The RNA Content of Fungal Extracellular Vesicles: At the “Cutting-Edge” of Pathophysiology Regulation
Abstract
1. Introduction
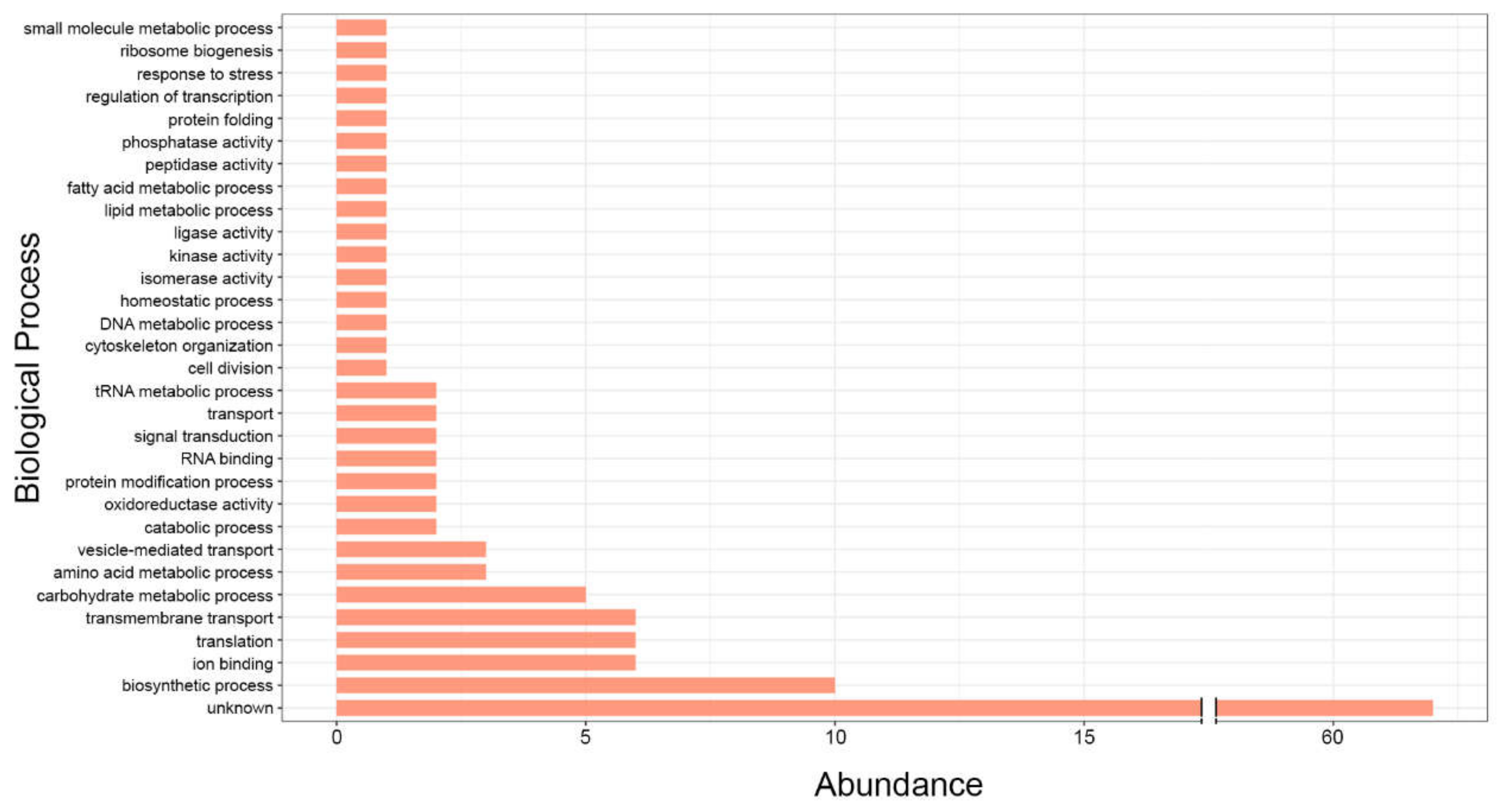
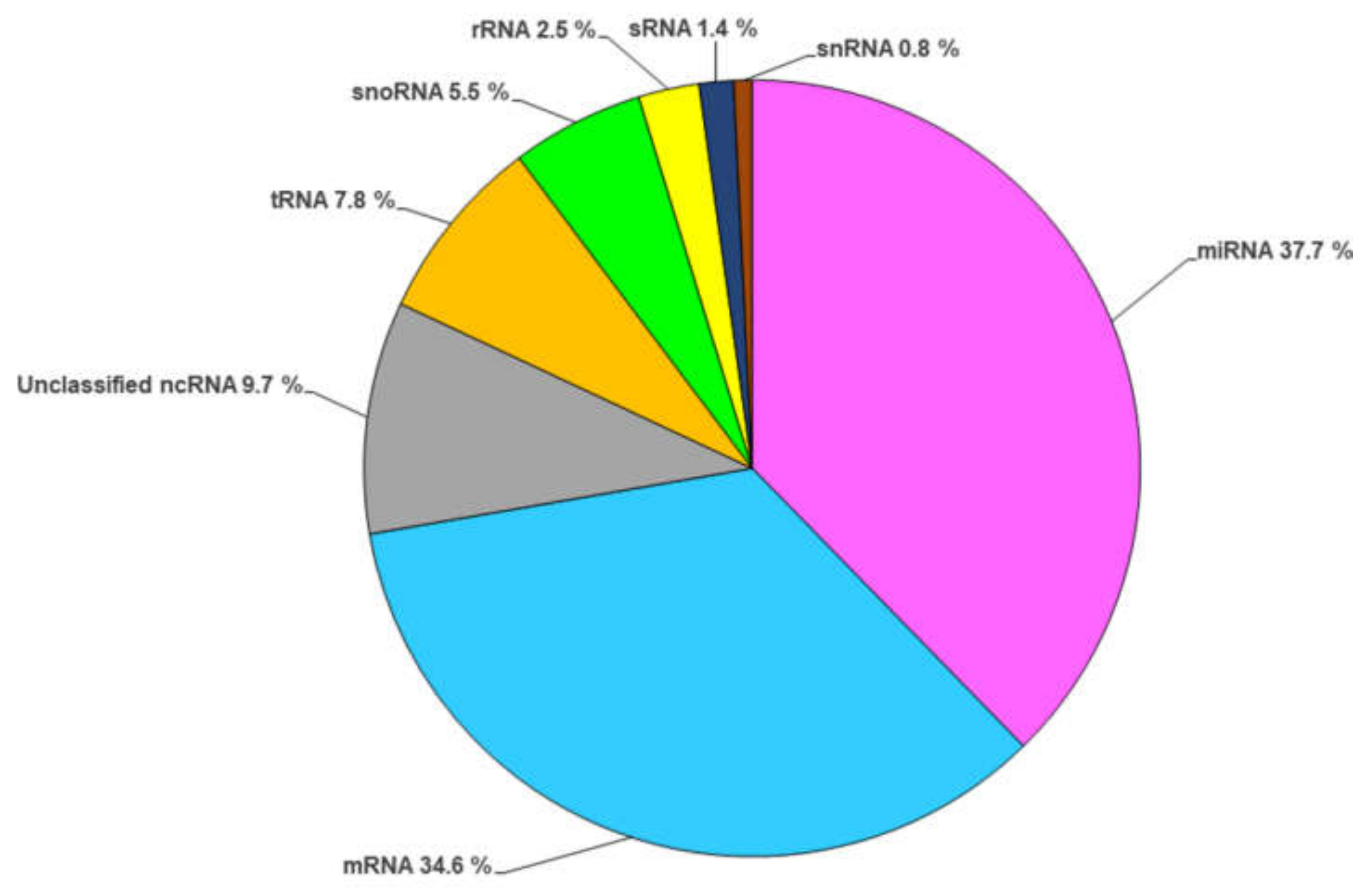
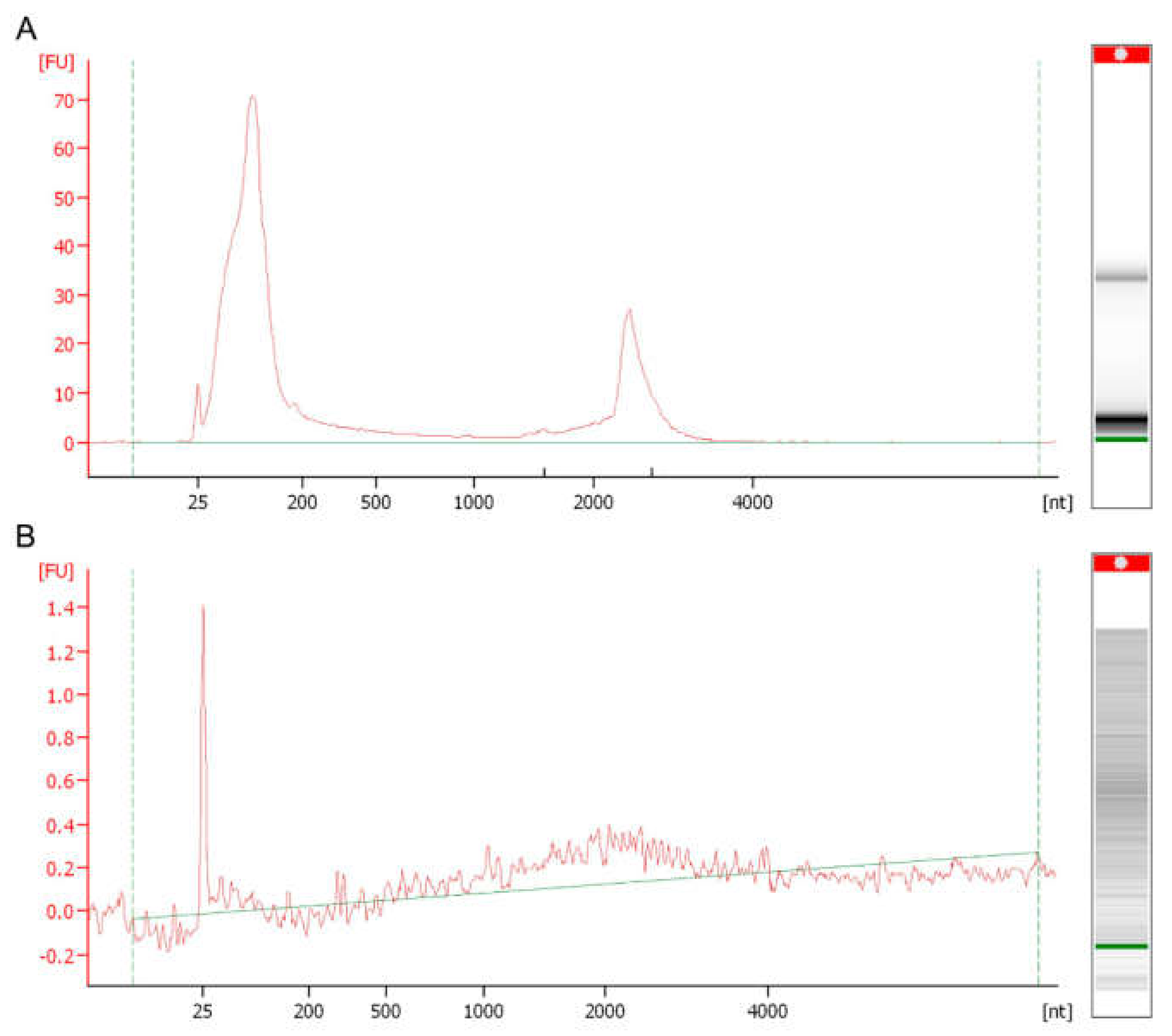
2. RNA Content of Fungal EVs
3. The Signaling Role of Fungal RNA-EVs during Host–Pathogen Interaction
4. Advancements in Fungal RNA-EV Research
5. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Toledo Martins, S.D.; Szwarc, P.; Goldenberg, S.; Alves, L.R. Extracellular Vesicles in Fungi: Composition and Functions. Curr. Top. Microbiol. Immunol. 2019, 422, 45–59. [Google Scholar] [PubMed]
- Bleackley, M.R.; Dawson, C.S.; Anderson, M.A. Fungal Extracellular Vesicles with a Focus on Proteomic Analysis. Proteomics 2019, 19, e1800232. [Google Scholar] [CrossRef] [PubMed]
- El Andaloussi, S.; Mäger, I.; Breakefield, X.O.; Wood, M.J. Extracellular vesicles: Biology and emerging therapeutic opportunities. Nat. Rev. Drug. Discov. 2013, 12, 347–357. [Google Scholar] [CrossRef] [PubMed]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Kalra, H.; Simpson, R.J.; Ji, H.; Aikawa, E.; Altevogt, P.; Askenase, P.; Bond, V.C.; Borràs, F.E.; Breakefield, X.; Budnik, V.; et al. Vesiclepedia: A Compendium for Extracellular Vesicles with Continuous Community Annotation. PLoS Biol. 2012, 10, e1001450. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef]
- Takeo, K.; Uesaka, I.; Uehira, K.; Nishiura, M. Fine structure of Cryptococcus neoformans grown in vivo as observed by freeze-etching. J. Bacteriol. 1973, 113, 1449–1454. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Nimrichter, L.; Oliveira, D.L.; Frases, S.; Miranda, K.; Zaragoza, O.; Alvarez, M.; Nakouzi, A.; Feldmesser, M.; Casadevall, A. Vesicular Polysaccharide Export in Cryptococcus neoformans Is a Eukaryotic Solution to the Problem of Fungal Trans-Cell Wall Transport. Eukaryot. Cell 2007, 6, 48–59. [Google Scholar] [CrossRef]
- Keller, S.; Ridinger, J.; Rupp, A.-K.; Janssen, J.W.G.; Altevogt, P. Body fluid derived exosomes as a novel template for clinical diagnostics. J. Transl. Med. 2011, 9, 86. [Google Scholar] [CrossRef]
