical Phenotype of 13 Patients from Six Kindreds. Am. J. Hum. Genet. 2002, 70, 336–348. [Google Scholar] [CrossRef]
- Zhang, J.; Zhou, Y. Identification of Key Genes and Pathways Associated with Age-Related Macular Degeneration. J. Ophthalmol. 2020, 2020, e2714746. [Google Scholar] [CrossRef]
- Chen, J.; Wang, W.; Li, Q. Increased Th1/Th17 Responses Contribute to Low-Grade Inflammation in Age-Related Macular Degeneration. CPB 2017, 44, 357–367. [Google Scholar] [CrossRef]
- Yan, Y.; Chen, R.; Wang, X.; Hu, K.; Huang, L.; Lu, M.; Hu, Q. CCL19 and CCR7 Expression, Signaling Pathways, and Adjuvant Functions in Viral Infection and Prevention. Front. Cell Dev. Biol. 2019, 7, 212. [Google Scholar] [CrossRef] [PubMed]
- Rutar, M.; Natoli, R.; Chia, R.; Valter, K.; Provis, J.M. Chemokine-Mediated Inflammation in the Degenerating Retina Is Coordinated by Müller Cells, Activated Microglia, and Retinal Pigment Epithelium. J. Neuroinflamm. 2015, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Faber, C.; Jehs, T.; Juel, H.B.; Singh, A.; Falk, M.K.; Sørensen, T.L.; Nissen, M.H. Early and Exudative Age-Related Macular Degeneration Is Associated with Increased Plasma Levels of Soluble TNF Receptor II. Acta Ophthalmol. 2015, 93, 242–247. [Google Scholar] [CrossRef]
- Wang, H.; Han, X.; Wittchen, E.S.; Hartnett, M.E. TNF-α Mediates Choroidal Neovascularization by Upregulating VEGF Expression in RPE through ROS-Dependent β-Catenin Activation. Mol. Vis. 2016, 22, 116–128. [Google Scholar] [PubMed]
- Shi, X.; Semkova, I.; Müther, P.S.; Dell, S.; Kociok, N.; Joussen, A.M. Inhibition of TNF-Alpha Reduces Laser-Induced Choroidal Neovascularization. Exp. Eye Res. 2006, 83, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Fritsche, L.G.; Chen, W.; Schu, M.; Yaspan, B.L.; Yu, Y.; Thorleifsson, G.; Zack, D.J.; Arakawa, S.; Cipriani, V.; Ripke, S.; et al. Seven New Loci Associated with Age-Related Macular Degeneration. Nat. Genet. 2013, 45, 433–439. [Google Scholar] [CrossRef]
- Markomichelakis, N.N.; Theodossiadis, P.G.; Sfikakis, P.P. Regression of Neovascular Age-Related Macular Degeneration Following Infliximab Therapy. Am. J. Ophthalmol. 2005, 139, 537–540. [Google Scholar] [CrossRef]
- Fernández-Vega, B.; Fernández-Vega, Á.; Rangel, C.M.; Nicieza, J.; Villota-Deleu, E.; Vega, J.A.; Sanchez-Avila, R.M. Blockade of Tumor Necrosis Factor-Alpha: A Role for Adalimumab in Neovascular Age-Related Macular Degeneration Refractory to Anti-Angiogenesis Therapy? Case Rep. Ophthalmol. 2016, 7, 154–162. [Google Scholar] [CrossRef]
- Toris, C.; Gulati, V. The Biology, Pathology and Therapeutic Use of Prostaglandins in the Eye. Clin. Lipidol. 2011, 6, 577–591. [Google Scholar] [CrossRef]
- Kim, S.J.; Toma, H.S. Inhibition of Choroidal Neovascularization by Intravitreal Ketorolac. Arch. Ophthalmol. 2010, 128, 596–600. [Google Scholar] [CrossRef]
