Mechanisms for Bile Acids CDCA- and DCA-Stimulated Hepatic Spexin Expression
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Test Substances
2.3. Measurement of SPX Promoter Activity in SMMC7721 and BEL-7402 Cells
2.4. Measurement of SPX mRNA Expression in Mice by Real-Time PCR
2.5. Western Blot Analysis
2.6. Statistical Analysis
3. Results
3.1. Screening of Bile Acid Monomers with Effects on SPX Promoter Activation
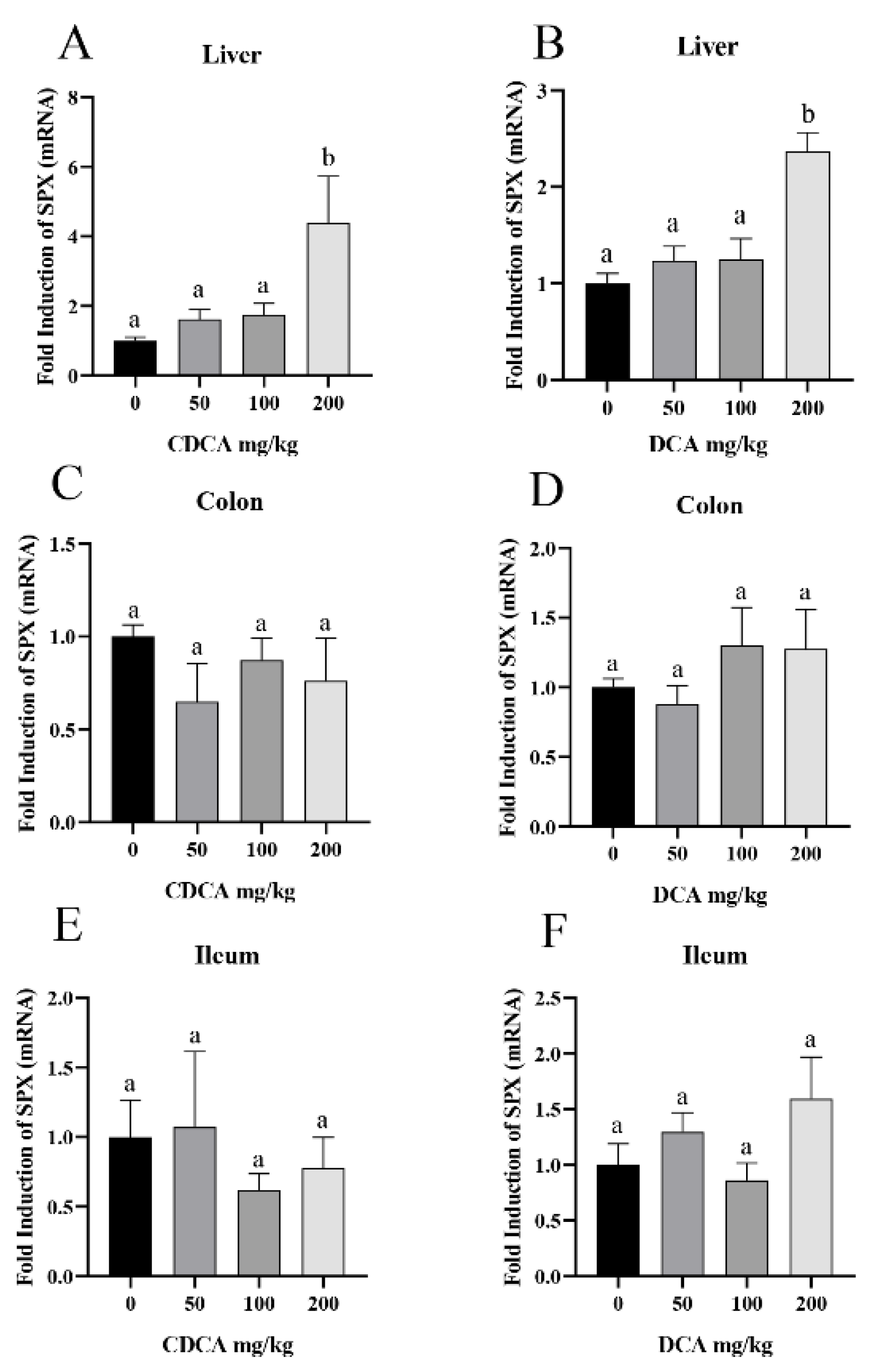
3.2. CDCA- and DCA-Induced SPX Expression in Mouse Liver
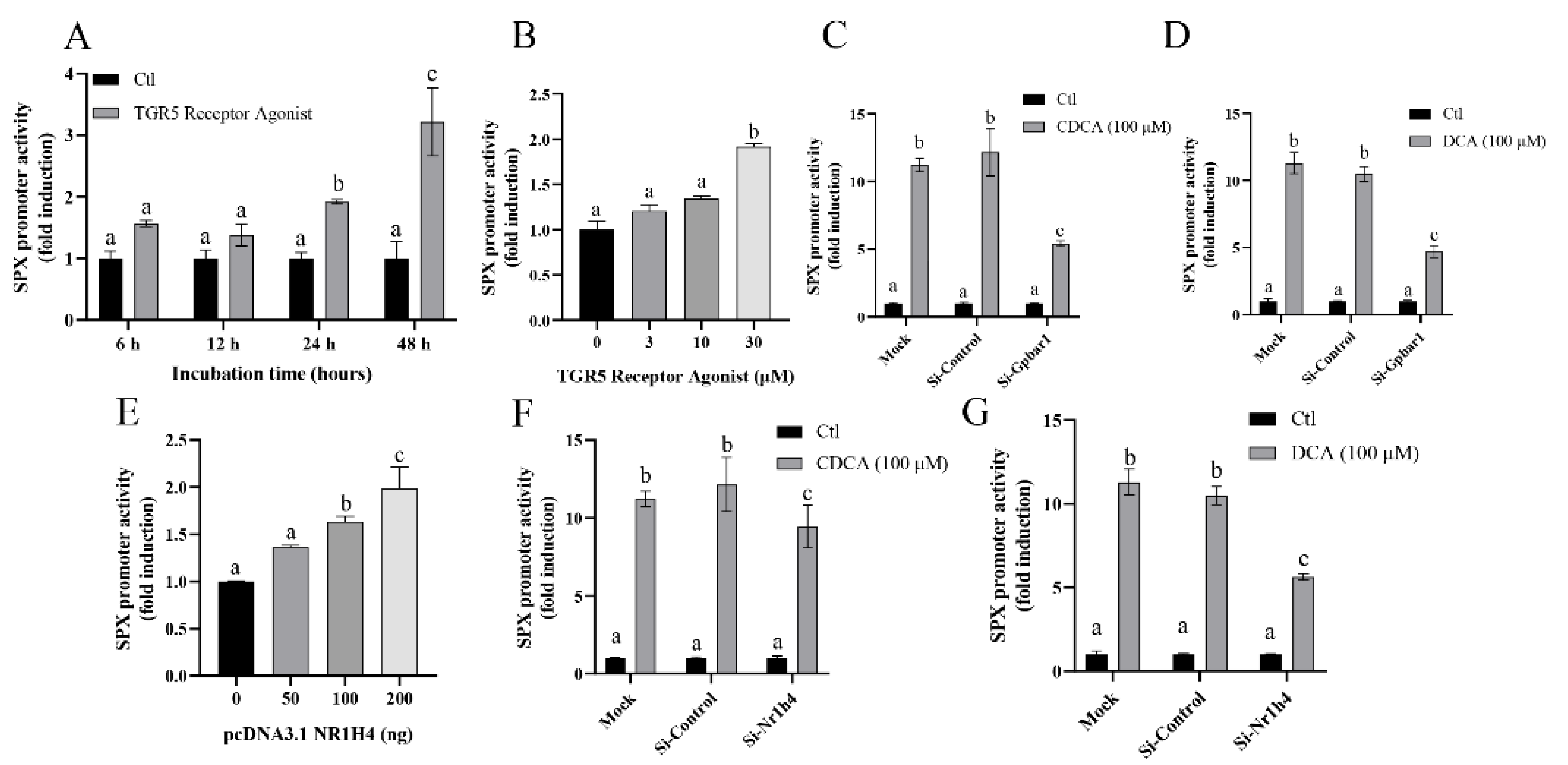
3.3. Bile Acid Receptor-Mediated CDCA- and DCA-Induced SPX Promoter Activity
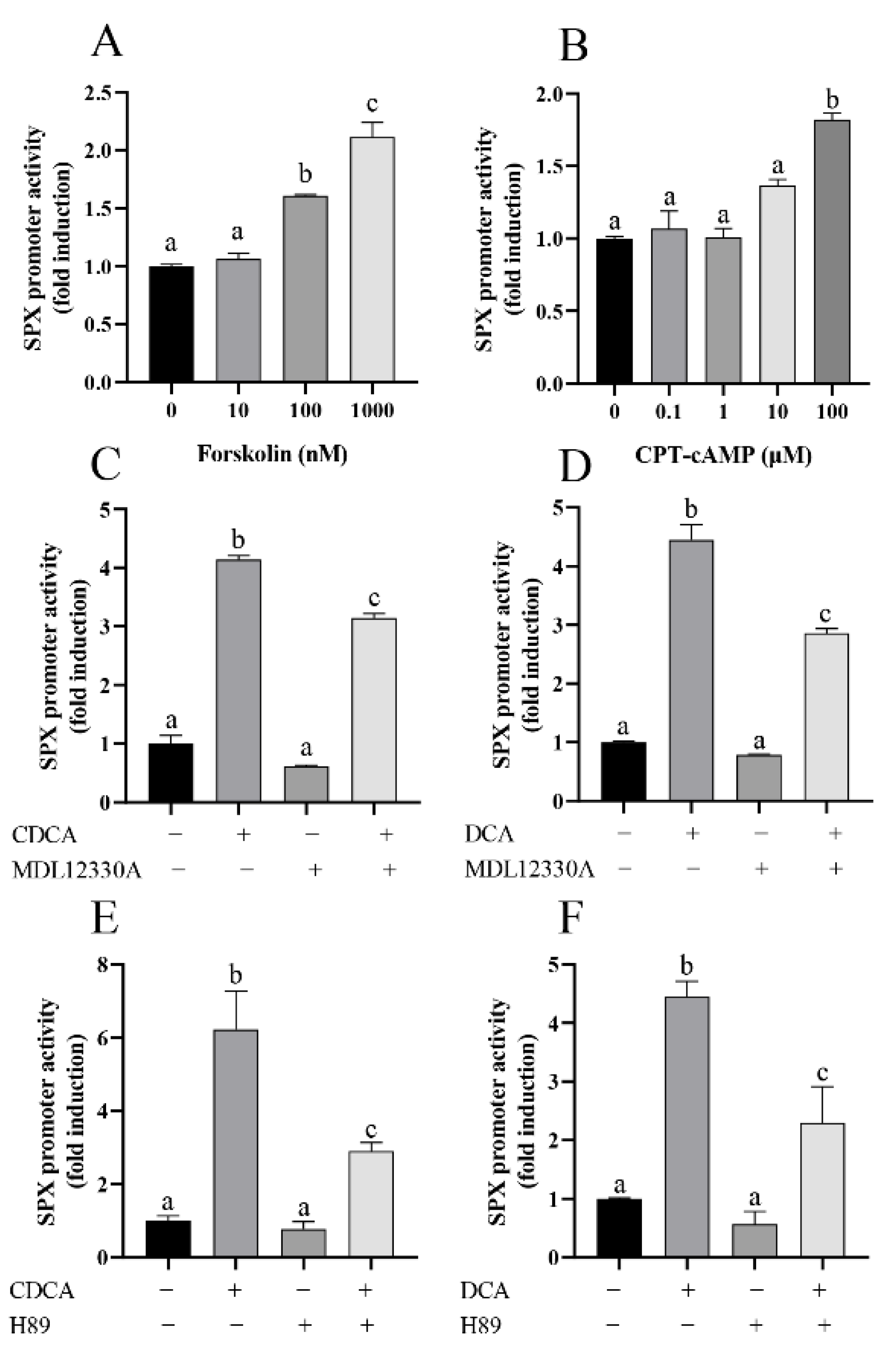
3.4. The Role of AC/cAMP/PKA Pathway in CDCA- and DCA-Induced SPX Promoter Activity
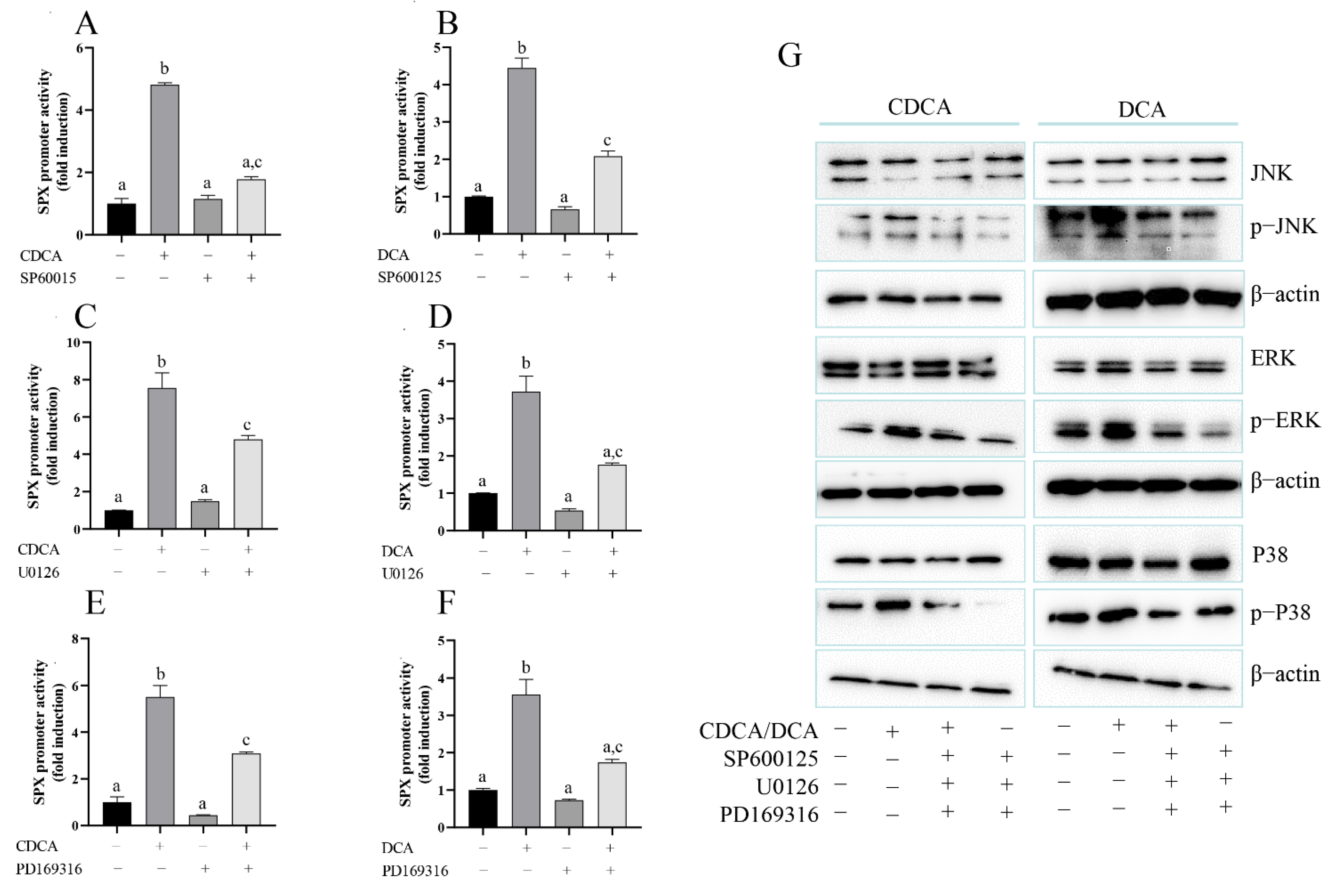
3.5. The Role of MAPK/ERK, MAPK/JNK, and MAPK/p38 Pathway in CDCA- and DCA-Induced SPX Promoter Activity
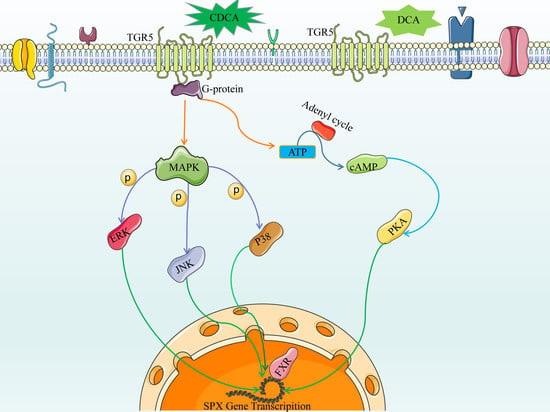
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, D.K.; Yun, S.; Son, G.H.; Hwang, J.I.; Park, C.R.; Kim, J.I.; Kim, K.; Vaudry, H.; Seong, J.Y. Coevolution of the Spexin/Galanin/Kisspeptin Family: Spexin Activates Galanin Receptor Type Ii and Iii. Endocrinology 2014, 155, 1864–1873. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mirabeau, O.; Perlas, E.; Severini, C.; Audero, E.; Gascuel, O.; Possenti, R.; Birney, E.; Rosenthal, N.; Gross, C. Identification of Novel Peptide Hormones in the Human Proteome by Hidden Markov Model Screening. Genome Res. 2007, 17, 320–327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, L.; Ma, Y.; Gu, M.; Zhang, Y.; Yan, S.; Li, N.; Wang, Y.; Ding, X.; Yin, J.; Fan, N.; et al. Spexin Peptide Is Expressed in Human Endocrine and Epithelial Tissues and Reduced after Glucose Load in Type 2 Diabetes. Peptides 2015, 71, 232–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Porzionato, A.; Rucinski, M.; Macchi, V.; Stecco, C.; Malendowicz, L.K.; De Caro, R. Spexin Expression in Normal Rat Tissues. J. Histochem. Cytochem. 