- Ikeda, M.A.K.; de Almeida, J.R.F.; Jannuzzi, G.P.; Cronemberger-Andrade, A.; Torrecilhas, A.C.T.; Moretti, N.S.; Da Cunha, J.P.C.; De Almeida, S.R.; Ferreira, K.S. Extracellular Vesicles from Sporothrix brasiliensis Are an Important Virulence Factor That Induce an Increase in Fungal Burden in Experimental Sporotrichosis. Front. Microbiol. 2018, 9, 2286. [Google Scholar] [CrossRef]
- Vargas, G.; Rocha, J.D.B.; Oliveira, D.L.; Albuquerque, P.C.; Frases, S.; Santos, S.S.; Nosanchuk, J.D.; Gomes, A.M.O.; Medeiros, L.C.A.S.; Miranda, K.; et al. Compositional and immunobiological analyses of extracellular vesicles released by Candida albicans. Cell. Microbiol. 2014, 17, 389–407. [Google Scholar] [CrossRef]
- Gehrmann, U.; Qazi, K.R.; Johansson, C.; Hultenby, K.; Karlsson, M.; Lundeberg, L.; Gabrielsson, S.; Scheynius, A. Nanovesicles from Malassezia sympodialis and host exosomes induce cytokine responses—Novel mechanisms for host-microbe interactions in atopic eczema. PLoS ONE 2011, 6, e21480. [Google Scholar] [CrossRef]
- Albuquerque, P.C.; Nakayasu, E.S.; Rodrigues, M.L.; Frases, S.; Casadevall, A.; Zancope-Oliveira, R.M.; Almeida, I.C.; Nosanchuk, J.D. Vesicular transport inHistoplasma capsulatum: An effective mechanism for trans-cell wall transfer of proteins and lipids in ascomycetes. Cell. Microbiol. 2008, 10, 1695–1710. [Google Scholar] [CrossRef]
- Oliveira, D.L.; Nakayasu, E.S.; Joffe, L.S.; Guimarães, A.J.; Sobreira, T.J.P.; Nosanchuk, J.D.; Cordero, R.J.B.; Frases, S.; Casadevall, A.; Almeida, I.C.; et al. Characterization of Yeast Extracellular Vesicles: Evidence for the Participation of Different Pathways of Cellular Traffic in Vesicle Biogenesis. PLoS ONE 2010, 5, e11113. [Google Scholar] [CrossRef]
- Vallejo, M.C.; Matsuo, A.L.; Ganiko, L.; Medeiros, L.C.S.; Miranda, K.; Silva, L.S.; Freymüller-Haapalainen, E.; Sinigaglia-Coimbra, R.; Almeida, I.C.; Puccia, R. The pathogenic fungus Paracoccidioides brasiliensis exports extracellular vesicles containing highly immunogenic alpha-Galactosyl epitopes. Eukaryot. Cell 2011, 10, 343–351. [Google Scholar] [CrossRef]