- Khandhadia, S.; Cipriani, V.; Yates, J.R.W.; Lotery, A.J. Age-Related Macular Degeneration and the Complement System. Immunobiology 2012, 217, 127–146. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.O.; Ritter, R.; Abel, K.J.; Manning, A.; Panhuysen, C.; Farrer, L.A. Complement Factor H Polymorphism and Age-Related Macular Degeneration. Science 2005, 308, 421–424. [Google Scholar] [CrossRef] [PubMed]
- Klein, R.J.; Zeiss, C.; Chew, E.Y.; Tsai, J.-Y.; Sackler, R.S.; Haynes, C.; Henning, A.K.; SanGiovanni, J.P.; Mane, S.M.; Mayne, S.T.; et al. Complement Factor H Polymorphism in Age-Related Macular Degeneration. Science 2005, 308, 385–389. [Google Scholar] [CrossRef]
- Chen, M.; Forrester, J.V.; Xu, H. Synthesis of Complement Factor H by Retinal Pigment Epithelial Cells Is Down-Regulated by Oxidized Photoreceptor Outer Segments. Exp. Eye Res. 2007, 84, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Chirco, K.R.; Sohn, E.H.; Stone, E.M.; Tucker, B.A.; Mullins, R.F. Structural and Molecular Changes in the Aging Choroid: Implications for Age-Related Macular Degeneration. Eye 2017, 31, 10–25. [Google Scholar] [CrossRef]
- Clark, S.J.; Bishop, P.N.; Day, A.J. Complement Factor H and Age-Related Macular Degeneration: The Role of Glycosaminoglycan Recognition in Disease Pathology. Biochem. Soc. Trans. 2010, 38, 1342–1348. [Google Scholar] [CrossRef]
- Perkins, S.; Fung, K.W.; Khan, S. Molecular Interactions between Complement Factor H and Its Heparin and Heparan Sulfate Ligands. Front. Immunol. 2014, 5, 126. [Google Scholar] [CrossRef]
- Gold, B.; Merriam, J.E.; Zernant, J.; Hancox, L.S.; Taiber, A.J.; Gehrs, K.; Cramer, K.; Neel, J.; Bergeron, J.; Barile, G.R.; et al. Variation in Factor B (BF) and Complement Component 2 (C2) Genes Is Associated with Age-Related Macular Degeneration. Nat. Genet. 2006, 38, 458–462. [Google Scholar] [CrossRef]
- Seddon, J.M.; Reynolds, R.; Maller, J.; Fagerness, J.A.; Daly, M.J.; Rosner, B. Prediction Model for Prevalence and Incidence of Advanced Age-Related Macular Degeneration Based on Genetic, Demographic, and Environmental Variables. Investig. Ophthalmol. Vis. Sci. 2009, 50, 2044–2053. [Google Scholar] [CrossRef]
- Seth, A.; Cui, J.; To, E.; Kwee, M.; Matsubara, J. Complement-Associated Deposits in the Human Retina. Investig. Ophthalmol. Vis. Sci. 2008, 49, 743–750. [Google Scholar] [CrossRef]
- Mullins, R.F.; Schoo, D.P.; Sohn, E.H.; Flamme-Wiese, M.J.; Workamelahu, G.; Johnston, R.M.; Wang, K.; Tucker, B.A.; Stone, E.M. The Membrane Attack Complex in Aging Human Choriocapillaris: Relationship to Macular Degeneration and Choroidal Thinning. Am. J. Pathol. 2014, 184, 3142–3153. [Google Scholar] [CrossRef] [PubMed]
- Chirco, K.R.; Tucker, B.A.; Stone, E.M.; Mullins, R.F. Selective Accumulation of the Complement Membrane Attack Complex in Aging Choriocapillaris. Exp. Eye Res. 2016, 146, 393–397. [Google Scholar] [CrossRef] [PubMed]
