A miR-9-5p/FOXO1/CPEB3 Feed-Forward Loop Drives the Progression of Hepatocellular Carcinoma
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Source
2.2. Differentially Expressed Genes
2.3. Enrichment Analysis
2.4. Survival Analysis
2.5. Regulatory Networks and Hub Analysis
2.6. Cell Lines and Cell Culture
2.7. Transfection
2.8. Invasion Assay
2.9. Cell Proliferation Assay
2.10. Cell Apoptosis Assay
2.11. Animals
2.12. Statistical Analysis
3. Results
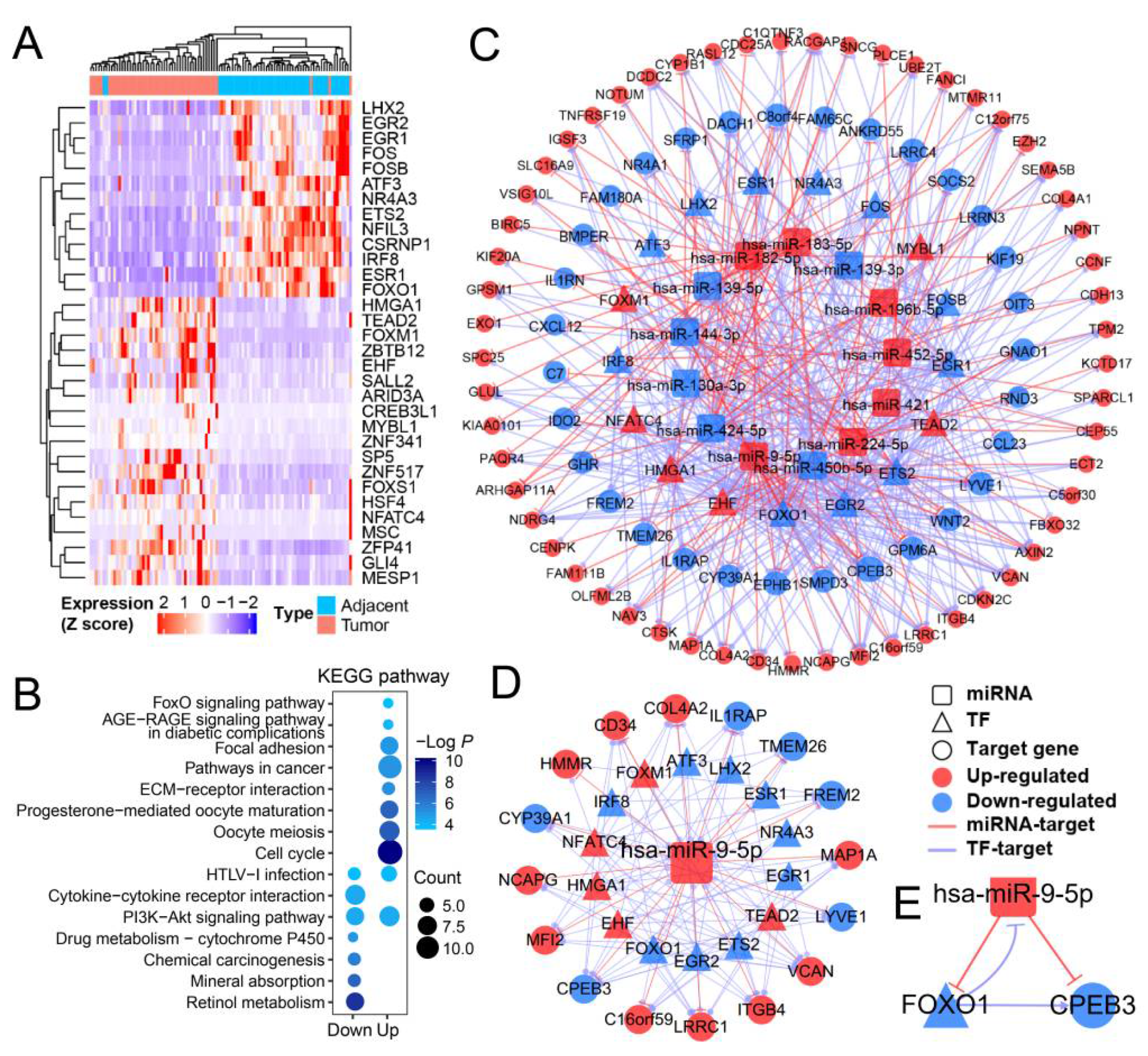
3.1. The Gene Function Analysis and Regulatory Network Analyses Revealed miR-9-5p/FOXO1/CPEB3 FFL in HCC
3.2. miR-9-5p Promotes Tumor Proliferation, while FOXO1 and CPEB3 Play Tumor-Suppressive Roles in HCC
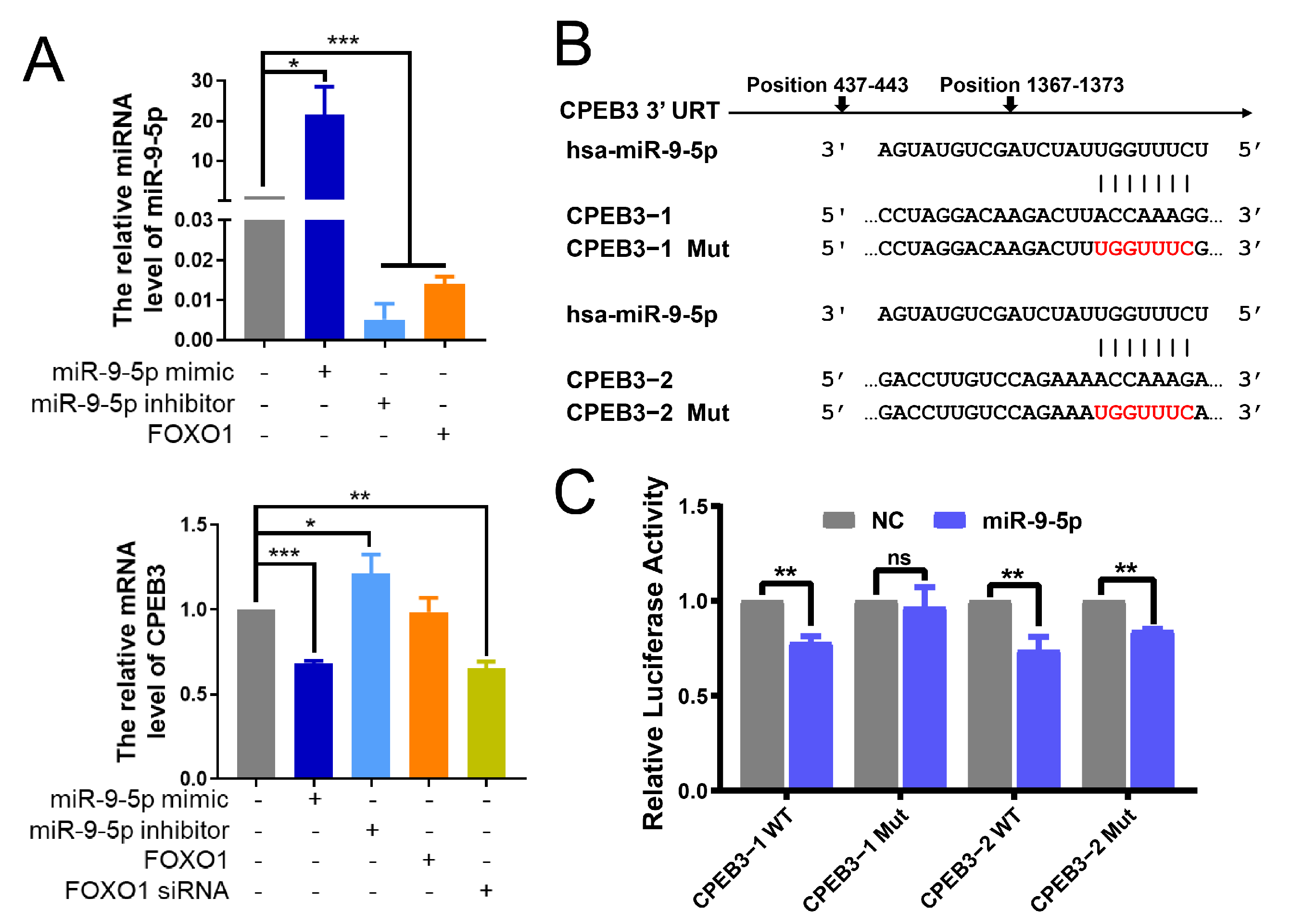
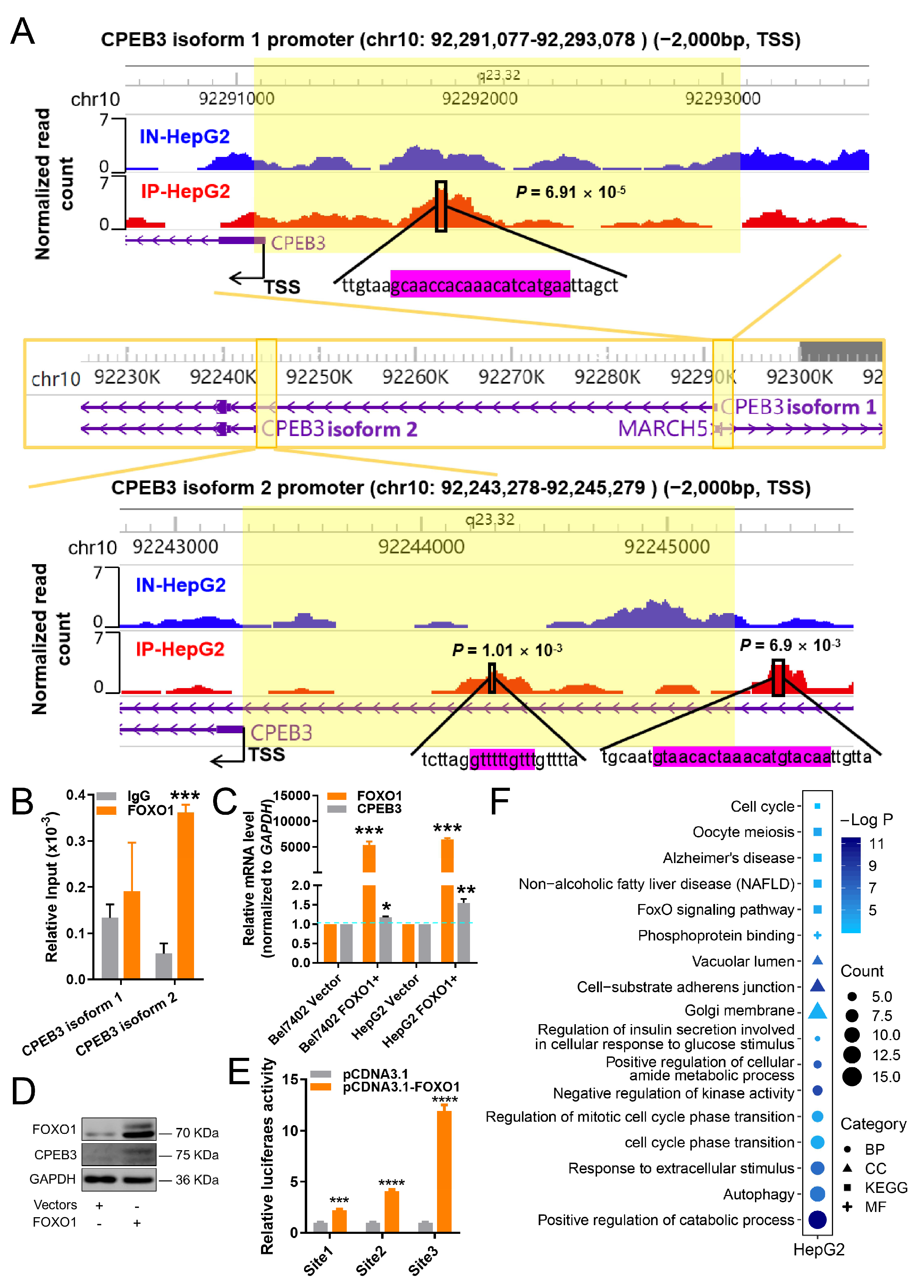
3.3. The Existence of FFL among miR-9-5p, FOXO1 and CPEB3 in HCC
3.4. The miR-9-5p/FOXO1/CPEB3 FFL May Promote Progression of HCC In Vivo
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rupaimoole, R.; Slack, F.J. MicroRNA Therapeutics: Towards a New Era for the Management of Cancer and Other Diseases. Nat. Rev. Drug Discov. 2017, 16, 203–222. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.-S.; Lim, J.W.C.; Richards, L.J.; Bunt, J. The Convergent Roles of the Nuclear Factor I Transcription Factors in Development and Cancer. Cancer Lett. 2017, 410, 124–138. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.-M.; Kuang, S.; Xiong, X.; Gao, T.; Liu, C.; Guo, A.-Y. Transcription Factor and MicroRNA Co-Regulatory Loops: Important Regulatory Motifs in Biological Processes and Diseases. Brief. Bioinform. 2015, 16, 45–58. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Liu, X.; Lv, M.; Wu, Y.; Kuang, S.; Gong, J.; Yuan, P.; Zhong, Z.; Li, Q.; Jia, H.; et al. MicroRNA and Transcription Factor Co-Regulatory Network Analysis Reveals MiR-19 Inhibits CYLD in T-Cell Acute Lymphoblastic Leukemia. Nucleic Acids Res. 2012, 40, 5201–5214. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Zhang, Q.; Xia, M.; Hu, F.; Ma, Z.; Chen, Z.; Guo, A.-Y. Differential Co-Expression and Regulatory Network Analysis Uncover the Relapse Factor and Mechanism of T Cell Acute Leukemia. Mol. Therm. Nucleic Acids 2018, 12, 184–194. [Google Scholar] [CrossRef]
- McInnes, N.; Sadlon, T.J.; Brown, C.Y.; Pederson, S.; Beyer, M.; Schultze, J.L.; McColl, S.; Goodall, G.J.; Barry, S.C. FOXP3 and FOXP3-Regulated MicroRNAs Suppress SATB1 in Breast Cancer Cells. Oncogene 2012, 31, 1045–1054. [Google Scholar] [CrossRef]
- Picon-Ruiz, M.; Pan, C.; Drews-Elger, K.; Jang, K.; Besser, A.H.; Zhao, D.; Morata-Tarifa, C.; Kim, M.; Ince, T.A.; Azzam, D.J.; et al. Interactions between Adipocytes and Breast Cancer Cells Stimulate Cytokine Production and Drive Src/Sox2/MiR-302b-Mediated Malignant Progression. Cancer Res. 2016, 76, 491–504. [Google Scholar] [CrossRef]
- Huang, W.; Hu, H.; Zhang, Q.; Wu, X.; Wei, F.; Yang, F.; Gan, L.; Wang, N.; Yang, X.; Guo, A.-Y. Regulatory Networks in Mechanotransduction Reveal Key Genes in Promoting Cancer Cell Stemness and Proliferation. Oncogene 2019, 38, 6818–6834. [Google Scholar] [CrossRef]
- Zhang, Q.; Hu, H.; Chen, S.-Y.; Liu, C.-J.; Hu, F.-F.; Yu, J.; Wu, Y.; Guo, A.-Y. Transcriptome and Regulatory Network Analyses of CD19-CAR-T Immunotherapy for B-ALL. Genom. Proteom. Bioinform. 2019, 17, 190–200. [Google Scholar] [CrossRef]
- Xie, G.-Y.; Xia, M.; Miao, Y.-R.; Luo, M.; Zhang, Q.; Guo, A.-Y. FFLtool: A Web Server for Transcription Factor and MiRNA Feed Forward Loop Analysis in Human. Bioinformatics 2020, 36, 2605–2607. [Google Scholar] [CrossRef]