- Leone, F.; Bellani, L.; Mucciflora, S.; Giorgetti, L.; Bongioanni, P.; Simili, M.; Maserti, B.; Del Carratore, R. Analysis of extracellular vesicles produced in the biofilm by the dimorphic yeast Pichia fermentans. J. Cell. Physiol. 2017, 233, 2759–2767. [Google Scholar] [CrossRef]
- Bielska, E.; Sisquella, M.A.; Aldeieg, M.; Birch, C.; O’Donoghue, E.J.; May, R.C. Pathogen-derived extracellular vesicles mediate virulence in the fatal human pathogen Cryptococcus gattii. Nat. Commun. 2018, 9, 1–9. [Google Scholar] [CrossRef]
- Peres da Silva, R.; Longo, L.G.; Cunha, J.P.D.; Sobreira, T.J.; Rodrigues, M.L.; Faoro, H.; Goldenberg, S.; Alves, L.R.; Puccia, R. Comparison of the RNA Content of Extracellular Vesicles Derived from Paracoccidioides brasiliensis and Paracoccidioides lutzii. Cells 2019, 8, 765. [Google Scholar] [CrossRef]
- Lavrin, T.; Konte, T.; Kostanjšek, R.; Sitar, S.; Sepčič, K.; Mihevc, S.P.; Žagar, E.; Župunski, V.; Lenassi, M.; Rogelj, B.; et al. The Neurotropic Black Yeast Exophiala dermatitidis Induces Neurocytotoxicity in Neuroblastoma Cells and Progressive Cell Death. Cells 2020, 9, 963. [Google Scholar] [CrossRef]
- Karkowska-Kuleta, J.; Kulig, K.; Karnas, E.; Zuba-Surma, E.; Woznicka, O.; Pyza, E.; Kuleta, P.; Osyczka, A.; Rapala-Kozik, M.; Kozik, A. Characteristics of Extracellular Vesicles Released by the Pathogenic Yeast-Like Fungi Candida glabrata, Candida parapsilosis and Candida tropicalis. Cells 2020, 9, 1722. [Google Scholar] [CrossRef]
- Yang, B.; Wang, J.; Jiang, H.; Lin, H.; Ou, Z.; Ullah, A.; Hua, Y.; Chen, J.; Lin, X.; Hu, X.; et al. Extracellular Vesicles Derived from Talaromyces marneffei Yeasts Mediate Inflammatory Response in Macrophage Cells by Bioactive Protein Components. Front. Microbiol. 2020, 11, 603183. [Google Scholar] [CrossRef] [PubMed]
- Zamith-Miranda, D.; Amatuzzi, R.F.; da Rocha, I.F.M.; Martins, S.T.; Lucena, A.C.; Vieira, A.Z.; Trentin, G.; Almeida, F.; Rodrigues, M.L.; Nakayasu, E.S.; et al. Transcriptional and translational landscape of Candida auris in response to caspofungin. Comput. Struct. Biotechnol. J. 2021, 19, 5264–5277. [Google Scholar] [CrossRef]
- Silva, B.M.; Prados-Rosales, R.; Espadas-Moreno, J.; Wolf, J.M.; Luque-Garcia, J.L.; Gonçalves, T.; Casadevall, A. Characterization of Alternaria infectoria extracellular vesicles. Med. Mycol. 2013, 52, 202–210. [Google Scholar] [CrossRef] [PubMed]
