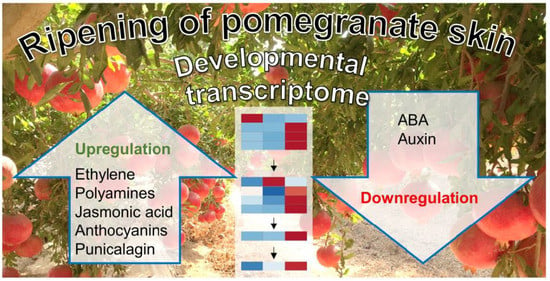
Ripening of Pomegranate Skin as Revealed by Developmental Transcriptomics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. RNA-Seq and Bioinformatics Analyses
3. Results and Discussion
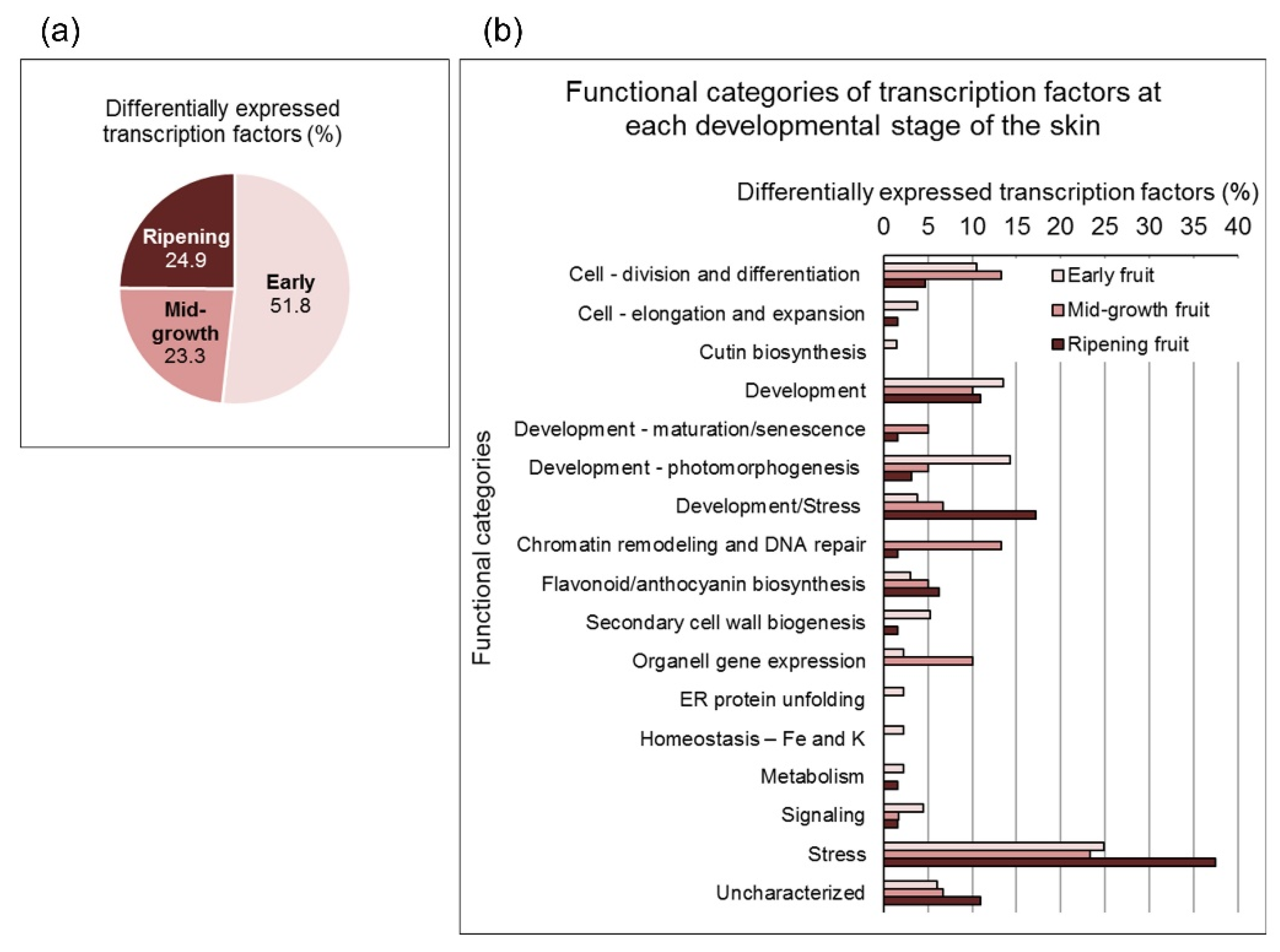
3.1. Data Validation and Characterization of Pomegranate Skin Developmental Stages
3.2. The Climacteric Nature of Pomegranate Skin
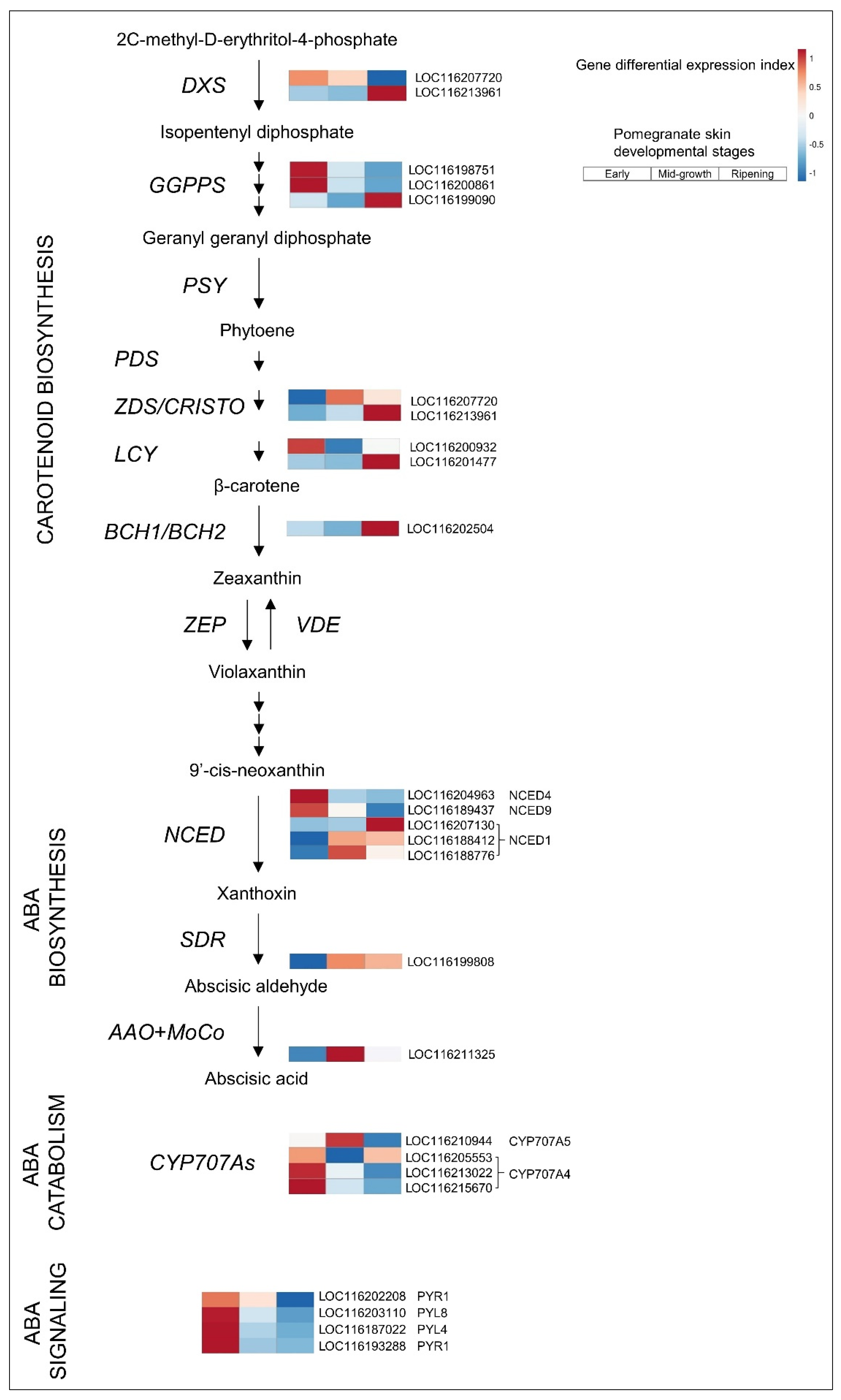
3.2.1. ABA
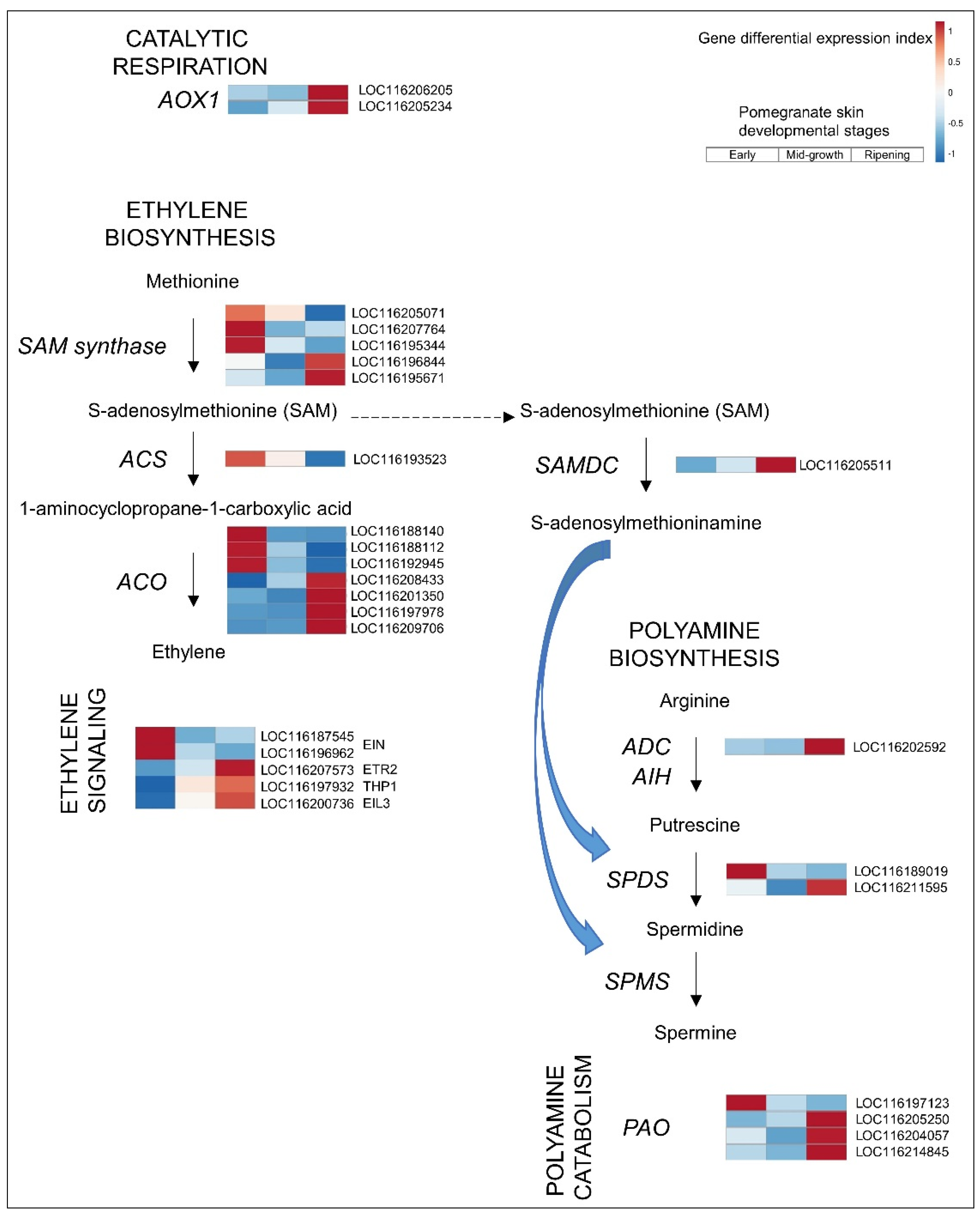
3.2.2. Ethylene
3.2.3. PAs
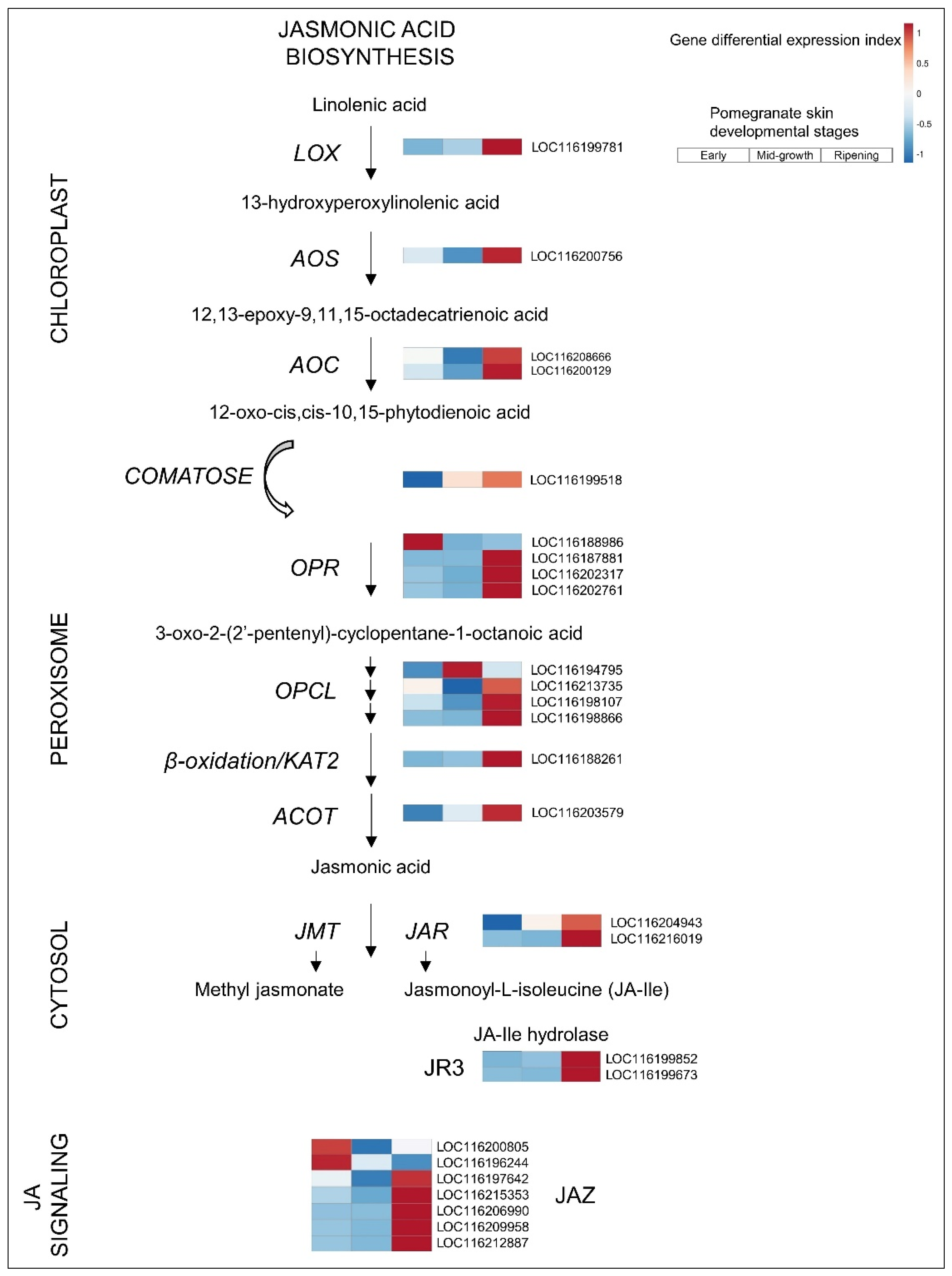
3.2.4. JA
3.2.5. GA, AUXIN, CK, and BR
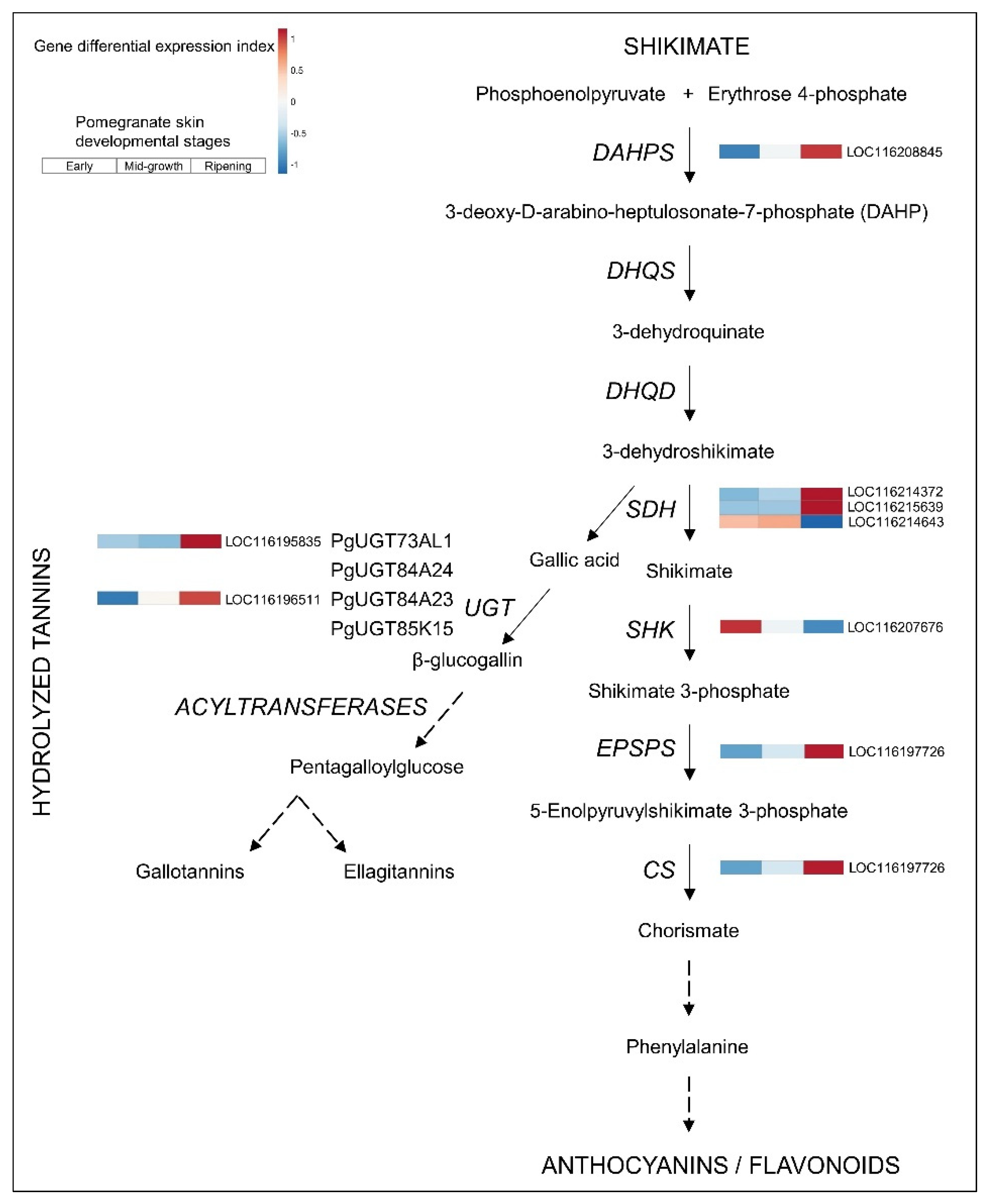
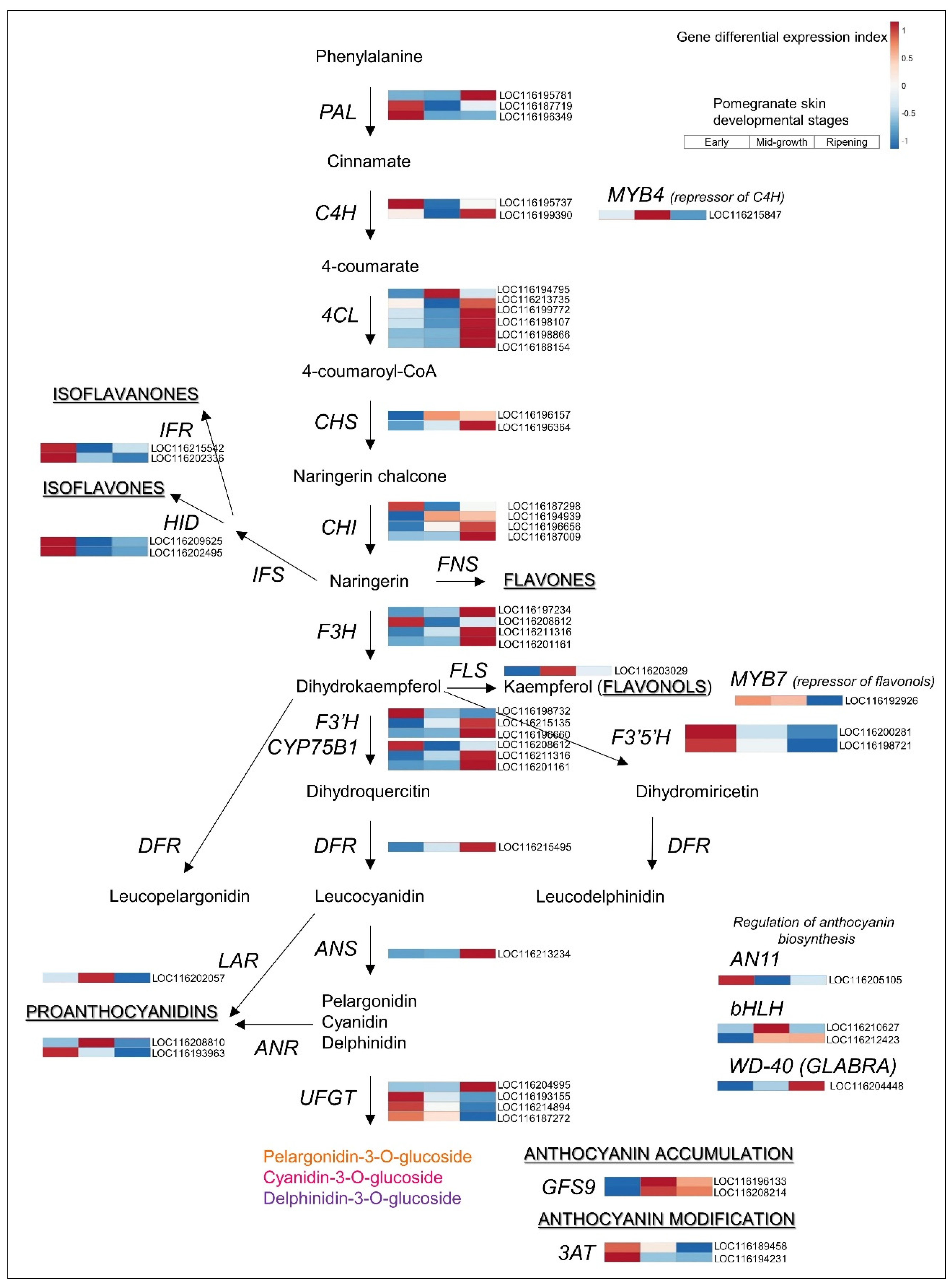
3.3. Upregulated Biosynthetic Pathways of Hydrolyzable Tannins and Anthocyanins
3.4. Strengthening Pomegranate Skin against Cracking: Cutin, Suberin, Wax, and Cell Wall
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Viuda-Martos, M.; Fernández-López, J.; Pérez-Álvarez, J.A. Pomegranate and its many functional components as related to human health: A review. Compr. Rev. Food Sci. Food Saf. 2010, 9, 635–654. [Google Scholar] [CrossRef] [PubMed]
- Holland, D.; Hatib, K.; Bar-Ya’akov, I. Pomegranate: Botany, horticulture, breeding. Hortic. Rev. 2009, 35, 127–191. [Google Scholar]
- Mphahlele, R.R.; Fawole, O.A.; Stander, M.A.; Opara, U.L. Preharvest and postharvest factors influencing bioactive compounds in pomegranate (Punica granatum L.)—A review. Sci. Hortic. 2014, 178, 114–123. [Google Scholar] [CrossRef]
- Singh, B.; Singh, J.P.; Kaur, A.; Singh, N. Antimicrobial potential of pomegranate peel: A review. Int. J. Food Sci. Technol. 2019, 54, 959–965. [Google Scholar] [CrossRef]
- Amir, R.; Borochov-Neori, H.; Tian, L.; Holland, D. The biodiversity of different traits of pomegranate fruit peels from a broad collection of diverse cultivars. Sci. Hortic. 2019, 246, 842–848. [Google Scholar] [CrossRef]
- Yuan, Z.; Fang, Y.; Zhang, T.; Fei, Z.; Han, F.; Liu, C.; Liu, M.; Xiao, W.; Zhang, W.; Wu, S.; et al. The pomegranate (Punica granatum L.) genome provides insights into fruit quality and ovule developmental biology. Plant Biotechnol. J. 2018, 16, 1363–1374. [Google Scholar] [CrossRef] [Green Version]
