Large-Scale Profiling on lncRNAs in Human Platelets: Correlation with Platelet Reactivity
Abstract
:1. Introduction
2. Materials and Methods
2.1. Healthy Participants, Patients and Blood Samples
2.2. Platelet Aggregation Assay
2.3. Platelet Isolation, Purification and RNA Extraction
2.4. Genome-Wide Large-Scale Next-Generation Deep Sequencing of lncRNAs and mRNAs
2.5. Quality Control and RNA-Seq Data Processing
2.6. Gene Ontology (GO) and Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis
2.7. LncRNAs Target Gene Prediction
2.8. Quantitative Reverse Transcription-Quantitative Polymerase Chain Reaction (qRT-qPCR) of Selected lncRNAs
2.9. Statistics
3. Results
3.1. lncRNAs Are Highly Expressed in Human Platelets
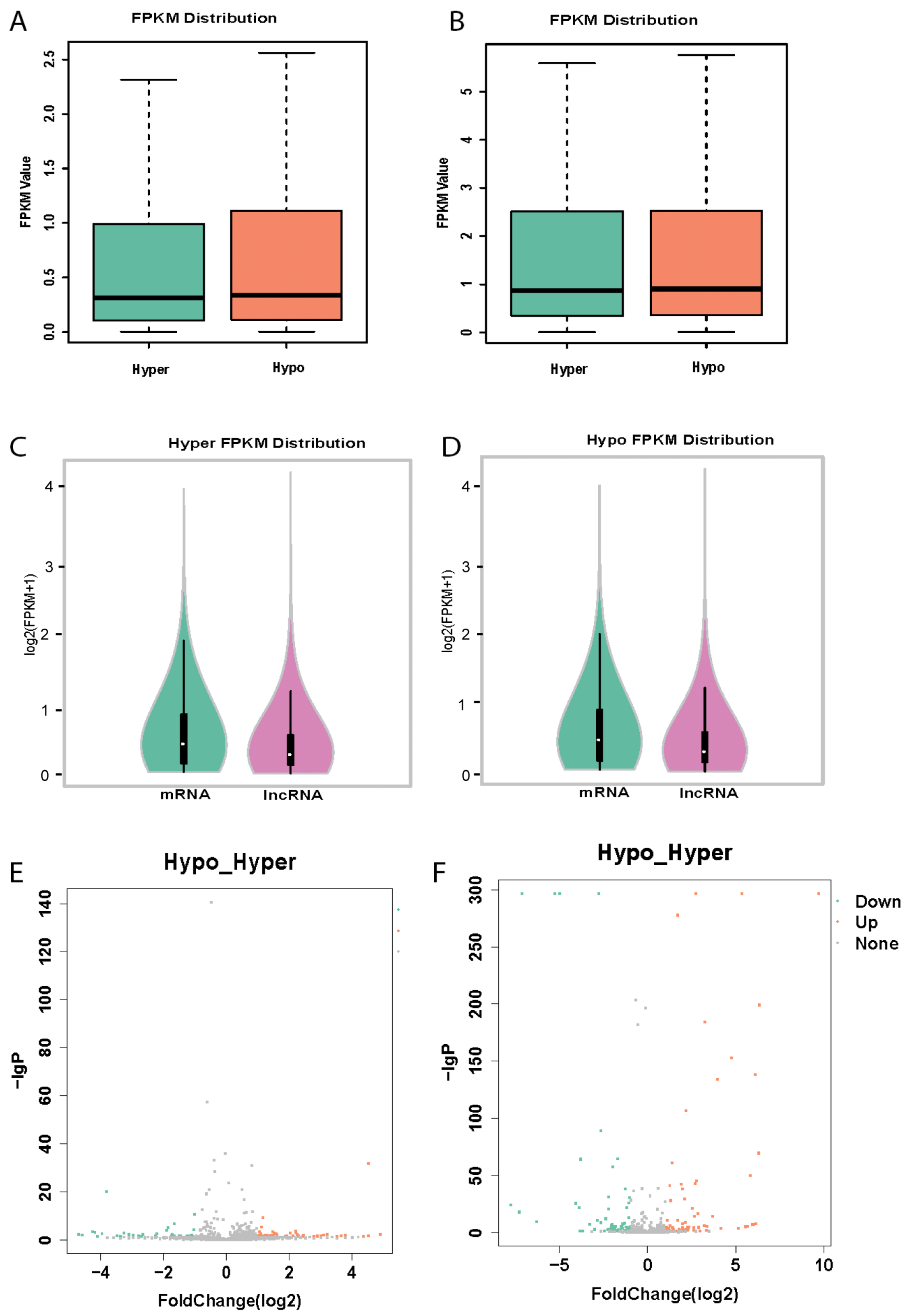
3.2. Platelet lncRNAs Are Differentially Expressed in Hyperreactive and Hyporeactive Platelets
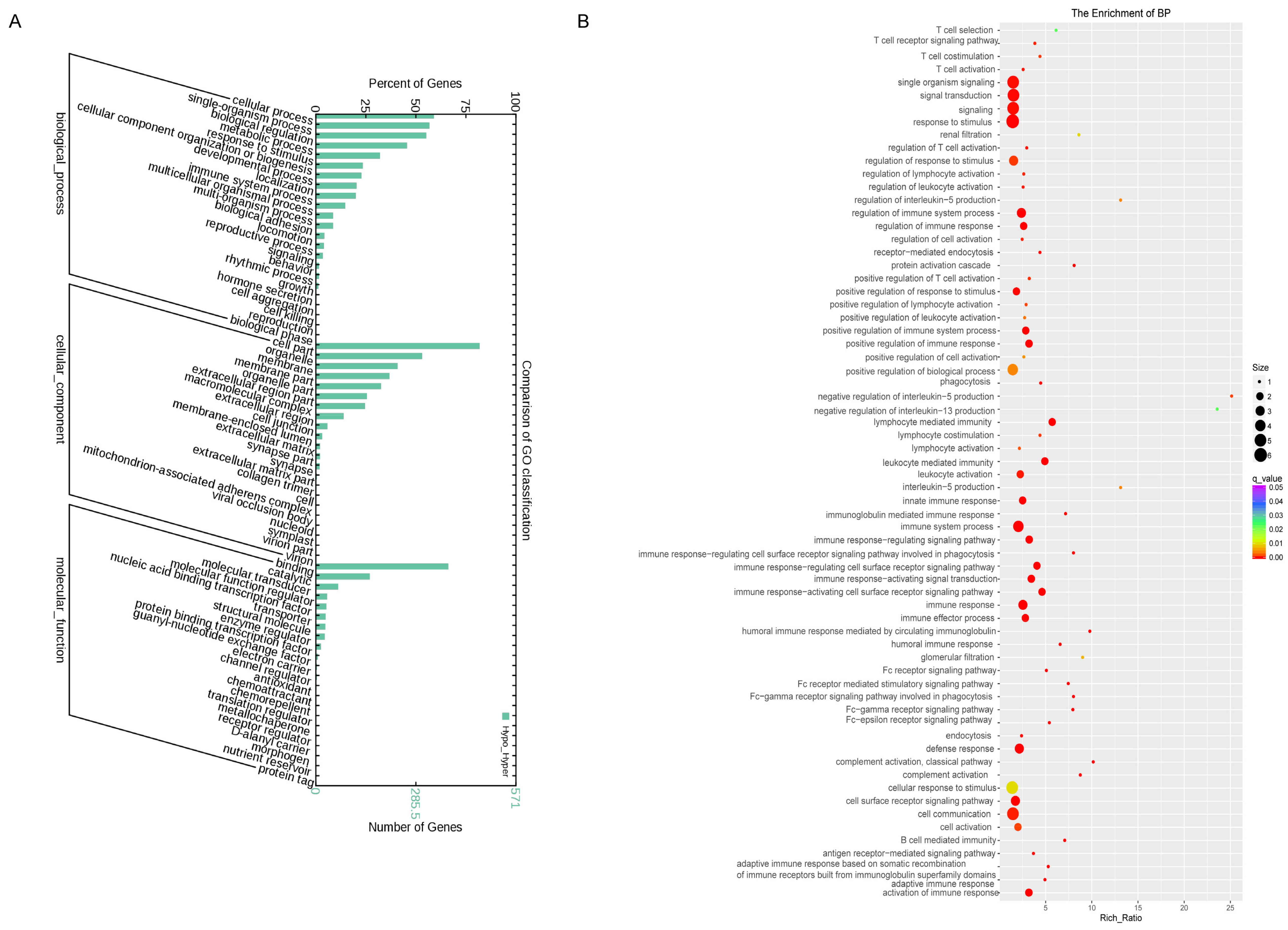
3.3. Gene Ontology Enrichment Analysis of Differentially Expressed Platelets mRNAs
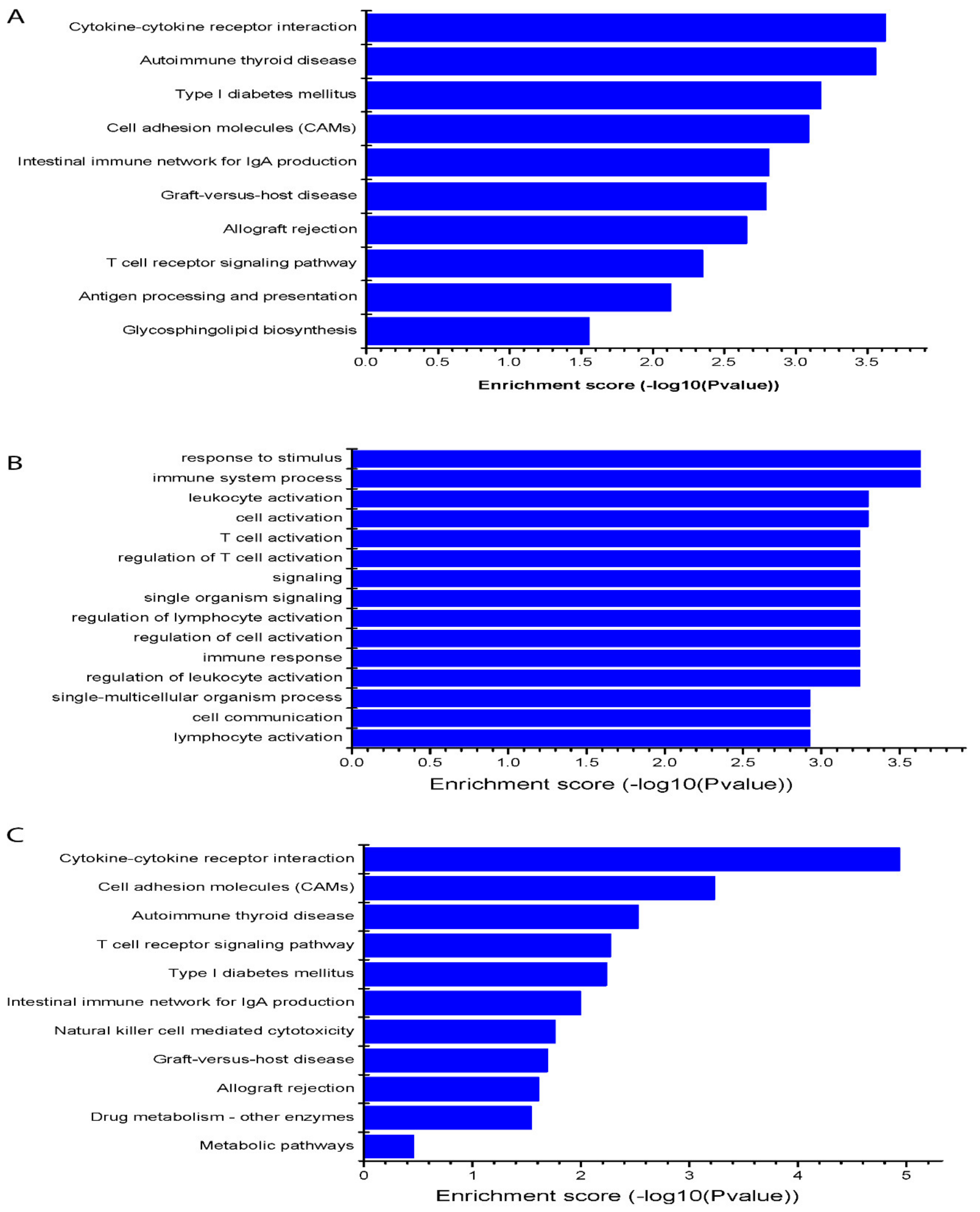
3.4. KEGG Target Pathway of the Differentially Expressed Platelets lncRNAs
3.5. LncRNA Regulated-mRNAs via Target Gene Prediction Are Involved in Platelets Aggregation and Other Functions
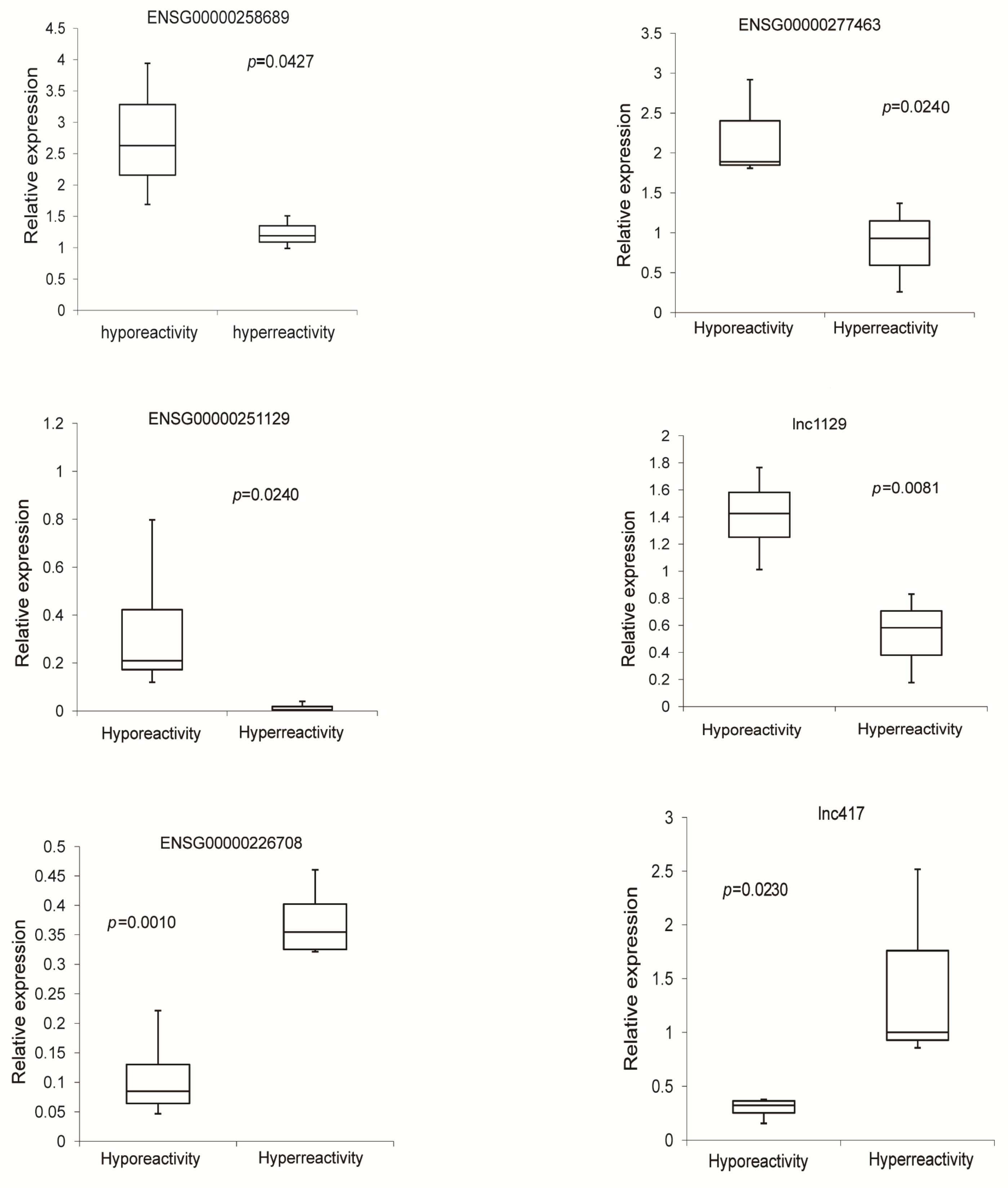
3.6. Validation of Differentially Expressed lncRNAs in Hyperreactive and Hyporeactive Platelets
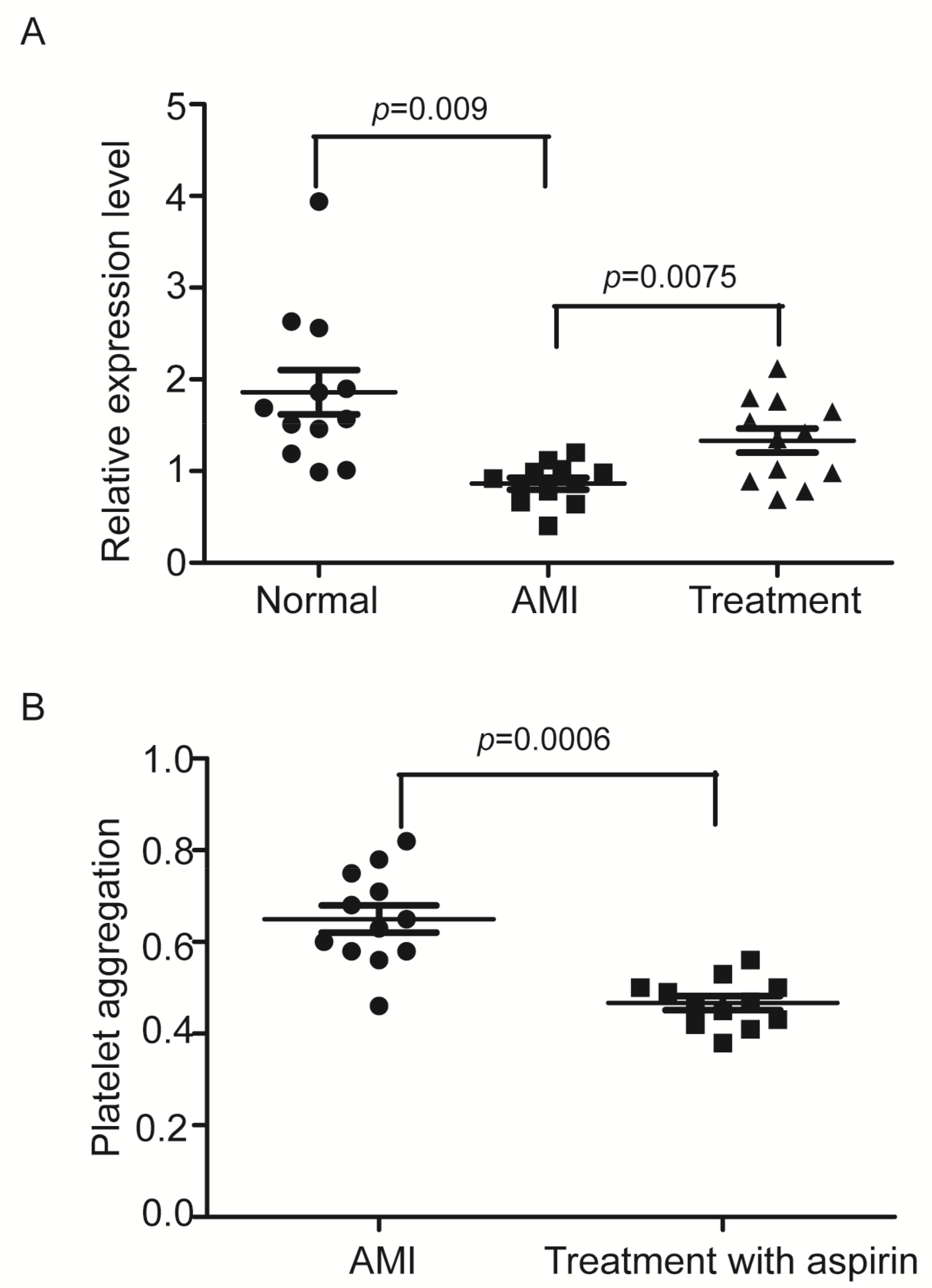
3.7. The Expression of ENSG00000258689, a Down-Regulated lncRNA in Hyperreactive Platelets Is Significantly Decreased in Platelets from Patients with AMI