- Bitencourt, T.A.; Rezende, C.P.; Quaresemin, N.R.; Moreno, P.; Hatanaka, O.; Rossi, A.; Martinez-Rossi, N.M.; Almeida, F. Extracellular Vesicles from the Dermatophyte Trichophyton interdigitale Modulate Macrophage and Keratinocyte Functions. Front. Immunol. 2018, 9, 2343. [Google Scholar] [CrossRef]
- Liu, M.; Bruni, G.O.; Taylor, C.M.; Zhang, Z.; Wang, P. Comparative genome-wide analysis of extracellular small RNAs from the mucormycosis pathogen Rhizopus delemar. Sci. Rep. 2018, 8, 5243. [Google Scholar] [CrossRef]
- Bleackley, M.R.; Samuel, M.; Garcia-Ceron, D.; McKenna, J.A.; Lowe, R.; Pathan, M.; Zhao, K.; Ang, C.-S.; Mathivanan, S.; Anderson, M.A. Extracellular Vesicles from the Cotton Pathogen Fusarium oxysporum f. sp. vasinfectum Induce a Phytotoxic Response in Plants. Front. Plant Sci. 2020, 10, 1610. [Google Scholar] [CrossRef]
- De Paula, R.G.; Antoniêto, A.C.C.; Nogueira, K.M.V.; Ribeiro, L.F.C.; Rocha, M.C.; Malavazi, I.; Almeida, F.; Silva, R.N. Extracellular vesicles carry cellulases in the industrial fungus Trichoderma reesei. Biotechnol. Biofuels 2019, 12, 146. [Google Scholar] [CrossRef]
- Souza, J.A.M.; Baltazar, L.D.M.; Carregal, V.M.; Gouveia-Eufrasio, L.; De Oliveira, A.G.; Dias, W.G.; Campos Rocha, M.; Rocha de Miranda, K.; Malavazi, I.; Santos, D.D.A.; et al. Characterization of Aspergillus fumigatus Extracellular Vesicles and Their Effects on Macrophages and Neutrophils Functions. Front. Microbiol. 2019, 10, 2008. [Google Scholar] [CrossRef]
- Brauer, V.S.; Pessoni, A.M.; Bitencourt, T.A.; de Paula, R.G.; Rocha, L.D.O.; Goldman, G.H.; Almeida, F. Extracellular Vesicles from Aspergillus flavus Induce M1 Polarization In Vitro. mSphere 2020, 5, e00190-20. [Google Scholar] [CrossRef]
- Costa, J.H.; Bazioli, J.M.; Barbosa, L.D.; Dos Santos, P.L.T., Jr.; Reis, F.C.G.; Klimeck, T.; Crnkovic, C.M.; Berlinck, R.G.S.; Sussulini, A.; Rodrigues, M.L.; et al. Phytotoxic Tryptoquialanines Produced In Vivo by Penicillium digitatum Are Exported in Extracellular Vesicles. mBio 2021, 12, e03393-20. [Google Scholar] [CrossRef]