2010, 58, 825–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhuang, M.; Lai, Q.; Yang, C.; Ma, Y.; Fan, B.; Bian, Z.; Lin, C.; Bai, J.; Zeng, G. Spexin as an Anxiety Regulator in Mouse Hippocampus: Mechanisms for Transcriptional Regulation of Spexin Gene Expression by Corticotropin Releasing Factor. Biochem. Biophys. Res. Commun. 2020, 525, 326–333. [Google Scholar] [CrossRef] [PubMed]
- Ma, A.; Bai, J.; He, M.; Wong, A.O. Spexin as a Neuroendocrine Signal with Emerging Functions. Gen. Comp. Endocrinol. 2018, 265, 90–96. [Google Scholar] [CrossRef]
- Kumar, S.; Hossain, M.J.; Javed, A.; Kullo, I.J.; Balagopal, P.B. Relationship of Circulating Spexin with Markers of Cardiovascular Disease: A Pilot Study in Adolescents with Obesity. Pediatr. Obes. 2018, 13, 374–380. [Google Scholar] [CrossRef]
- Al-Daghri, N.M.; Alenad, A.; Al-Hazmi, H.A.; Amer, O.E.; Hussain, S.D.; Alokail, M.S. Spexin Levels Are Associated with Metabolic Syndrome Components. Dis. Markers 2018, 2018, 1679690. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Zhang, M.; Huang, T.; Yang, L.-L.; Fu, H.-B.; Zhao, L.; Zhong, L.L.; Mu, H.-X.; Shi, X.-K.; Leung, C.F.; et al. Spexin Enhances Bowel Movement through Activating L-Type Voltage-Dependent Calcium Channel Via Galanin Receptor 2 in Mice. Sci. Rep. 2015, 5, 12095. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Li, G.; She, Y.; Zhang, Z. Low Levels of Spexin and Adiponectin May Predict Insulin Resistance in Patients with Non-Alcoholic Fatty Liver. Pract. Lab. Med. 2021, 24, e00207. [Google Scholar] [CrossRef]
- Lin, C.; Zhao, L.; Huang, T.; Lu, L.; Khan, M.; Liu, J.; Zhong, L.L.D.; Cai, Z.-W.; Fan, B.-M.; Wong, A.O.L.; et al. Spexin Acts as Novel Regulator for Bile Acid Synthesis. Front. Physiol. 2018, 9, 378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vallim, T.Q.D.A.; Tarling, E.J.; Edwards, P.A. Edwards. Pleiotropic Roles of Bile Acids in Metabolism. Cell Metab. 2013, 17, 657–669. [Google Scholar]