- Zeng, S.; Whitmore, S.S.; Sohn, E.H.; Riker, M.J.; Wiley, L.A.; Scheetz, T.E.; Stone, E.M.; Tucker, B.A.; Mullins, R.F. Molecular Response of Chorioretinal Endothelial Cells to Complement Injury: Implications for Macular Degeneration. J. Pathol. 2016, 238, 446–456. [Google Scholar] [CrossRef] [PubMed]
- Doerner, S.K.; Reis, E.S.; Leung, E.S.; Ko, J.S.; Heaney, J.D.; Berger, N.A.; Lambris, J.D.; Nadeau, J.H. High-Fat Diet-Induced Complement Activation Mediates Intestinal Inflammation and Neoplasia, Independent of Obesity. Mol. Cancer Res. 2016, 14, 953–965. [Google Scholar] [CrossRef]
- Toomey, C.B.; Kelly, U.; Saban, D.R.; Bowes Rickman, C. Regulation of Age-Related Macular Degeneration-like Pathology by Complement Factor H. Proc. Natl. Acad. Sci. USA 2015, 112, E3040–E3049. [Google Scholar] [CrossRef]
- Liao, D.S.; Grossi, F.V.; El Mehdi, D.; Gerber, M.R.; Brown, D.M.; Heier, J.S.; Wykoff, C.C.; Singerman, L.J.; Abraham, P.; Grassmann, F.; et al. Complement C3 Inhibitor Pegcetacoplan for Geographic Atrophy Secondary to Age-Related Macular Degeneration: A Randomized Phase 2 Trial. Ophthalmology 2020, 127, 186–195. [Google Scholar] [CrossRef]
- Wu, J.; Sun, X. Complement System and Age-Related Macular Degeneration: Drugs and Challenges. Drug Des. Dev. Ther. 2019, 13, 2413–2425. [Google Scholar] [CrossRef]
- Klaver, C.C.; Kliffen, M.; van Duijn, C.M.; Hofman, A.; Cruts, M.; Grobbee, D.E.; van Broeckhoven, C.; de Jong, P.T. Genetic Association of Apolipoprotein E with Age-Related Macular Degeneration. Am. J. Hum. Genet. 1998, 63, 200–206. [Google Scholar] [CrossRef]
- Hu, M.L.; Quinn, J.; Xue, K. Interactions between Apolipoprotein E Metabolism and Retinal Inflammation in Age-Related Macular Degeneration. Life 2021, 11, 635. [Google Scholar] [CrossRef]
- Li, C.-M.; Clark, M.E.; Chimento, M.F.; Curcio, C.A. Apolipoprotein Localization in Isolated Drusen and Retinal Apolipoprotein Gene Expression. Investig. Ophthalmol. Vis. Sci. 2006, 47, 3119–3128. [Google Scholar] [CrossRef]
- Yang, P.; Skiba, N.P.; Tewkesbury, G.M.; Treboschi, V.M.; Baciu, P.; Jaffe, G.J. Complement-Mediated Regulation of Apolipoprotein E in Cultured Human RPE Cells. Investig. Ophthalmol. Vis. Sci. 2017, 58, 3073–3085. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Gallo, N.B.; Hancox, L.S.; Miller, N.J.; Radeke, C.M.; Maloney, M.A.; Cooper, J.B.; Hageman, G.S.; Anderson, D.H.; Johnson, L.V.; et al. Systems-Level Analysis of Age-Related Macular Degeneration Reveals Global Biomarkers and Phenotype-Specific Functional Networks. Genome Med. 2012, 4, 16. [Google Scholar] [CrossRef]
- Yu, Y.; Bhangale, T.R.; Fagerness, J.; Ripke, S.; Thorleifsson, G.; Tan, P.L.; Souied, E.H.; Richardson, A.J.; Merriam, J.E.; Buitendijk, G.H.S.; et al. Common Variants near FRK/COL10A1 and VEGFA Are Associated with Advanced Age-Related Macular Degeneration. Hum. Mol. Genet. 2011, 20, 3699–3709. [Google Scholar] [CrossRef] [PubMed]