- Llovet, J.M.; Montal, R.; Sia, D.; Finn, R.S. Molecular Therapies and Precision Medicine for Hepatocellular Carcinoma. Nat. Rev. Clin. Oncol. 2018, 15, 599–616. [Google Scholar] [CrossRef]
- Wu, H.; Tao, J.; Li, X.; Zhang, T.; Zhao, L.; Wang, Y.; Zhang, L.; Xiong, J.; Zeng, Z.; Zhan, N.; et al. MicroRNA-206 Prevents the Pathogenesis of Hepatocellular Carcinoma by Modulating Expression of Met Proto-Oncogene and Cyclin-Dependent Kinase 6 in Mice. Hepatology 2017, 66, 1952–1967. [Google Scholar] [CrossRef]
- Sandbothe, M.; Buurman, R.; Reich, N.; Greiwe, L.; Vajen, B.; Gürlevik, E.; Schäffer, V.; Eilers, M.; Kühnel, F.; Vaquero, A.; et al. The MicroRNA-449 Family Inhibits TGF-β-Mediated Liver Cancer Cell Migration by Targeting SOX4. J. Hepatol. 2017, 66, 1012–1021. [Google Scholar] [CrossRef]
- Han, D.; Li, J.; Wang, H.; Su, X.; Hou, J.; Gu, Y.; Qian, C.; Lin, Y.; Liu, X.; Huang, M.; et al. Circular RNA CircMTO1 Acts as the Sponge of MicroRNA-9 to Suppress Hepatocellular Carcinoma Progression. Hepatology 2017, 66, 1151–1164. [Google Scholar] [CrossRef]
- Dong, T.; Zhang, Y.; Chen, Y.; Liu, P.; An, T.; Zhang, J.; Yang, H.; Zhu, W.; Yang, X. FOXO1 Inhibits the Invasion and Metastasis of Hepatocellular Carcinoma by Reversing ZEB2-Induced Epithelial-Mesenchymal Transition. Oncotarget 2017, 8, 1703–1713. [Google Scholar] [CrossRef]
- Cosimo, E.; Tarafdar, A.; Moles, M.W.; Holroyd, A.K.; Malik, N.; Catherwood, M.A.; Hay, J.; Dunn, K.M.; Macdonald, A.M.; Guichard, S.M.; et al. AKT/MTORC2 Inhibition Activates FOXO1 Function in CLL Cells Reducing B-Cell Receptor-Mediated Survival. Clin. Cancer Res. 2019, 25, 1574–1587. [Google Scholar] [CrossRef]