- Ding, L.; Yang, L.; Wang, Z.; Huang, W. Bile Acid Nuclear Receptor Fxr and Digestive System Diseases. Acta Pharm. Sin. B 2015, 5, 135–144. [Google Scholar] [CrossRef] [Green Version]
- Deng, F.; Bae, Y.H. Bile Acid Transporter-Mediated Oral Drug Delivery. J. Control Release 2020, 327, 100–116. [Google Scholar] [CrossRef]
- Shapiro, H.; Kolodziejczyk, A.A.; Halstuch, D.; Elinav, E. Bile Acids in Glucose Metabolism in Health and Disease. J. Exp. Med. 2018, 215, 383–396. [Google Scholar] [CrossRef]
- Jia, W.; Wei, M.; Rajani, C.; Zheng, X. Targeting the Alternative Bile Acid Synthetic Pathway for Metabolic Diseases. Protein Cell 2021, 12, 411–425. [Google Scholar] [CrossRef] [PubMed]
- Zollner, G.; Trauner, M. Mechanisms of Cholestasis. Clin. Liver Dis. 2008, 12, 1–26. [Google Scholar] [CrossRef]
- Carey, E.J.; Ali, A.H.; Lindor, K.D. Primary Biliary Cirrhosis. Lancet 2015, 386, 1565–1575. [Google Scholar] [CrossRef]
- Düfer, M.; Hörth, K.; Wagner, R.; Schittenhelm, B.; Prowald, S.; Wagner, T.F.; Oberwinkler, J.; Lukowski, R.; Gonzalez, F.J.; Krippeit-Drews, P.; et al. Bile Acids Acutely Stimulate Insulin Secretion of Mouse—Cells Via Farnesoid X Receptor Activation and Katp Channel Inhibition. Diabetes 2012, 61, 1479–1489. [Google Scholar] [CrossRef] [Green Version]
- Brighton, C.; Rievaj, J.; Kuhre, R.E.; Glass, L.; Schoonjans, K.; Holst, J.J.; Gribble, F.; Reimann, F. Bile Acids Trigger Glp-1 Release Predominantly by Accessing Basolaterally Located G Protein-Coupled Bile Acid Receptors. Endocrinology 2015, 156, 3961–3970. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, D.R.; Schmidt, S.; Holmstrom, S.R.; Makishima, M.; Yu, R.T.; Cummins, C.L.; Mangelsdorf, D.J.; Kliewer, S.A. Akr1b7 Is Induced by the Farnesoid X Receptor and Metabolizes Bile Acids. J. Biol. Chem. 2011, 286, 2425–2432. [Google Scholar] [CrossRef] [Green Version]