- Boutin, T.S.; Charteris, D.G.; Chandra, A.; Campbell, S.; Hayward, C.; Campbell, A.; UK Biobank Eye & Vision Consortium; Nandakumar, P.; Hinds, D.; 23andMe Research Team; et al. Insights into the Genetic Basis of Retinal Detachment. Hum. Mol. Genet. 2020, 29, 689–702. [Google Scholar] [CrossRef] [PubMed]
- Stone, E.M.; Braun, T.A.; Russell, S.R.; Kuehn, M.H.; Lotery, A.J.; Moore, P.A.; Eastman, C.G.; Casavant, T.L.; Sheffield, V.C. Missense Variations in the Fibulin 5 Gene and Age-Related Macular Degeneration. N. Engl. J. Med. 2004, 351, 346–353. [Google Scholar] [CrossRef]
- Rojo Arias, J.E.; Jászai, J. Gene Expression Profile of the Murine Ischemic Retina and Its Response to Aflibercept (VEGF-Trap). Sci. Rep. 2021, 11, 15313. [Google Scholar] [CrossRef]
- Adams, J.N.; Raffield, L.M.; Martelle, S.E.; Freedman, B.I.; Langefeld, C.D.; Carr, J.J.; Cox, A.J.; Bowden, D.W. Genetic Analysis of Advanced Glycation End Products in the DHS MIND Study. Gene 2016, 584, 173–179. [Google Scholar] [CrossRef][Green Version]
- Ishibashi, T.; Murata, T.; Hangai, M.; Nagai, R.; Horiuchi, S.; Lopez, P.F.; Hinton, D.R.; Ryan, S.J. Advanced Glycation End Products in Age-Related Macular Degeneration. Arch. Ophthalmol. 1998, 116, 1629–1632. [Google Scholar] [CrossRef]
- Fernandez-Godino, R.; Pierce, E.A.; Garland, D.L. Extracellular Matrix Alterations and Deposit Formation in AMD. Adv. Exp. Med. Biol. 2016, 854, 53–58. [Google Scholar] [CrossRef]
- Butler, J.M.; Supharattanasitthi, W.; Yang, Y.C.; Paraoan, L. RNA-Seq Analysis of Ageing Human Retinal Pigment Epithelium: Unexpected up-Regulation of Visual Cycle Gene Transcription. J. Cell. Mol. Med. 2021, 25, 5572–5585. [Google Scholar] [CrossRef]
- Gollapalli, D.R.; Rando, R.R. The Specific Binding of Retinoic Acid to RPE65 and Approaches to the Treatment of Macular Degeneration. Proc. Natl. Acad. Sci. USA 2004, 101, 10030–10035. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Bai, Y.; Huang, L.; Qi, Y.; Zhang, Q.; Li, S.; Wu, Y.; Li, X. Protective Effect of Autophagy on Human Retinal Pigment Epithelial Cells against Lipofuscin Fluorophore A2E: Implications for Age-Related Macular Degeneration. Cell Death Dis. 2015, 6, e1972. [Google Scholar] [CrossRef] [PubMed]
- Speakman, J.R. Use of High-Fat Diets to Study Rodent Obesity as a Model of Human Obesity. Int. J. Obes. 2019, 43, 1491–1492. [Google Scholar] [CrossRef] [PubMed]
- Hintze, K.J.; Benninghoff, A.D.; Cho, C.E.; Ward, R.E. Modeling the Western Diet for Preclinical Investigations. Adv. Nutr. 2018, 9, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Hintze, K.J.; Benninghoff, A.D.; Ward, R.E. Formulation of the Total Western Diet (TWD) as a Basal Diet for Rodent Cancer Studies. J. Agric. Food Chem. 2012, 60, 6736–6742. [Google Scholar] [CrossRef]
- Keeling, E.; Lynn, S.A.; Koh, Y.M.; Scott, J.A.; Kendall, A.; Gatherer, M.; Page, A.; Cagampang, F.R.; Lotery, A.J.; Ratnayaka, J.A. A High Fat “Western-Style” Diet Induces AMD-Like Features in Wildtype Mice. Mol. Nutr. Food Res. 2022, 66, e2100823. [Google Scholar] [CrossRef]