- Yang, N.; Zhou, J.; Li, Q.; Han, F.; Yu, Z. MiR-96 Exerts Carcinogenic Effect by Activating AKT/GSK-3β/β-Catenin Signaling Pathway through Targeting Inhibition of FOXO1 in Hepatocellular Carcinoma. Cancer Cell Int. 2019, 19, 38. [Google Scholar] [CrossRef]
- Huang, W.; Chen, Z.; Shang, X.; Tian, D.; Wang, D.; Wu, K.; Fan, D.; Xia, L. Sox12, a Direct Target of FoxQ1, Promotes Hepatocellular Carcinoma Metastasis through up-Regulating Twist1 and FGFBP1. Hepatology 2015, 61, 1920–1933. [Google Scholar] [CrossRef]
- Levrero, M.; Zucman-Rossi, J. Mechanisms of HBV-Induced Hepatocellular Carcinoma. J. Hepatol. 2016, 64, S84–S101. [Google Scholar] [CrossRef]
- Tarazona, S.; Furió-Tarí, P.; Turrà, D.; Pietro, A.D.; Nueda, M.J.; Ferrer, A.; Conesa, A. Data Quality Aware Analysis of Differential Expression in RNA-Seq with NOISeq R/Bioc Package. Nucleic Acids Res. 2015, 43, e140. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhou, B.; Pache, L.; Chang, M.; Khodabakhshi, A.H.; Tanaseichuk, O.; Benner, C.; Chanda, S.K. Metascape Provides a Biologist-Oriented Resource for the Analysis of Systems-Level Datasets. Nat. Commun. 2019, 10, 1523. [Google Scholar] [CrossRef]
- Vlachos, I.S.; Zagganas, K.; Paraskevopoulou, M.D.; Georgakilas, G.; Karagkouni, D.; Vergoulis, T.; Dalamagas, T.; Hatzigeorgiou, A.G. DIANA-MiRPath v3.0: Deciphering MicroRNA Function with Experimental Support. Nucleic Acids Res. 2015, 43, W460–W466. [Google Scholar] [CrossRef]
- Hu, H.; Miao, Y.-R.; Jia, L.-H.; Yu, Q.-Y.; Zhang, Q.; Guo, A.-Y. AnimalTFDB 3.0: A Comprehensive Resource for Annotation and Prediction of Animal Transcription Factors. Nucleic Acids Res. 2019, 47, D33–D38. [Google Scholar] [CrossRef]