- Park, S.-J.; Lee, S.-B.; Lee, N.-S.; Ryu, Y.-J.; Lee, G.; Cho, J. Direct Effect of Chenodeoxycholic Acid on Differentiation of Mouse Embryonic Stem Cells Cultured under Feeder-Free Culture Conditions. BioMed Res. Int. 2013, 2013, 375076. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulkarni, O.P.; Hartter, I.; Mulay, S.R.; Hagemann, J.; Darisipudi, M.N.; Vr, S.K.; Romoli, S.; Thomasova, D.; Ryu, M.; Kobold, S.; et al. Toll-Like Receptor 4-Induced Il-22 Accelerates Kidney Regeneration. J. Am. Soc. Nephrol. 2014, 25, 978–989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakahara, M.; Fujii, H.; Maloney, P.R.; Shimizu, M.; Sato, R. Bile Acids Enhance Low Density Lipoprotein Receptor Gene Expression Via a Mapk Cascade-Mediated Stabilization of Mrna. J. Biol. Chem. 2002, 277, 37229–37234. [Google Scholar] [CrossRef] [Green Version]
- Jeong, B.; Kim, K.-K.; Lee, T.-H.; Kim, H.-R.; Park, B.-S.; Park, J.-W.; Jeong, J.-K.; Seong, J.-Y.; Lee, B.-J. Spexin Regulates Hypothalamic Leptin Action on Feeding Behavior. Biomolecules 2022, 12, 236. [Google Scholar] [CrossRef]
- Lv, S.; Zhou, Y.; Feng, Y.; Zhang, X.; Wang, X.; Yang, Y.; Wang, X. Peripheral Spexin Inhibited Food Intake in Mice. Int. J. Endocrinol. 2020, 2020, 4913785. [Google Scholar] [CrossRef] [PubMed]
- Walewski, J.L.; Ge, F.; Iv, H.L.; Levin, N.; Schwartz, G.J.; Vasselli, J.R.; Pomp, A.; Dakin, G.; Berk, P.D. Spexin Is a Novel Human Peptide That Reduces Adipocyte Uptake of Long Chain Fatty Acids and Causes Weight Loss in Rodents with Diet-Induced Obesity. Obesity 2014, 22, 1643–1652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Sun, L.; Zheng, L.; Su, M.; Liu, H.; Wei, Y.; Li, D.; Wang, Y.; Dai, C.; Gong, Y.; et al. Spexin Protects Cardiomyocytes from Hypoxia-Induced Metabolic and Mitochondrial Dysfunction. Naunyn Schmiedebergs Arch Pharmacol. 