- Teixeira da Silva, J.A.; Rana, T.S.; Narzary, D.; Verma, N.; Meshram, D.T.; Ranade, S.A. Pomegranate biology and biotechnology: A review. Sci. Hortic. 2013, 160, 85–107. [Google Scholar] [CrossRef]
- Joshi, M.; Schmilovitch, Z.e.; Ginzberg, I. Pomegranate fruit growth and skin characteristics in hot and dry climate. Front. Plant Sci. 2021, 12, 725479. [Google Scholar] [CrossRef]
- Drogoudi, P.; Pantelidis, G.E.; Vekiari, S.A. Physiological disorders and fruit quality attributes in pomegranate: Effects of meteorological parameters, canopy position and acetylsalicylic acid foliar sprays. Front. Plant Sci. 2021, 12, 645547. [Google Scholar] [CrossRef]
- Singh, A.; Shukla, A.K.; Meghwal, P.R. Fruit cracking in pomegranate: Extent, cause, and management—A review. Int. J. Fruit Sci. 2020, 20, S1234–S1253. [Google Scholar] [CrossRef]
- Winkel-Shirley, B. Flavonoid biosynthesis. a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001, 126, 485–493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ben-Simhon, Z.; Judeinstein, S.; Nadler-Hassar, T.; Trainin, T.; Bar-Ya’akov, I.; Borochov-Neori, H.; Holland, D. A pomegranate (Punica granatum L.) WD40-repeat gene is a functional homologue of Arabidopsis TTG1 and is involved in the regulation of anthocyanin biosynthesis during pomegranate fruit development. Planta 2011, 234, 865–881. [Google Scholar] [CrossRef] [PubMed]
- Habashi, R.; Hacham, Y.; Dhakarey, R.; Matityahu, I.; Holland, D.; Tian, L.; Amir, R. Elucidating the role of shikimate dehydrogenase in controlling the production of anthocyanins and hydrolysable tannins in the outer peels of pomegranate. BMC Plant Biol. 2019, 19, 476. [Google Scholar] [CrossRef]
- Fuentes, L.; Figueroa, C.R.; Valdenegro, M. Recent advances in hormonal regulation and cross-talk during non-climacteric fruit development and ripening. Horticulturae 2019, 5, 45. [Google Scholar] [CrossRef] [Green Version]