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Santosh, B.; Varshney, A.; Yadava, P.K. Non-coding RNAs: Biological functions and applications. Cell Biochem. Funct. 2015, 33, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Philippen, L.E.; Dirkx, E.; da Costa-Martins, P.A.; De Windt, L.J. Non-coding RNA in control of gene regulatory programs in cardiac development and disease. J. Mol. Cell Cardiol. 2015, 89, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Jiang, X.; Ning, Q. The emerging roles of long noncoding RNAs in common cardiovascular diseases. Hypertens. Res. 2015, 38, 375–379. [Google Scholar] [CrossRef] [PubMed]
- Uchida, S.; Dimmeler, S. Long noncoding RNAs in cardiovascular diseases. Circ. Res. 2015, 116, 737–750. [Google Scholar] [CrossRef] [Green Version]
- Archer, K.; Broskova, Z.; Bayoumi, A.S.; Teoh, J.P.; Davila, A.; Tang, Y.; Su, H.; Kim, I.M. Long non-coding RNAs as master regulators in cardiovascular disease. Int. J. Mol. Sci. 2015, 16, 23651–23667. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.; Kraus, W.L. From discovery to function: The expanding roles of long noncoding RNAs in physiology and disease. Endocr. Rev. 2015, 36, 25–64. [Google Scholar] [CrossRef] [Green Version]
- Gremmel, T.; Frelinger, A.L., III; Michelson, A.D. Platelet Physiology. Semin. Thromb Hemost. 2016, 42, 191–204. [Google Scholar]