2020, 393, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Behrooz, M.; Vaghef-Mehrabany, E.; Ostadrahimi, A. Different Spexin Level in Obese Vs Normal Weight Children and Its Relationship with Obesity Related Risk Factors. Nutr. Metab. Cardiovasc. Dis. 2020, 30, 674–682. [Google Scholar] [CrossRef]
- Kumar, S.; Hossain, J.; Nader, N.; Aguirre, R.; Sriram, S.; Balagopal, P.B. Decreased Circulating Levels of Spexin in Obese Children. J. Clin. Endocrinol. Metab. 2016, 101, 2931–2936. [Google Scholar] [CrossRef] [Green Version]
- Chavez-Talavera, O.; Tailleux, A.; Lefebvre, P.; Staels, B. Bile Acid Control of Metabolism and Inflammation in Obesity, Type 2 Diabetes, Dyslipidemia, and Nonalcoholic Fatty Liver Disease. Gastroenterology 2017, 152, 1679–1694 e3. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Chiang, J.Y. Bile Acids as Metabolic Regulators. Curr. Opin. Gastroenterol. 2015, 31, 159–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiang, J.Y. Bile Acids: Regulation of Synthesis. J. Lipid Res. 2009, 50, 1955–1966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winston, J.A.; Theriot, C.M. Diversification of Host Bile Acids by Members of the Gut Microbiota. Gut Microbes 2020, 11, 158–171. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B. Bile Salt Biotransformations by Human Intestinal Bacteria. J. Lipid Res. 2006, 47, 241–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, K.; Kakiyama, G.; Suzuki, M.; Naritaka, N.; Takei, H.; Sato, H.; Kimura, A.; Murai, T.; Kurosawa, T.; Pandak, W.M.; et al. Changes in Conjugated Urinary Bile Acids across Age Groups. Steroids 2020, 164, 108730. [Google Scholar] [CrossRef] [PubMed]
- Suga, T.; Yamaguchi, H.; Sato, T.; Maekawa, M.; Goto, J.; Mano, N. Preference of Conjugated Bile Acids over Unconjugated Bile Acids as Substrates for Oatp1b1 and Oatp1b3. PLoS ONE 2017, 12, e0169719. [Google Scholar] [CrossRef] [Green Version]