- Qin, G.; Xu, C.; Ming, R.; Tang, H.; Guyot, R.; Kramer, E.M.; Hu, Y.; Yi, X.; Qi, Y.; Xu, X.; et al. The pomegranate (Punica granatum L.) genome and the genomics of punicalagin biosynthesis. Plant J. 2017, 91, 1108–1128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ono, N.N.; Britton, M.T.; Fass, J.N.; Nicolet, C.M.; Lin, D.; Tian, L. Exploring the transcriptome landscape of pomegranate fruit peel for natural product biosynthetic gene and SSR marker discovery. J. Integr. Plant Biol. 2011, 53, 800–813. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Cao, D.; Li, H.; Zhao, D.; Xue, H.; Niu, J.; Chen, L.; Zhang, F.; Cao, S. Complementary iTRAQ-based proteomic and RNA sequencing-based transcriptomic analyses reveal a complex network regulating pomegranate (Punica granatum L.) fruit peel colour. Sci. Rep. 2018, 8, 12362. [Google Scholar] [CrossRef] [Green Version]
- Ginzberg, I.; Barel, G.; Ophir, R.; Tzin, E.; Tanami, Z.; Muddarangappa, T.; De Jong, W.; Fogelman, E. Transcriptomic profiling of heat-stress response in potato periderm. J. Exp. Bot. 2009, 60, 4411–4421. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2012, 29, 15–21. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gentleman, R.C.; Carey, V.J.; Bates, D.M.; Bolstad, B.; Dettling, M.; Dudoit, S.; Ellis, B.; Gautier, L.; Ge, Y.; Gentry, J. Bioconductor: Open software development for computational biology and bioinformatics. Genome Biol. 2004, 5, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Metsalu, T.; Vilo, J. ClustVis: A web tool for visualizing clustering of multivariate data using Principal Component Analysis and heatmap. Nucleic Acids Res. 2015, 43, W566–W570. [Google Scholar] [CrossRef]
- Buchfink, B.; Xie, C.; Huson, D.H. Fast and sensitive protein alignment using DIAMOND. Nat. Methods 2015, 12, 59–60. [Google Scholar] [CrossRef]
- Conesa, A.; Götz, S.; García-Gómez, J.M.; Terol, J.; Talón, M.; Robles, M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [Green Version]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Daminato, M.; Guzzo, F.; Casadoro, G. A SHATTERPROOF-like gene controls ripening in non-climacteric strawberries, and auxin and abscisic acid antagonistically affect its expression. J. Exp. Bot. 2013, 64, 3775–3786. [Google Scholar] [CrossRef] [Green Version]