- Lindsay, C.R.; Edelstein, L.C. MicroRNAs in Platelet Physiology and Function. Semin. Thromb Hemost. 2016, 42, 215–222. [Google Scholar] [CrossRef]
- Bijak, M.; Dzieciol, M.; Rywaniak, J.; Saluk, J.; Zielinska, M. Platelets miRNA as a Prediction Marker of Thrombotic Episodes. Dis. Markers 2016, 2872507. [Google Scholar] [CrossRef]
- Gutmann, C.; Joshi, A.; Zampetaki, A.; Mayr, M. The Landscape of Coding and Noncoding RNAs in Platelets. Antioxid. Redox Signal. 2021, 34, 1200–1216. [Google Scholar] [CrossRef]
- Yan, S.; Liu, X.; Ke, X.; Xian, Z.; Peng, C.; Wang, X.; Chen, M. Screening on platelet LncRNA expression profile discloses novel residual platelet reactivity biomarker. Int. J. Lab. Hematol. 2020, 42, 661–668. [Google Scholar] [PubMed]
- Zhou, M.; Gao, M.; Luo, Y.; Gui, R.; Ji, H. Long non-coding RNA metallothionein 1 pseudogene 3 promotes p2y12 expression by sponging miR-126 to activate platelet in diabetic animal model. Platelets 2019, 30, 452–459. [Google Scholar] [PubMed]
- Kondkar, A.A.; Bray, M.S.; Leal, S.M.; Nagalla, S.; Liu, D.J.; Jin, Y.; Dong, J.F.; Ren, Q.; Whiteheart, S.W.; Shaw, C.; et al. VAMP8/endobrevin is overexpressed in hyperreactive human platelets: Suggested role for platelet microRNA. J. Thromb. Haemost. 2010, 8, 369–378. [Google Scholar] [PubMed] [Green Version]
- Yee, D.L.; Sun, C.W.; Bergeron, A.L.; Dong, J.F.; Bray, P.F. Aggregometry detects platelet hyperreactivity in healthy individuals. Blood 2005, 106, 2723–2729. [Google Scholar] [PubMed] [Green Version]