- Jadhav, K.; Xu, Y.; Xu, Y.; Li, Y.; Xu, J.; Zhu, Y.; Adorini, L.; Lee, Y.K.; Kasumov, T.; Yin, L.; et al. Reversal of Metabolic Disorders by Pharmacological Activation of Bile Acid Receptors Tgr5 and Fxr. Mol. Metab. 2018, 9, 131–140. [Google Scholar] [CrossRef]
- Schaap, F.G.; Trauner, M.; Jansen, P.L.M. Bile Acid Receptors as Targets for Drug Development. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 55–67. [Google Scholar] [CrossRef]
- Makishima, M.; Okamoto, A.Y.; Repa, J.J.; Tu, H.; Learned, R.M.; Luk, A.; Hull, M.V.; Lustig, K.D.; Mangelsdorf, D.J.; Shan, B. Identification of a Nuclear Receptor for Bile Acids. Science 1999, 284, 1362–1365. [Google Scholar] [CrossRef]
- Fiorucci, S.; Distrutti, E. The Pharmacology of Bile Acids and Their Receptors. Handb. Exp. Pharmacol. 2019, 256, 3–18. [Google Scholar] [PubMed]
- Lew, J.-L.; Zhao, A.; Yu, J.; Huang, L.; de Pedro, N.; Peláez, F.; Wright, S.D.; Cui, J. The Farnesoid X Receptor Controls Gene Expression in a Ligand- and Promoter-Selective Fashion. J. Biol. Chem. 2004, 279, 8856–8861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, Y.Y.; Sheng, L. Regulation of Bile Acid Receptor Activity. Liver Res. 2018, 2, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Bertolini, A.; Fiorotto, R.; Strazzabosco, M. Bile Acids and Their Receptors: Modulators and Therapeutic Targets in Liver Inflammation. Semin. Immunopathol. 2022, 44, 547–564. [Google Scholar] [CrossRef] [PubMed]
- Fiorucci, S.; Mencarelli, A.; Palladino, G.; Cipriani, S. Bile-Acid-Activated Receptors: Targeting Tgr5 and Farnesoid-X-Receptor in Lipid and Glucose Disorders. Trends Pharmacol. Sci. 2009, 30, 570–580. [Google Scholar] [CrossRef] [PubMed]
- Bao, L.; Hao, D.; Wang, X.; He, X.; Mao, W.; Li, P. Transcriptome Investigation of Anti-Inflammation and Immuno-Regulation Mechanism of Taurochenodeoxycholic Acid. BMC Pharmacol. Toxicol. 2021, 22, 23. [Google Scholar] [CrossRef]