- Dighe, S.; Zhao, J.; Steffen, L.; Mares, J.A.; Meuer, S.M.; Klein, B.E.K.; Klein, R.; Millen, A.E. Diet Patterns and the Incidence of Age-Related Macular Degeneration in the Atherosclerosis Risk in Communities (ARIC) Study. Br. J. Ophthalmol. 2020, 104, 1070–1076. [Google Scholar] [CrossRef]
- Wang, L.; Clark, M.E.; Crossman, D.K.; Kojima, K.; Messinger, J.D.; Mobley, J.A.; Curcio, C.A. Abundant Lipid and Protein Components of Drusen. PLoS ONE 2010, 5, e10329. [Google Scholar] [CrossRef]
- Sasaki, M.; Kawasaki, R.; Rogers, S.; Man, R.E.K.; Itakura, K.; Xie, J.; Flood, V.; Tsubota, K.; Lamoureux, E.; Wang, J.J. The Associations of Dietary Intake of Polyunsaturated Fatty Acids With Diabetic Retinopathy in Well-Controlled Diabetes. Investig. Ophthalmol. Vis. Sci. 2015, 56, 7473–7479. [Google Scholar] [CrossRef]
- Hu, P.; Uddin, M.N.; Neuringer, M.; Renner, L.; Stoddard, J.; Boulton, M.E.; McGill, T.J.; Chan-Ling, T.; Grant, M. Evidence of Diabetic Retinopathy in a Western (High Fat) Diet-Induced Non-Human Primate Model of Type 2 Diabetes (T2D). Investig. Ophthalmol. Vis. Sci. 2018, 59, 3589. [Google Scholar]
- Reynolds, R.; Rosner, B.; Seddon, J.M. Serum Lipid Biomarkers and Hepatic Lipase Gene Associations with Age-Related Macular Degeneration. Ophthalmology 2010, 117, 1989–1995. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.K.M.; Simpson, J.A.; Richardson, A.J.; English, D.R.; Aung, K.Z.; Makeyeva, G.A.; Guymer, R.H.; Giles, G.G.; Hopper, J.; Robman, L.D.; et al. Apolipoprotein E Gene Associations in Age-Related Macular Degeneration: The Melbourne Collaborative Cohort Study. Am. J. Epidemiol. 2012, 175, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Hodge, W.G.; Schachter, H.M.; Barnes, D.; Pan, Y.; Lowcock, E.C.; Zhang, L.; Sampson, M.; Morrison, A.; Tran, K.; Miguelez, M.; et al. Efficacy of Omega-3 Fatty Acids in Preventing Age-Related Macular Degeneration: A Systematic Review. Ophthalmology 2006, 113, 1165–1172; quiz 1172–1173, 1178. [Google Scholar] [CrossRef] [PubMed]
- Chong, E.W.-T.; Robman, L.D.; Simpson, J.A.; Hodge, A.M.; Aung, K.Z.; Dolphin, T.K.; English, D.R.; Giles, G.G.; Guymer, R.H. Fat Consumption and Its Association with Age-Related Macular Degeneration. Arch. Ophthalmol. 2009, 127, 674–680. [Google Scholar] [CrossRef] [PubMed]
- Agrón, E.; Mares, J.; Clemons, T.E.; Swaroop, A.; Chew, E.Y.; Keenan, T.D.L.; AREDS and AREDS2 Research Groups. Dietary Nutrient Intake and Progression to Late Age-Related Macular Degeneration in the Age-Related Eye Disease Studies 1 and 2. Ophthalmology 2021, 128, 425–442. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-Y.; Tie, L.-J.; Wu, S.-S.; Lv, P.-L.; Huang, H.-W.; Wang, W.-Q.; Wang, H.; Ma, L. Overweight, Obesity, and Risk of Age-Related Macular Degeneration. Investig. Ophthalmol. Vis. Sci. 2016, 57, 1276–1283. [Google Scholar] [CrossRef]