- Wei, S.; Wang, H.; Zhang, G.; Lu, Y.; An, X.; Ren, S.; Wang, Y.; Chen, Y.; White, J.G.; Zhang, C.; et al. Platelet IκB kinase-β deficiency increases mouse arterial neointima formation via delayed glycoprotein Ibα shedding. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 241–248. [Google Scholar]
- Shan, Z.; Qin, S.; Li, W.; Wu, W.; Yang, J.; Chu, M.; Li, X.; Huo, Y.; Schaer, G.L.; Wang, S.; et al. An Endocrine Genetic Signal Between Blood Cells and Vascular Smooth Muscle Cells: Role of MicroRNA-223 in Smooth Muscle Function and Atherogenesis. J. Am. Coll. Cardiol. 2015, 65, 2526–2537. [Google Scholar]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar]
- Wang, Q.; Wang, N.; Cai, R.; Zhao, F.; Xiong, Y.; Li, X.; Wang, A.; Lin, P.; Jin, Y. Genome-wide analysis and functional prediction of long non-coding RNAs in mouse uterus during the implantation window. Oncotarget 2017, 8, 84360–84372. [Google Scholar]
- Ibeagha-Awemu, E.M.; Li, R.; Dudemaine, P.L.; Do, D.N.; Bissonnette, N. Transcriptome Analysis of Long Non-Coding RNA in the Bovine Mammary Gland Following Dietary Supplementation with Linseed Oil and Safflower Oil. Int. J. Mol. Sci. 2018, 19, 3610. [Google Scholar]
- Zhao, P.; Liu, S.; Zhong, Z.; Jiang, T.; Weng, R.; Xie, M.; Yang, S.; Xia, X. Analysis of expression profiles of long noncoding RNAs and mRNAs in brains of mice infected by rabies virus by RNA sequencing. Sci. Rep. 2018, 8, 11858. [Google Scholar]