- Vrebalov, J.; Pan, I.L.; Arroyo, A.J.M.; McQuinn, R.; Chung, M.; Poole, M.; Rose, J.; Seymour, G.; Grandillo, S.; Giovannoni, J. Fleshy fruit expansion and ripening are regulated by the tomato SHATTERPROOF gene TAGL1. Plant Cell 2009, 21, 3041–3062. [Google Scholar] [CrossRef] [Green Version]
- Ferrándiz, C.; Liljegren, S.J.; Yanofsky, M.F. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 2000, 289, 436–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karlova, R.; Chapman, N.; David, K.; Angenent, G.C.; Seymour, G.B.; de Maagd, R.A. Transcriptional control of fleshy fruit development and ripening. J. Exp. Bot. 2014, 65, 4527–4541. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cherian, S.; Figueroa, C.R.; Nair, H. ‘Movers and shakers’ in the regulation of fruit ripening: A cross-dissection of climacteric versus non-climacteric fruit. J. Exp. Bot. 2014, 65, 4705–4722. [Google Scholar] [CrossRef] [Green Version]
- Chervin, C. Should starch metabolism be a key point of the climacteric vs. Non-climacteric fruit definition? Front. Plant Sci. 2020, 11, 609189. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Li, M.; Liu, B.; Yan, M.; Yu, X.; Zi, H.; Liu, R.; Yamamuro, C. Interlinked regulatory loops of ABA catabolism and biosynthesis coordinate fruit growth and ripening in woodland strawberry. Proc. Natl. Acad. Sci. USA 2018, 115, E11542–E11550. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Lee, J.G.; Hong, Y.; Lee, E.J. Analysis of eight phytohormone concentrations, expression levels of ABA biosynthesis genes, and ripening-related transcription factors during fruit development in strawberry. J. Plant Physiol. 2019, 239, 52–60. [Google Scholar] [CrossRef]
- Jia, H.-F.; Chai, Y.-M.; Li, C.-L.; Lu, D.; Luo, J.-J.; Qin, L.; Shen, Y.-Y. Abscisic acid plays an important role in the regulation of strawberry fruit ripening. Plant Physiol. 2011, 157, 188–199. [Google Scholar] [CrossRef] [Green Version]