- Yáñez-Mó, M.; Siljander, P.R.-M.; Andreu, Z.; Bedina Zavec, A.; Borràs, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Wolf, P. The Nature and Significance of Platelet Products in Human Plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef]
- Takahashi, A.; Okada, R.; Nagao, K.; Kawamata, Y.; Hanyu, A.; Yoshimoto, S.; Takasugi, M.; Watanabe, S.; Kanemaki, M.T.; Obuse, C.; et al. Exosomes maintain cellular homeostasis by excreting harmful DNA from cells. Nat. Commun. 2017, 8, 15287. [Google Scholar] [CrossRef]
- Cleare, L.G.; Zamith, D.; Heyman, H.M.; Couvillion, S.P.; Nimrichter, L.; Rodrigues, M.L.; Nakayasu, E.S.; Nosanchuk, J.D. Media matters! Alterations in the loading and release of Histoplasma capsulatum extracellular vesicles in response to different nutritional milieus. Cell. Microbiol. 2020, 22, e13217. [Google Scholar] [CrossRef]
- Bahn, Y.-S.; Xue, C.; Idnurm, A.; Rutherford, J.C.; Heitman, J.; E Cardenas, M. Sensing the environment: Lessons from fungi. Nat. Rev. Genet. 2007, 5, 57–69. [Google Scholar] [CrossRef]
- Reis, F.C.G.; Borges, B.S.; Jozefowicz, L.J.; Sena, B.A.G.; Garcia, A.W.A.; Medeiros, L.C.; Martins, S.T.; Honorato, L.; Schrank, A.; Vainstein, M.H.; et al. A Novel Protocol for the Isolation of Fungal Extracellular Vesicles Reveals the Participation of a Putative Scramblase in Polysaccharide Export and Capsule Construction in Cryptococcus gattii. mSphere 2019, 4, e00080-19. [Google Scholar] [CrossRef]
- García-Rodas, R.; Cordero, R.J.B.; Trevijano-Contador, N.; Janbon, G.; Moyrand, F.; Casadevall, A.; Zaragoza, O. Capsule Growth in Cryptococcus neoformans Is Coordinated with Cell Cycle Progression. mBio 2014, 5, e00945-14. [Google Scholar] [CrossRef]
- Zhao, K.; Bleackley, M.; Chisanga, D.; Gangoda, L.; Fonseka, P.; Liem, M.; Kalra, H.; Al Saffar, H.; Keerthikumar, S.; Ang, C.-S.; et al. Extracellular vesicles secreted by Saccharomyces cerevisiae are involved in cell wall remodelling. Commun. Biol. 2019, 2, 1–13. [Google Scholar] [CrossRef]
- Honorato, L.; Demetrio, J.F.; Ellis, C.C.; Piffer, A.; Pereira, Y.; Frases, S.; de Sousa Araújo, G.R.; Pontes, B.; Mendes, M.T.; Pereira, M.D.; et al. Extracellular vesicles regulate yeast growth, biofilm formation, and yeast-to-hypha differentiation in Candida albicans. mBio 2021, 13, e00301-22. [Google Scholar] [CrossRef]