- Chatterjee, M.; Rath, D.; Gawaz, M. Role of chemokine receptors CXCR4 and CXCR7 for platelet function. Biochem. Soc. Trans. 2015, 43, 720–726. [Google Scholar] [PubMed]
- Schubert, P.; Coupland, D.; Nombalais, M.M.; Walsh, G.; Devine, D.V. RhoA/ROCK signaling contributes to sex differences in the activation of human platelets. Thromb. Res. 2016, 139, 50–55. [Google Scholar] [PubMed]
- Suresh, A.; Sanji, N.; Kamath, P.M.; Devendrappa, S.L.; Hanumanthareddy, S.G.; Maniyar, I.; Rudrappa, S.S. A Pilot Study on the Effect of Angiotensin Receptor Blockers on Platelet Aggregation in Hypertensive Patients—A Prospective Observational Study. J. Clin. Diagn. Res. 2016, 10, FC14–FC16. [Google Scholar] [PubMed]
- Zhi, H.; Rauova, L.; Hayes, V.; Gao, C.; Boylan, B.; Newman, D.K.; McKenzie, S.E.; Cooley, B.C.; Poncz, M.; Newman, P.J. Cooperative integrin/ITAM signaling in platelets enhances thrombus formation in vitro and in vivo. Blood 2013, 121, 1858–1867. [Google Scholar] [PubMed] [Green Version]
- Ali, R.A.; Wuescher, L.M.; Worth, R.G. Platelets: Essential components of the immune system. Curr. Trends Immunol. 2015, 16, 65–78. [Google Scholar]
- Xu, X.R.; Carrim, N.; Neves, M.A.; McKeown, T.; Stratton, T.W.; Coelho, R.M.; Lei, X.; Chen, P.; Xu, J.; Dai, X.; et al. Platelets and platelet adhesion molecules: Novel mechanisms of thrombosis and anti-thrombotic therapies. Thromb. J. 2016, 14, S29. [Google Scholar]
- Heililahong, H.; Jin, P.; Lei, H.; Gu, H.; Qian, B.; Wang, X.; Dai, J.; Cai, X. Whole transcriptome analysis of platelet concentrates during storage. Blood Transfus. 2022. [Google Scholar] [CrossRef]