- Age-Related Eye Disease Study Research Group. A Randomized, Placebo-Controlled, Clinical Trial of High-Dose Supplementation with Vitamins C and E, Beta Carotene, and Zinc for Age-Related Macular Degeneration and Vision Loss: AREDS Report No. 8. Arch. Ophthalmol. 2001, 119, 1417–1436. [Google Scholar] [CrossRef]
- Age-Related Eye Disease Study 2 Research Group. Lutein + Zeaxanthin and Omega-3 Fatty Acids for Age-Related Macular Degeneration: The Age-Related Eye Disease Study 2 (AREDS2) Randomized Clinical Trial. JAMA 2013, 309, 2005–2015. [Google Scholar] [CrossRef]

| Gene | LogFC | Adjusted p-Value | Protein |
|---|---|---|---|
| Cd244a | 6.27 | 1.62 × 10−3 | Natural Killer Cell Receptor 2B4 |
| Ripply1 | 5.52 | 9.98 × 10−3 | Ripply Transcriptional Repressor 1 |
| Lilrb4a | 5.28 | 4.81 × 10−2 | Leukocyte Immunoglobulin-Like Receptor Subfamily B member 4 |
| Fcer1a | 5.26 | 2.85 × 10−2 | Fc Epsilon Receptor Ia |
| Dnajc22 | 5.24 | 1.44 × 10−2 | DnaJ Heat Shock Protein Family (Hsp40) Member C22 |
| Alkal2 | 5.11 | 3.11 × 10−2 | ALK And LTK Ligand 2 |
| Ncr1 | 5.04 | 1.16 × 10−2 | Natural Cytotoxicity Triggering Receptor 1 |
| Ccl19 | 4.94 | 7.60 × 10−3 | C-C Motif Chemokine Ligand 19 |
| Slc38a11 | 4.81 | 3.85 × 10−2 | Solute Carrier Family 38 Member 11 |
| Ces2e | 4.71 | 4.62 × 10−2 | Pyrethroid Hydrolase Ces2e |
| Lpar3 | 4.66 | 1.73 × 10−2 | Lysophosphatidic Acid Receptor 3 |
| Sele | 4.62 | 4.49 × 10−2 | Selectin E |
| Sirpb1a | 4.61 | 9.02 × 10−3 | Signal-Regulatory Protein Beta 1A |
| Efhb | 4.57 | 1.33 × 10−2 | EF-Hand Domain-Containing Family Member B |
| Pgpep1l | 4.56 | 2.15 × 10−2 | Pyroglutamyl-Peptidase 1-Like Protein |
| Tnfrsf13b | 4.54 | 2.94 × 10−2 | Tumor Necrosis Factor Receptor Superfamily Member 13B |
| Il12b | 4.49 | 2.65 × 10−2 | Interleukin-12 Subunit Beta |
| Tmem232 | 4.46 | 2.53 × 10−2 | Transmembrane Protein 232 |
| Trbc1 | 4.41 | 1.51 × 10−2 | T Cell Receptor Beta Constant 1 |
| Slc4a1 | 4.34 | 3.36 × 10−2 | Solute Carrier Family 4 Member 1 |
| Olfr574 | 4.34 | 1.60 × 10−2 | Olfactory Receptor Family 51 Subfamily T Member 1 |
| Xlr | 4.28 | 5.51 × 10−3 | X-Linked Lymphocyte-Regulated Protein PM1 |
| Gpr141 | 4.25 | 4.73 × 10−2 | G Protein-Coupled Receptor 141 |
| Cnr2 | 4.25 | 3.48 × 10−2 | Cannabinoid Receptor 2 |
| Mael | 4.23 | 4.08 × 10−2 | Maelstrom Spermatogenic Transposon Silencer |
| Lao1 | 4.22 | 4.06 × 10−2 | Amine Oxidase |
| Mcoln2 | 4.16 | 1.72 × 10−2 | Mucolipin TRP Cation Channel 2 |
| Ccl22 | 4.16 | 3.77 × 10−2 | C-C Motif Chemokine Ligand 22 |
| Rnase1 | 4.11 | 2.66 × 10−2 | Ribonuclease A Family Member 1, Pancreatic |
| Ptgs2os | 4.11 | 3.37 × 10−2 | Prostaglandin-Endoperoxide Synthase 2, Opposite Strand |
| Biological Processes | p-Value | Adjusted p-Value | Gene Ratio | Genes |
|---|---|---|---|---|
| Melanocyte differentiation | 1.79 × 10−7 | 9.49 × 10−5 | 0.35 | Edn3, Ednrb, Mitf, Mlph, Rab27a, Slc24a5, Sox10, and Tyrp1 |