- Cramer, G.R.; Ghan, R.; Schlauch, K.A.; Tillett, R.L.; Heymann, H.; Ferrarini, A.; Delledonne, M.; Zenoni, S.; Fasoli, M.; Pezzotti, M. Transcriptomic analysis of the late stages of grapevine (Vitis vinifera cv. Cabernet Sauvignon) berry ripening reveals significant induction of ethylene signaling and flavor pathways in the skin. BMC Plant Biol. 2014, 14, 370. [Google Scholar] [CrossRef] [Green Version]
- Raghavendra, A.S.; Gonugunta, V.K.; Christmann, A.; Grill, E. ABA perception and signalling. Trends Plant Sci. 2010, 15, 395–401. [Google Scholar] [CrossRef]
- Chai, Y.-m.; Jia, H.-f.; Li, C.-l.; Dong, Q.-h.; Shen, Y.-y. FaPYR1 is involved in strawberry fruit ripening. J. Exp. Bot. 2011, 62, 5079–5089. [Google Scholar] [CrossRef] [Green Version]
- Pareek, S.; Valero, D.; Serrano, M. Postharvest biology and technology of pomegranate. J. Sci. Food Agric. 2015, 95, 2360–2379. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, L.; Monsalve, L.; Morales-Quintana, L.; Valdenegro, M.; Martínez, J.-P.; Defilippi, B.G.; González-Agüero, M. Differential expression of ethylene biosynthesis genes in drupelets and receptacle of raspberry (Rubus idaeus). J. Plant Physiol. 2015, 179, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Trainotti, L.; Pavanello, A.; Casadoro, G. Different ethylene receptors show an increased expression during the ripening of strawberries: Does such an increment imply a role for ethylene in the ripening of these non-climacteric fruits? J. Exp. Bot. 2005, 56, 2037–2046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Grimplet, J.; David, K.; Castellarin, S.D.; Terol, J.; Wong, D.C.J.; Luo, Z.; Schaffer, R.; Celton, J.-M.; Talon, M.; et al. Ethylene receptors and related proteins in climacteric and non-climacteric fruits. Plant Sci. 2018, 276, 63–72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hewitt, S.; Dhingra, A. Beyond ethylene: New insights regarding the role of alternative oxidase in the respiratory climacteric. Front. Plant Sci. 2020, 11, 543958. [Google Scholar] [CrossRef]
- Xu, F.; Yuan, S.; Zhang, D.-W.; Lv, X.; Lin, H.-H. The role of alternative oxidase in tomato fruit ripening and its regulatory interaction with ethylene. J. Exp. Bot. 2012, 63, 5705–5716. [Google Scholar] [CrossRef]