- Breet, N.J.; van Werkum, J.W.; Bouman, H.J.; Kelder, J.C.; Ruven, H.J.; Bal, E.T.; Deneer, V.H.; Harmsze, A.M.; van der Heyden, J.A.; Rensing, B.J.; et al. Comparison of platelet function tests in predicting clinical outcome in patients undergoing coronary stent implantation. JAMA 2010, 303, 754–762. [Google Scholar]
- Edelstein, L.C.; Simon, L.M.; Montoya, R.T.; Holinstat, M.; Chen, E.S.; Bergeron, A.; Kong, X.; Nagalla, S.; Mohandas, N.; Cohen, D.E.; et al. Racial differences in human platelet PAR4 reactivity reflect expression of PCTP and miR-376c. Nat. Med. 2013, 19, 1609–1616. [Google Scholar]
- Ye, B.; Li, F.; Chen, M.; Weng, Y.; Qi, C.; Xie, Y.; Zhang, Q.; Ding, H.; Zhang, J.; Gao, X. A panel of platelet-associated circulating long non-coding RNAs as potential biomarkers for colorectal cancer. Genomics 2022, 114, 31–37. [Google Scholar]
| Name | Gene Name | Biotype | Position | Length (bp) |
|---|---|---|---|---|
| ENSG00000281344 | HELLPAR | macro_lncRNA | chr12:102197585-102402596:+ | 205,012 |
| ENSG00000245532 | NEAT1 | lincRNA | chr11:65422774-65445540:+ | 22,679 |
| ENSG00000251562 | MALAT1 | lincRNA | chr11:65497762-65506516:+ | 8708 |
| ENSG00000253352 | TUG1 | antisense | chr22:30970677-30979395:+ | 5673 |
| ENSG00000260032 | LINC00657 | lincRNA | chr20:36045622-36050960:− | 5339 |
| ENSG00000261026 | CTD-3247F14.2 | sense_overlapping | chr8:22679013-22684009:− | 4997 |