| Angiogenesis | 2.08 × 10−13 | 5.51 × 10−10 | 0.13 | Ackr3, Angpt1, Angpt2, Angptl2, Apold1, Calcrl, Cfh, Clic4, Col18a1, Col8a1, Col8a2, Cxcr3, Cyp1b1, Ecscr, Ephb4, Esm1, Fap, Fzd8, Htatip2, Mcam, Nrp2, Pik3r6, Plxnd1, Ptgs2, Ptprb, Rapgef3, Rhoj, Rspo3, Tbx4, Tek, Tgfbr3, Tie1, Tnfaip2, and Vegfc |
| Cell surface receptor signaling pathway | 1.50 × 10−9 | 1.50 × 10−6 | 0.13 | Adgra3, Adgrf5, Adgrg6, Calcrl, Cd22, Cd86, Cxcr3, Cysltr1, Edn3, Fcer1a, Fzd2, Fzd4, Fzd7, Fzd8, Gpr157, Il12b, Itgal, Itpkb, Npr1, Osmr, Ostn, Pth1r, Tnfrsf1b, and Tshr |
| Positive regulation of angiogenesis | 5.68 × 10−7 | 2.15 × 10−4 | 0.12 | Angpt2, Brca1, Chil1, Cxcr3, Cybb, Cyp1b1, Cysltr1, Ets1, Itgb3, Itgb8, Pik3r6, Ptgis, Rapgef3, Tek, Tgfbr2, Thbs1, Tie1, and Vegfc |
| Inflammatory response | 1.14 × 10−9 | 1.50 × 10−6 | 0.10 | Agtr1a, Axl, Bmp2, Bmp6, Ccl19, Ccl22, Ccl4, Cfh, Chil1, Cnr2, Csf1r, Cxcl10, Cxcr3, Cyba, Cybb, Cysltr1, Gbp5, Il25, Lilrb4a, Lipa, Ly86, P2rx7, Pla2g2e, Prkcq, Ptgs1, Ptgs2, Rarres2, Sele, Selp, Slc11a1, Thbs1, Themis2, Tlr13, and Tnfrsf1b |
| Response to bacterium | 1.29 × 10−6 | 3.56 × 10−4 | 0.09 | Adamts9, Bank1, Bmp2, Cxcl10, Fkbp5, Gbp5, Gpc3, Gzma, Ifi211, Ifit3, Iigp1, Lrat, Ms4a1, Myo1f, Naaladl2, Nexn, Ociad2, P2rx7, Rnase1, Serpina3f, Serpinb9, Slc11a1, Tgtp1, and Trf |
| Immune response | 8.75 × 10−7 | 2.90 × 10−4 | 0.08 | Ackr3, Azgp1, B2m, Bmp6, Ccl19, Ccl22, Ccl4, Cd28, Cd86, Cfh, Colec12, Ctsk, Ctss, Cxcl10, Cxcl12, Cxcr3, Endou, Enpp2, H2-Ab1, H2-Eb1, H2-M3, Itgb8, Ly86, Serpinb9, Tgfbr3, Tgtp1, Tnfrsf1b, Tnfsf10, Tnfsf14, Vav1 |
| Cell adhesion | 1.48 × 10−8 | 9.80 × 10−6 | 0.08 | Ackr3, Azgp1, Cd22, Cd33, Cd84, Cldn1, Cldn2, Cntnap4, Col12a1, Col18a1, Col8a1, Col8a2, Cyp1b1, Dpp4, Ephb4, Fap, Fblim1, Fbln5, Gpnmb, Hpse, Icam2, Itga9, Itgal, Itgb3, Itgb8, Jcad, Kitl, Lgals3bp, Ly9, Mcam, Mybpc2, Nid2, Pcdh12, Plpp3, Sele, Sell, Selp, Siglecf, Spp1, Svep1, Thbs1, Vcam1, and Vwf |
| Positive regulation of cell population proliferation | 1.98 × 10−7 | 9.49 × 10−5 | 0.07 | Adora2b, Agtr1a, Aldh1a2, Bambi, Calcrl, Cd38, Cdk2, Clec7a, Col18a1, Csf1r, Cxcl10, Cxcl12, Cxcr3, Dpp4, Edn3, Ednra, Ednrb, Enpp2, Esm1, Ets1, Fgf7, Gab2, Gcnt2, Gli1, Kitl, Lrp5, Nog, Ntn1, Osmr, Pax3, Pdgfc, Pdgfd, Ptgs2, Pth1r, S100b, Stox1, Tgfbr3, Thbs1, Tshr, and Vegfc |
| Multicellular organism development | 1.21 × 10−6 | 3.56 × 10−4 | 0.06 | Ackr3, Angpt1, Angpt2, Ano1, Axl, B2m, Bmp2, Bmp6, Cdh19, Csf1r, Ecscr, Eda2r, Ephb4, Eya1, Eya2, Fhl1, Foxd1, Foxd3, Fst, Fzd2, Fzd4, Fzd7, Fzd8, Gli1, Gpr157, Gsx2, Htatip2, Krt8, Lbx1, Lrp5, Mael, Mertk, Met, Mgp, Mitf, Nog, Nrp2, Ostn, Pax3, Pdgfc, Pdgfd, Pitx2, Plpp3, Plxnd1, Ripply1, Sema3b, Sema3c, Sema6d, Serpine2, Sfrp5, Shisa2, Smoc1, Sox6, Stpg4, Tbx4, Tek, Tie1, Tmem88, Tnfaip2, Vegfc, and Wipf3 |
| Molecular Pathways | p-Value | Adjusted p-Value | Gene Ratio | Genes |
|---|---|---|---|---|
| Hemoglobin binding | 2.43 × 10−5 | 3.96 × 10−3 | 0.57 | Hbb-bs, Hbb-bt, Lrp2, Slc4a1 |