- Turchinovich, A.; Drapkina, O.; Tonevitsky, A. Transcriptome of Extracellular Vesicles: State-of-the-Art. Front. Immunol. 2019, 10, 202. [Google Scholar] [CrossRef]
- Munhoz da Rocha, I.F.; Amatuzzi, R.F.; Lucena, A.C.R.; Faoro, H.; Alves, L.R. Cross-Kingdom Extracellular Vesicles EV-RNA Communication as a Mechanism for Host-Pathogen Interaction. Front. Cell Infect. Microbiol. 2020, 10, 593160. [Google Scholar] [CrossRef]
- Rodrigues, M.L.; Casadevall, A. A two-way road: Novel roles for fungal extracellular vesicles. Mol. Microbiol. 2018, 110, 11–15. [Google Scholar] [CrossRef]
- Zarnowski, R.; Sanchez, H.; Covelli, A.S.; Dominguez, E.; Jaromin, A.; Bernhardt, J.; Mitchell, K.F.; Heiss, C.; Azadi, P.; Mitchell, A.; et al. Candida albicans biofilm–induced vesicles confer drug resistance through matrix biogenesis. PLoS Biol. 2018, 16, e2006872. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Cosmopoulos, K.; Thorley-Lawson, D.A.; van Eijndhoven, M.A.J.; Hopmans, E.S.; Lindenberg, J.L.; de Gruijl, T.D.; Würdinger, T.; Middeldorp, J.M. Functional delivery of viral miRNAs via exosomes. Proc. Natl. Acad. Sci. USA 2010, 107, 6328–6333. [Google Scholar] [CrossRef]
- Peres da Silva, R.; Puccia, R.; Rodrigues, M.L.; Oliveira, D.L.; Joffe, L.S.; César, G.V.; Nimrichter, L.; Goldenberg, S.; Alves, L.R. Extracellular vesicle-mediated export of fungal RNA. Sci. Rep. 2015, 5, 7763. [Google Scholar] [CrossRef]
- Bitencourt, T.A.; Oliveira, F.B.; Sanches, P.R.; Rossi, A.; Martinez-Rossi, N.M. The prp4 kinase gene and related spliceosome factor genes in Trichophyton rubrum respond to nutrients and antifungals. J. Med. Microbiol. 2019, 68, 591–599. [Google Scholar] [CrossRef]
- Sieber, P.; Voigt, K.; Kämmer, P.; Brunke, S.; Schuster, S.; Linde, J. Comparative Study on Alternative Splicing in Human Fungal Pathogens Suggests Its Involvement During Host Invasion. Front. Microbiol. 2018, 9, 2313. [Google Scholar] [CrossRef]