| ENSG00000215458 | AATBC | antisense | chr21:43805758-43812567:− | 4598 |
| ENSG00000225733 | FGD5-AS1 | antisense | chr3:14920347-14948424:− | 3792 |
| ENSG00000271614 | LINC00936 | lincRNA | chr12:89708959-89712590:+ | 3632 |
| ENSG00000231721 | LINC-PINT | antisense | chr7:130941760-131110176:− | 3505 |
| ENSG00000231607 | DLEU2 | antisense | chr13:49982552-50125720:− | 3068 |
| ENSG00000251022 | THAP9-AS1 | antisense | chr4:82893009-82900960:− | 2770 |
| ENSG00000227165 | WDR11-AS1 | antisense | chr10:120761812-120851345:− | 2500 |
| ENSG00000270055 | CTD-3092A11.2 | sense_intronic | chr15:30487963-30490313:+ | 2351 |
| ENSG00000265148 | BZRAP1-AS1 | antisense | chr17:58325450-58415766:+ | 2337 |
| ENSG00000250334 | LINC00989 | lincRNA | chr4:79492416-79576460:+ | 2080 |
| ENSG00000254614 | AP003068.23 | antisense | chr11:65177606-65181834:− | 1662 |
| ENSG00000234883 | MIR155HG | lincRNA | chr21:25561909-25575168:+ | 1600 |
| ENSG00000236304 | AP001189.4 | antisense | chr11:76657056-76663866:+ | 1426 |
| ENSG00000229124 | VIM-AS1 | antisense | chr10:17214239-17229985:− | 1351 |
| ENSG00000272053 | RP11-367G6.3 | lincRNA | chr6:25014952-25042170:− | 1030 |
| ENSG00000253535 | RP11-624C23.1 | antisense | chr8:24295814-24912073:− | 741 |
| ENSG00000263934 | SNORD3A | snoRNA | chr17:19188016-19188714:+ | 699 |
| ENSG00000237781 | RP11-54A4.2 | antisense | chr1:150548562-150557724:− | 665 |
| ENSG00000262202 | RP11-160E2.6 | lincRNA | chr17:19112000-19112636:− | 637 |