- Wada, T.; Stepniak, E.; Hui, L.; Leibbrandt, A.; Katada, T.; Nishina, H.; Wagner, E.F.; Penninger, J. Antagonistic Control of Cell Fates by Jnk and P38-Mapk Signaling. Cell Death Differ. 2008, 15, 89–93. [Google Scholar] [CrossRef]
- Chappell, W.H.; Steelman, L.S.; Long, J.M.; Kempf, R.C.; Abrams, S.L.; Franklin, R.A.; Bäsecke, J.; Stivala, F.; Donia, M.; Fagone, P.; et al. Ras/Raf/Mek/Erk and Pi3k/Pten/Akt/Mtor Inhibitors: Rationale and Importance to Inhibiting These Pathways in Human Health. Oncotarget 2011, 2, 135–164. [Google Scholar] [CrossRef] [Green Version]
- Chen, A.; Zheng, S. Curcumin Inhibits Connective Tissue Growth Factor Gene Expression in Activated Hepatic Stellate Cells in Vitro by Blocking Nf-Kappab and Erk Signalling. Br. J. Pharmacol. 2008, 153, 557–567. [Google Scholar] [CrossRef] [Green Version]
- van de Peppel, I.P.; Bodewes, F.A.; Verkade, H.J.; Jonker, J.W. Bile Acid Homeostasis in Gastrointestinal and Metabolic Complications of Cystic Fibrosis. J. Cyst. Fibros. 2019, 18, 313–320. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Shu, T.; Liu, G.; Mei, H.; Zhu, X.; Huang, X.; Zhang, L.; Jiang, Z. Quantitative Profiling of 19 Bile Acids in Rat Plasma, Liver, Bile and Different Intestinal Section Contents to Investigate Bile Acid Homeostasis and the Application of Temporal Variation of Endogenous Bile Acids. J. Steroid Biochem. Mol. Biol. 2017, 172, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Lee, Y.-K.; Bundman, D.; Han, Y.; Thevananther, S.; Kim, C.-S.; Chua, S.S.; Wei, P.; Heyman, R.A.; Karin, M.; et al. Redundant Pathways for Negative Feedback Regulation of Bile Acid Production. Dev. Cell 2002, 2, 721–731. [Google Scholar] [CrossRef] [Green Version]
- Meng, Z.; Liu, N.; Fu, X.; Wang, X.; Wang, Y.D.; Chen, W.D.; Zhang, L.; Forman, B.M.; Huang, W. Insufficient Bile Acid Signaling Impairs Liver Repair in Cyp27(−/−) Mice. J. Hepatol. 2011, 55, 885–895. [Google Scholar] [CrossRef]