- Alves, L.R.; da Silva, R.P.; Sanchez, D.A.; Zamith-Miranda, D.; Rodrigues, M.L.; Goldenberg, S.; Puccia, R.; Nosanchuk, J.D. Extracellular Vesicle-Mediated RNA Release in Histoplasma capsulatum. mSphere 2019, 4, e00176-19. [Google Scholar] [CrossRef]
- Rayner, S.; Bruhn, S.; Vallhov, H.; Andersson, A.; Billmyre, R.B.; Scheynius, A. Identification of small RNAs in extracellular vesicles from the commensal yeast Malassezia sympodialis. Sci. Rep. 2017, 7, 39742. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Cui, K.; Jothi, R.; Zhao, D.-M.; Jing, X.; Zhao, K.; Xue, H.-H. GABP controls a critical transcription regulatory module that is essential for maintenance and differentiation of hematopoietic stem/progenitor cells. Blood 2011, 117, 2166–2178. [Google Scholar] [CrossRef] [PubMed]
- Modica, L.; Iotti, G.; D’Avola, A.; Blasi, F. Prep1 (pKnox1) Regulates Mouse Embryonic HSC Cycling and Self-Renewal Affecting the Stat1-Sca1 IFN-Dependent Pathway. PLoS ONE 2014, 9, e107916. [Google Scholar] [CrossRef] [PubMed]
- Bitencourt, T.A.; Hatanaka, O.; Pessoni, A.M.; Freitas, M.S.; Trentin, G.; Santos, P.; Rossi, A.; Martinez-Rossi, N.M.; Alves, L.L.; Casadevall, A.; et al. Fungal Extracellular Vesicles Are Involved in Intraspecies Intracellular Communication. mBio 2022, 13, e03272-21. [Google Scholar] [CrossRef]
- Zamith-Miranda, D.; Heyman, H.M.; Couvillion, S.P.; Cordero, R.J.B.; Rodrigues, M.L.; Nimrichter, L.; Casadevall, A.; Amatuzzi, R.F.; Alves, L.R.; Nakayasu, E.S.; et al. Comparative Molecular and Immunoregulatory Analysis of Extracellular Vesicles from Candida albicans and Candida auris. mSystems 2021, 6, e00822-21. [Google Scholar] [CrossRef] [PubMed]
- Castillo-González, C.; Zhang, X. The Trojan Horse of the Plant Kingdom. Cell Host Microbe 2018, 24, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Buck, A.H.; Coakley, G.; Simbari, F.; McSorley, H.J.; Quintana, J.F.; Le Bihan, T.; Kumar, S.; Abreu-Goodger, C.; Lear, M.; Harcus, Y.; et al. Exosomes secreted by nematode parasites transfer small RNAs to mammalian cells and modulate innate immunity. Nat. Commun. 2014, 5, 5488. [Google Scholar] [CrossRef]
- Cai, Q.; Qiao, L.; Wang, M.; He, B.; Lin, F.M.; Palmquist, J.; Huang, S.D.; Jin, H. Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 2018, 360, 1126–1129. [Google Scholar] [CrossRef]