- Gao, F.; Mei, X.; Li, Y.; Guo, J.; Shen, Y. Update on the roles of polyamines in fleshy fruit ripening, senescence, and quality. Front. Plant Sci. 2021, 12, 610313. [Google Scholar] [CrossRef]
- Agudelo-Romero, P.; Bortolloti, C.; Pais, M.S.; Tiburcio, A.F.; Fortes, A.M. Study of polyamines during grape ripening indicate an important role of polyamine catabolism. Plant Physiol. Biochem. 2013, 67, 105–119. [Google Scholar] [CrossRef]
- Guo, J.; Wang, S.; Yu, X.; Dong, R.; Li, Y.; Mei, X.; Shen, Y. Polyamines regulate strawberry fruit ripening by abscisic acid, auxin, and ethylene. Plant Physiol. 2018, 177, 339–351. [Google Scholar] [CrossRef] [Green Version]
- Garrido-Bigotes, A.; Figueroa, P.M.; Figueroa, C.R. Jasmonate metabolism and its relationship with abscisic acid during strawberry fruit development and ripening. J. Plant Growth Regul. 2018, 37, 101–113. [Google Scholar] [CrossRef]
- Jia, H.; Zhang, C.; Pervaiz, T.; Zhao, P.; Liu, Z.; Wang, B.; Wang, C.; Zhang, L.; Fang, J.; Qian, J. Jasmonic acid involves in grape fruit ripening and resistant against Botrytis cinerea. Funct. Integr. Genom. 2016, 16, 79–94. [Google Scholar] [CrossRef] [PubMed]
- Chini, A.; Fonseca, S.; Fernández, G.; Adie, B.; Chico, J.M.; Lorenzo, O.; García-Casado, G.; López-Vidriero, I.; Lozano, F.M.; Ponce, M.R.; et al. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 2007, 448, 666–671. [Google Scholar] [CrossRef] [PubMed]
- Woldemariam, M.G.; Onkokesung, N.; Baldwin, I.T.; Galis, I. Jasmonoyl-l-isoleucine hydrolase 1 (JIH1) regulates jasmonoyl-l-isoleucine levels and attenuates plant defenses against herbivores. Plant J. 2012, 72, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Symons, G.M.; Davies, C.; Shavrukov, Y.; Dry, I.B.; Reid, J.B.; Thomas, M.R. Grapes on Steroids. Brassinosteroids Are Involved in Grape Berry Ripening. Plant Physiol. 2005, 140, 150–158. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Llorca, M.; Muñoz, P.; Müller, M.; Munné-Bosch, S. Biosynthesis, metabolism and function of auxin, salicylic acid and melatonin in climacteric and non-climacteric fruits. Front. Plant Sci. 2019, 10, 136. [Google Scholar] [CrossRef] [Green Version]