- Tang, Y.; Li, M.; Wang, J.; Pan, Y.; Wu, F.-X. CytoNCA: A Cytoscape Plugin for Centrality Analysis and Evaluation of Protein Interaction Networks. BioSystems 2015, 127, 67–72. [Google Scholar] [CrossRef]
- Li, G.; Xu, W.; Zhang, L.; Liu, T.; Jin, G.; Song, J.; Wu, J.; Wang, Y.; Chen, W.; Zhang, C.; et al. Development and Validation of a CIMP-Associated Prognostic Model for Hepatocellular Carcinoma. EBioMedicine 2019, 47, 128–141. [Google Scholar] [CrossRef]
- Kitagawa, K.; Nakajima, G.; Kuramochi, H.; Ariizumi, S.-I.; Yamamoto, M. Lymphatic Vessel Endothelial Hyaluronan Receptor-1 Is a Novel Prognostic Indicator for Human Hepatocellular Carcinoma. Mol. Clin. Oncol. 2013, 1, 1039–1048. [Google Scholar] [CrossRef]
- Naboulsi, W.; Megger, D.A.; Bracht, T.; Kohl, M.; Turewicz, M.; Eisenacher, M.; Voss, D.M.; Schlaak, J.F.; Hoffmann, A.-C.; Weber, F.; et al. Quantitative Tissue Proteomics Analysis Reveals Versican as Potential Biomarker for Early-Stage Hepatocellular Carcinoma. J. Proteome Res. 2016, 15, 38–47. [Google Scholar] [CrossRef]
- Zou, C.-D.; Zhao, W.-M.; Wang, X.-N.; Li, Q.; Huang, H.; Cheng, W.-P.; Jin, J.-F.; Zhang, H.; Wu, M.-J.; Tai, S.; et al. MicroRNA-107: A Novel Promoter of Tumor Progression That Targets the CPEB3/EGFR Axis in Human Hepatocellular Carcinoma. Oncotarget 2016, 7, 266–278. [Google Scholar] [CrossRef]
- Lee, S.-Y.; Lee, G.R.; Woo, D.-H.; Park, N.H.; Cha, H.J.; Moon, Y.-H.; Han, I.-S. Depletion of Aurora A Leads to Upregulation of FoxO1 to Induce Cell Cycle Arrest in Hepatocellular Carcinoma Cells. Cell Cycle 2013, 12, 67–75. [Google Scholar] [CrossRef][Green Version]
- Dong, X.; Wang, F.; Xue, Y.; Lin, Z.; Song, W.; Yang, N.; Li, Q. MicroRNA-9-5p Downregulates Klf4 and Influences the Progression of Hepatocellular Carcinoma via the AKT Signaling Pathway. Int. J. Mol. Med. 2019, 43, 1417–1429. [Google Scholar] [CrossRef]