- Zhang, T.; Zhao, Y.-L.; Zhao, J.-H.; Wang, S.; Jin, Y.; Chen, Z.-Q.; Fang, Y.-Y.; Zhao, J.-H.; Ding, S.-W.; Guo, H.-S. Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2016, 2, 16153. [Google Scholar] [CrossRef]
- Regente, M.; Pinedo, M.; Clemente, H.S.; Balliau, T.; Jamet, E.; De La Canal, L. Plant extracellular vesicles are incorporated by a fungal pathogen and inhibit its growth. J. Exp. Bot. 2017, 68, 5485–5495. [Google Scholar] [CrossRef]
- Weiberg, A.; Wang, M.; Lin, F.M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.D.; Jin, H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 342, 118–123. [Google Scholar] [CrossRef]
- Wang, M.; Weiberg, A.; Lin, F.-M.; Thomma, B.P.H.J.; Huang, H.-D.; Jin, H. Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants 2016, 2, 1–10. [Google Scholar] [CrossRef]
- Cui, C.; Wang, Y.; Liu, J.; Zhao, J.; Sun, P.; Wang, S. A fungal pathogen deploys a small silencing RNA that attenuates mosquito immunity and facilitates infection. Nat. Commun. 2019, 10, 1–10. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bitencourt, T.A.; Pessoni, A.M.; Oliveira, B.T.M.; Alves, L.R.; Almeida, F. The RNA Content of Fungal Extracellular Vesicles: At the “Cutting-Edge” of Pathophysiology Regulation. Cells 2022, 11, 2184. https://doi.org/10.3390/cells11142184
Bitencourt TA, Pessoni AM, Oliveira BTM, Alves LR, Almeida F. The RNA Content of Fungal Extracellular Vesicles: At the “Cutting-Edge” of Pathophysiology Regulation. Cells. 2022; 11(14):2184. https://doi.org/10.3390/cells11142184
Chicago/Turabian StyleBitencourt, Tamires A., André M. Pessoni, Bianca T. M. Oliveira, Lysangela R. Alves, and Fausto Almeida. 2022. "The RNA Content of Fungal Extracellular Vesicles: At the “Cutting-Edge” of Pathophysiology Regulation" Cells 11, no. 14: 2184. https://doi.org/10.3390/cells11142184
APA StyleBitencourt, T. A., Pessoni, A. M., Oliveira, B. T. M., Alves, L. R., & Almeida, F. (2022). The RNA Content of Fungal Extracellular Vesicles: At the “Cutting-Edge” of Pathophysiology Regulation. Cells, 11(14), 2184. https://doi.org/10.3390/cells11142184