| ENSG00000259330 | INAFM2 | antisense | chr15:40325216-40326715:+ | 594 |
| ENSG00000254281 | KB-1507C5.4 | lincRNA | chr8:102978785-103000184:+ | 563 |
| ENSG00000238201 | AC114752.3 | lincRNA | chr2:64338067-64341647:− | 404 |
| ENSG00000276216 | CH17-373J23.1 | lincRNA | chr1:145281116-145281462:+ | 347 |
| ENSG00000273338 | RP11-386I14.4 | antisense | chr1:78004346-78004554:− | 209 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, Y.; Liu, R.; Xia, X.; Xing, L.; Jiang, J.; Bian, W.; Zhang, W.; Wang, C.; Zhang, C. Large-Scale Profiling on lncRNAs in Human Platelets: Correlation with Platelet Reactivity. Cells 2022, 11, 2256. https://doi.org/10.3390/cells11142256
Sun Y, Liu R, Xia X, Xing L, Jiang J, Bian W, Zhang W, Wang C, Zhang C. Large-Scale Profiling on lncRNAs in Human Platelets: Correlation with Platelet Reactivity. Cells. 2022; 11(14):2256. https://doi.org/10.3390/cells11142256
Chicago/Turabian StyleSun, Yeying, Rongrong Liu, Xiangwen Xia, Luchuan Xing, Jing Jiang, Weihua Bian, Wendy Zhang, Chunhua Wang, and Chunxiang Zhang. 2022. "Large-Scale Profiling on lncRNAs in Human Platelets: Correlation with Platelet Reactivity" Cells 11, no. 14: 2256. https://doi.org/10.3390/cells11142256
APA StyleSun, Y., Liu, R., Xia, X., Xing, L., Jiang, J., Bian, W., Zhang, W., Wang, C., & Zhang, C. (2022). Large-Scale Profiling on lncRNAs in Human Platelets: Correlation with Platelet Reactivity. Cells, 11(14), 2256. https://doi.org/10.3390/cells11142256