- Aishanjiang, A.; Rouzi, N.; Jiao, Z.; Wang, L.; Wusainahong, K.; Wumanjiang, N.; Musha, M.; Niyazi, M. MicroRNA-9 Enhances Invasion and Migration of Cervical Carcinomas by Directly Targeting FOXO1. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 2253–2260. [Google Scholar] [CrossRef]
- Shu, Z.; Gao, F.; Xia, Q.; Zhang, M. MiR-9-5p Promotes Cell Proliferation and Migration of Hepatocellular Carcinoma by Targeting CPEB3. Biomark. Med. 2021, 15, 97–108. [Google Scholar] [CrossRef]
- Hartke, J.; Johnson, M.; Ghabril, M. The Diagnosis and Treatment of Hepatocellular Carcinoma. Semin. Diagn. Pathol. 2017, 34, 153–159. [Google Scholar] [CrossRef]
- Llovet, J.M.; Di Bisceglie, A.M.; Bruix, J.; Kramer, B.S.; Lencioni, R.; Zhu, A.X.; Sherman, M.; Schwartz, M.; Lotze, M.; Talwalkar, J.; et al. Design and Endpoints of Clinical Trials in Hepatocellular Carcinoma. J. Natl. Cancer Inst. 2008, 100, 698–711. [Google Scholar] [CrossRef]
- Wang, Q.; Tang, Q.; Zhao, L.; Zhang, Q.; Wu, Y.; Hu, H.; Liu, L.; Liu, X.; Zhu, Y.; Guo, A.; et al. Time Serial Transcriptome Reveals Cyp2c29 as a Key Gene in Hepatocellular Carcinoma Development. Cancer Biol. Med. 2020, 17, 401–417. [Google Scholar] [CrossRef]
- Deng, R.; Cui, X.; Dong, Y.; Tang, Y.; Tao, X.; Wang, S.; Wang, J.; Chen, L. Construction of CircRNA-Based CeRNA Network to Reveal the Role of CircRNAs in the Progression and Prognosis of Hepatocellular Carcinoma. Front. Genet. 2021, 12, 626764. [Google Scholar] [CrossRef]
- Si, T.; Ning, X.; Zhao, H.; Zhang, M.; Huang, P.; Hu, Z.; Yang, L.; Lin, L. MicroRNA-9-5p Regulates the Mitochondrial Function of Hepatocellular Carcinoma Cells through Suppressing PDK4. Cancer Gene Therm. 2021, 28, 706–718. [Google Scholar] [CrossRef]