- Harel-Beja, R.; Tian, L.; Freilich, S.; Habashi, R.; Borochov-Neori, H.; Lahav, T.; Trainin, T.; Doron-Faigenboim, A.; Ophir, R.; Bar-Ya’akov, I.; et al. Gene expression and metabolite profiling analyses of developing pomegranate fruit peel reveal interactions between anthocyanin and punicalagin production. Tree Genet. Genomes 2019, 15, 22. [Google Scholar] [CrossRef]
- Tzulker, R.; Glazer, I.; Bar-Ilan, I.; Holland, D.; Aviram, M.; Amir, R. Antioxidant activity, polyphenol content, and related compounds in different fruit juices and homogenates prepared from 29 different pomegranate accessions. J. Agric. Food Chem. 2007, 55, 9559–9570. [Google Scholar] [CrossRef]
- Niemetz, R.; Gross, G.G. Enzymology of gallotannin and ellagitannin biosynthesis. Phytochemistry 2005, 66, 2001–2011. [Google Scholar] [CrossRef]
- Ono, N.N.; Qin, X.; Wilson, A.E.; Li, G.; Tian, L. Two UGT84 family glycosyltransferases catalyze a critical reaction of hydrolyzable tannin biosynthesis in pomegranate (Punica granatum). PLoS ONE 2016, 11, e0156319. [Google Scholar] [CrossRef]
- Du, C.; Wang, P.; Francis, F. Anthocyanins of pomegranate, Punica granatum. J. Food Sci. 1975, 40, 417–418. [Google Scholar] [CrossRef]
- Gil, M.I.; García-Viguera, C.; Artés, F.; Tomás-Barberán, F.A. Changes in pomegranate juice pigmentation during ripening. J. Sci. Food Agric. 1995, 68, 77–81. [Google Scholar] [CrossRef]
- Fanali, C.; Belluomo, M.G.; Cirilli, M.; Cristofori, V.; Zecchini, M.; Cacciola, F.; Russo, M.; Muleo, R.; Dugo, L. Antioxidant activity evaluation and HPLC-photodiode array/MS polyphenols analysis of pomegranate juice from selected italian cultivars: A comparative study. Electrophoresis 2016, 37, 1947–1955. [Google Scholar] [CrossRef] [PubMed]
- García-Pastor, M.E.; Zapata, P.J.; Castillo, S.; Martínez-Romero, D.; Guillén, F.; Valero, D.; Serrano, M. The effects of salicylic acid and its derivatives on increasing pomegranate fruit quality and bioactive compounds at harvest and during storage. Front. Plant Sci. 2020, 11, 668. [Google Scholar] [CrossRef] [PubMed]