| Sialic acid binding | 2.30 × 10−5 | 3.96 × 10−3 | 0.38 | Cd22, Cd33, Sele, Selp, Siglecf |
| Extracellular matrix binding | 2.09 × 10−5 | 3.96 × 10−3 | 0.23 | Adamts15, Clec14a, Dcn, Itgb3, Smoc1, Spp1, and Thbs1 |
| Fibronectin binding | 2.64 × 10−5 | 3.96 × 10−3 | 0.23 | Ctsk, Ctss, Fbln1, Igfbp3, Igfbp5, Itgb3, Thbs1 |
| Integrin binding | 2.81 × 10−7 | 2.12 × 10−4 | 0.13 | Cxcl12, Esm1, Fap, Fbln1, Fbln5, Fbn1, Gpnmb, Icam2, Itgb3, Itgb8, Lcp1, Lilrb4a, Plpp3, Spp1, Thbs1, Vcam1, and Vwf |
| Extracellular matrix structural constituent | 5.40 × 10−5 | 4.08 × 10−3 | 0.11 | Col10a1, Col18a1, Col8a1, Col8a2, Col9a3, Fbln1, Fbn1, Fbn2, Matn2, Nid2, Ntn1, Thbs1, Vwf |
| Signaling receptor activity | 4.79 × 10−6 | 1.21 × 10−3 | 0.10 | Cd48, Colec12, Cxcr3, Eda2r, Fzd4, Itgb8, Klrk1, Lrp2, Mrc2, P2rx7, Paqr6, Stra6, Tek, Tgfbr2, Tlr13, Tnfrsf19, Trem2, and Tshr |
| Carbohydrate binding | 2.04 × 10−6 | 7.73 × 10−4 | 0.10 | Agl, C4b, Cd22, Cd33, Clec12a, Clec14a, Clec1a, Clec4a2, Clec4d, Clec4n, Colec12, Galm, Galnt6, Klrk1, Man2a1, Mrc2, Sele, Sell, Selp, Siglecf |
| Heparin binding | 4.12 × 10−5 | 3.96 × 10−3 | 0.10 | Adamts1, Adamts15, Apoe, Cfh, Cxcl10, Fbn1, Fgf7, Gpnmb, Nrp2, Rspo3, Selp, Serpine2, Smoc1, Tgfbr3, and Thbs1 |
| Protein homodimerization activity | 2.24 × 10−5 | 3.96 × 10−3 | 0.06 | Ano1, Ano6, Apoe, Atp2a1, B2m, Cat, Csf1r, Dpp4, Dpyd, Fap, Fbln5, Fzd4, Galm, Gbp3, Gbp5, Gzma, H2-M3, Hnf4a, Impa2, Man2a1, Mgll, Nog, Npr3, Pdgfc, Pitx2, Pon1, Pon3, Ptgs2, Pth1r, Rdh5, S100b, Slc11a1, Slc4a1, St6gal1, Tpd52l1, Trim21, Trim30d, Tyr, Tyrobp, and Tyrp1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiao, J.; Xie, B.; Dao, D.; Spedale, M.; D’Souza, M.; Theriault, B.; Hariprasad, S.M.; Sulakhe, D.; Chang, E.B.; Skondra, D. High-Fat Diet Alters the Retinal Pigment Epithelium and Choroidal Transcriptome in the Absence of Gut Microbiota. Cells 2022, 11, 2076. https://doi.org/10.3390/cells11132076
Xiao J, Xie B, Dao D, Spedale M, D’Souza M, Theriault B, Hariprasad SM, Sulakhe D, Chang EB, Skondra D. High-Fat Diet Alters the Retinal Pigment Epithelium and Choroidal Transcriptome in the Absence of Gut Microbiota. Cells. 2022; 11(13):2076. https://doi.org/10.3390/cells11132076
Chicago/Turabian StyleXiao, Jason, Bingqing Xie, David Dao, Melanie Spedale, Mark D’Souza, Betty Theriault, Seenu M. Hariprasad, Dinanath Sulakhe, Eugene B. Chang, and Dimitra Skondra. 2022. "High-Fat Diet Alters the Retinal Pigment Epithelium and Choroidal Transcriptome in the Absence of Gut Microbiota" Cells 11, no. 13: 2076. https://doi.org/10.3390/cells11132076
APA StyleXiao, J., Xie, B., Dao, D., Spedale, M., D’Souza, M., Theriault, B., Hariprasad, S. M., Sulakhe, D., Chang, E. B., & Skondra, D. (2022). High-Fat Diet Alters the Retinal Pigment Epithelium and Choroidal Transcriptome in the Absence of Gut Microbiota. Cells, 11(13), 2076. https://doi.org/10.3390/cells11132076