- Yang, X.-W.; Shen, G.-Z.; Cao, L.-Q.; Jiang, X.-F.; Peng, H.-P.; Shen, G.; Chen, D.; Xue, P. MicroRNA-1269 Promotes Proliferation in Human Hepatocellular Carcinoma via Downregulation of FOXO1. BMC Cancer 2014, 14, 909. [Google Scholar] [CrossRef]
- Yang, J.; Li, T.; Gao, C.; Lv, X.; Liu, K.; Song, H.; Xing, Y.; Xi, T. FOXO1 3′UTR Functions as a CeRNA in Repressing the Metastases of Breast Cancer Cells via Regulating MiRNA Activity. FEBS Lett. 2014, 588, 3218–3224. [Google Scholar] [CrossRef]
- D’Ambrogio, A.; Nagaoka, K.; Richter, J.D. Translational Control of Cell Growth and Malignancy by the CPEBs. Nat. Rev. Cancer 2013, 13, 283–290. [Google Scholar] [CrossRef]
- Xu, H.; Cao, H.; Xiao, G. Signaling via PINCH: Functions, Binding Partners and Implications in Human Diseases. Gene 2016, 594, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Peng, S.-C.; Lai, Y.-T.; Huang, H.-Y.; Huang, H.-D.; Huang, Y.-S. A Novel Role of CPEB3 in Regulating EGFR Gene Transcription via Association with Stat5b in Neurons. Nucleic Acids Res. 2010, 38, 7446–7457. [Google Scholar] [CrossRef]
- Wang, Z. ErbB Receptors and Cancer. Methods Mol. Biol. 2017, 1652, 3–35. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Wang, Q.; Li, W.; Ma, X.; Wu, L.; Guo, F.; Zhao, S.; Huang, F.; Wang, H.; Qin, G. Overexpression of FOXO1 Ameliorates the Podocyte Epithelial-Mesenchymal Transition Induced by High Glucose in Vitro and in Vivo. Biochem. Biophys. Res. Commun. 2016, 471, 416–422. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.; Ko, F.C.F.; Yeung, Y.-S.; Ng, I.O.