- Baghel, R.S.; Keren-Keiserman, A.; Ginzberg, I. Metabolic changes in pomegranate fruit skin following cold storage promote chilling injury of the peel. Sci. Rep. 2021, 11, 1–13. [Google Scholar] [CrossRef]
- Philippe, G.; Sørensen, I.; Jiao, C.; Sun, X.; Fei, Z.; Domozych, D.S.; Rose, J.K.C. Cutin and suberin: Assembly and origins of specialized lipidic cell wall scaffolds. Curr. Opin. Plant Biol. 2020, 55, 11–20. [Google Scholar] [CrossRef]
- Ursache, R.; De Jesus Vieira Teixeira, C.; Dénervaud Tendon, V.; Gully, K.; De Bellis, D.; Schmid-Siegert, E.; Grube Andersen, T.; Shekhar, V.; Calderon, S.; Pradervand, S.; et al. GDSL-domain proteins have key roles in suberin polymerization and degradation. Nat. Plants 2021, 7, 353–364. [Google Scholar] [CrossRef]
- Woolfson, K.N.; Esfandiari, M.; Bernards, M.A. Suberin biosynthesis, assembly, and regulation. Plants 2022, 11, 555. [Google Scholar] [CrossRef]
- Zhao, Q.; Dixon, R.A. Transcriptional networks for lignin biosynthesis: More complex than we thought? Trends Plant Sci. 2011, 16, 227–233. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.-L.; Chen, S.-Y.; Liu, G.-T.; Jia, X.-Y.; Haq, S.u.; Deng, Z.-J.; Luo, D.-X.; Li, R.; Gong, Z.-H. Morphological, physiochemical, and transcriptome analysis and CaEXP4 identification during pepper (Capsicum annuum L.) fruit cracking. Sci. Hortic. 2022, 297, 110982. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ginzberg, I.; Faigenboim, A. Ripening of Pomegranate Skin as Revealed by Developmental Transcriptomics. Cells 2022, 11, 2215. https://doi.org/10.3390/cells11142215
Ginzberg I, Faigenboim A. Ripening of Pomegranate Skin as Revealed by Developmental Transcriptomics. Cells. 2022; 11(14):2215. https://doi.org/10.3390/cells11142215
Chicago/Turabian StyleGinzberg, Idit, and Adi Faigenboim. 2022. "Ripening of Pomegranate Skin as Revealed by Developmental Transcriptomics" Cells 11, no. 14: 2215. https://doi.org/10.3390/cells11142215
APA StyleGinzberg, I., & Faigenboim, A. (2022). Ripening of Pomegranate Skin as Revealed by Developmental Transcriptomics. Cells, 11(14), 2215. https://doi.org/10.3390/cells11142215