- Li, T.; Chiang, J.Y. Bile Acid Signaling in Liver Metabolism and Diseases. J. Lipids 2012, 2012, 754067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grant, S.M.; DeMorrow, S. Bile Acid Signaling in Neurodegenerative and Neurological Disorders. Int. J. Mol. Sci. 2020, 21, 5982. [Google Scholar] [CrossRef] [PubMed]
- Vallim, T.Q.D.A.; Tarling, E.J.; Ahn, H.; Hagey, L.R.; Romanoski, C.E.; Lee, R.G.; Graham, M.J.; Motohashi, H.; Yamamoto, M.; Edwards, P.A. Mafg Is a Transcriptional Repressor of Bile Acid Synthesis and Metabolism. Cell Metab. 2015, 21, 298–311. [Google Scholar] [CrossRef]

| Stage | Temperature (°C) | Time | Cycles |
|---|---|---|---|
| Initial denaturation Denaturation Annealing | 95 95 60 60–95 | 5 min 10 s 30 s | 1 35 |
| Melt curve | 1.6 °C/s | 1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, Q.; Ma, Y.; Bai, J.; Zhuang, M.; Pei, S.; He, N.; Yin, J.; Fan, B.; Bian, Z.; Zeng, G.; et al. Mechanisms for Bile Acids CDCA- and DCA-Stimulated Hepatic Spexin Expression. Cells 2022, 11, 2159. https://doi.org/10.3390/cells11142159
Lai Q, Ma Y, Bai J, Zhuang M, Pei S, He N, Yin J, Fan B, Bian Z, Zeng G, et al. Mechanisms for Bile Acids CDCA- and DCA-Stimulated Hepatic Spexin Expression. Cells. 2022; 11(14):2159. https://doi.org/10.3390/cells11142159
Chicago/Turabian StyleLai, Qi, Yanhua Ma, Jin Bai, Min Zhuang, Shaofei Pei, Ni He, Junlin Yin, Baomin Fan, Zhaoxiang Bian, Guangzhi Zeng, and et al. 2022. "Mechanisms for Bile Acids CDCA- and DCA-Stimulated Hepatic Spexin Expression" Cells 11, no. 14: 2159. https://doi.org/10.3390/cells11142159
APA StyleLai, Q., Ma, Y., Bai, J., Zhuang, M., Pei, S., He, N., Yin, J., Fan, B., Bian, Z., Zeng, G., & Lin, C. (2022). Mechanisms for Bile Acids CDCA- and DCA-Stimulated Hepatic Spexin Expression. Cells, 11(14), 2159. https://doi.org/10.3390/cells11142159