-L.; Yam, J.W.P. Integrin-Linked Kinase Overexpression and Its Oncogenic Role in Promoting Tumorigenicity of Hepatocellular Carcinoma. PLoS ONE 2011, 6, e16984. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dougherty, G.W.; Jose, C.; Gimona, M.; Cutler, M.L. The Rsu-1-PINCH1-ILK Complex Is Regulated by Ras Activation in Tumor Cells. Eur. J. Cell Biol. 2008, 87, 721–734. [Google Scholar] [CrossRef]
- Donthamsetty, S.; Bhave, V.S.; Mars, W.M.; Bowen, W.C.; Orr, A.; Haynes, M.M.; Wu, C.; Michalopoulos, G.K. Role of PINCH and Its Partner Tumor Suppressor Rsu-1 in Regulating Liver Size and Tumorigenesis. PLoS ONE 2013, 8, e74625. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, H.; Huang, W.; Zhang, H.; Li, J.; Zhang, Q.; Miao, Y.-R.; Hu, F.-F.; Gan, L.; Su, Z.; Yang, X.; et al. A miR-9-5p/FOXO1/CPEB3 Feed-Forward Loop Drives the Progression of Hepatocellular Carcinoma. Cells 2022, 11, 2116. https://doi.org/10.3390/cells11132116
Hu H, Huang W, Zhang H, Li J, Zhang Q, Miao Y-R, Hu F-F, Gan L, Su Z, Yang X, et al. A miR-9-5p/FOXO1/CPEB3 Feed-Forward Loop Drives the Progression of Hepatocellular Carcinoma. Cells. 2022; 11(13):2116. https://doi.org/10.3390/cells11132116
Chicago/Turabian StyleHu, Hui, Wei Huang, Hong Zhang, Jianye Li, Qiong Zhang, Ya-Ru Miao, Fei-Fei Hu, Lu Gan, Zhenhong Su, Xiangliang Yang, and et al. 2022. "A miR-9-5p/FOXO1/CPEB3 Feed-Forward Loop Drives the Progression of Hepatocellular Carcinoma" Cells 11, no. 13: 2116. https://doi.org/10.3390/cells11132116
APA StyleHu, H., Huang, W., Zhang, H., Li, J., Zhang, Q., Miao, Y.-R., Hu, F.-F., Gan, L., Su, Z., Yang, X., & Guo, A.-Y. (2022). A miR-9-5p/FOXO1/CPEB3 Feed-Forward Loop Drives the Progression of Hepatocellular Carcinoma. Cells, 11(13), 2116. https://doi.org/10.3390/cells11132116







