A Genome-Wide CRISPR Screen Identifies Factors Regulating Pluripotency Exit in Mouse Embryonic Stem Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. NANOG-GFP Reporter mESCs
2.3. Establishment of Single-Gene Knockouts in NANOG-GFP mESCs
2.4. Library Construction and Screening
2.5. Quantitative RT-RCR Analysis
2.6. RNA-seq
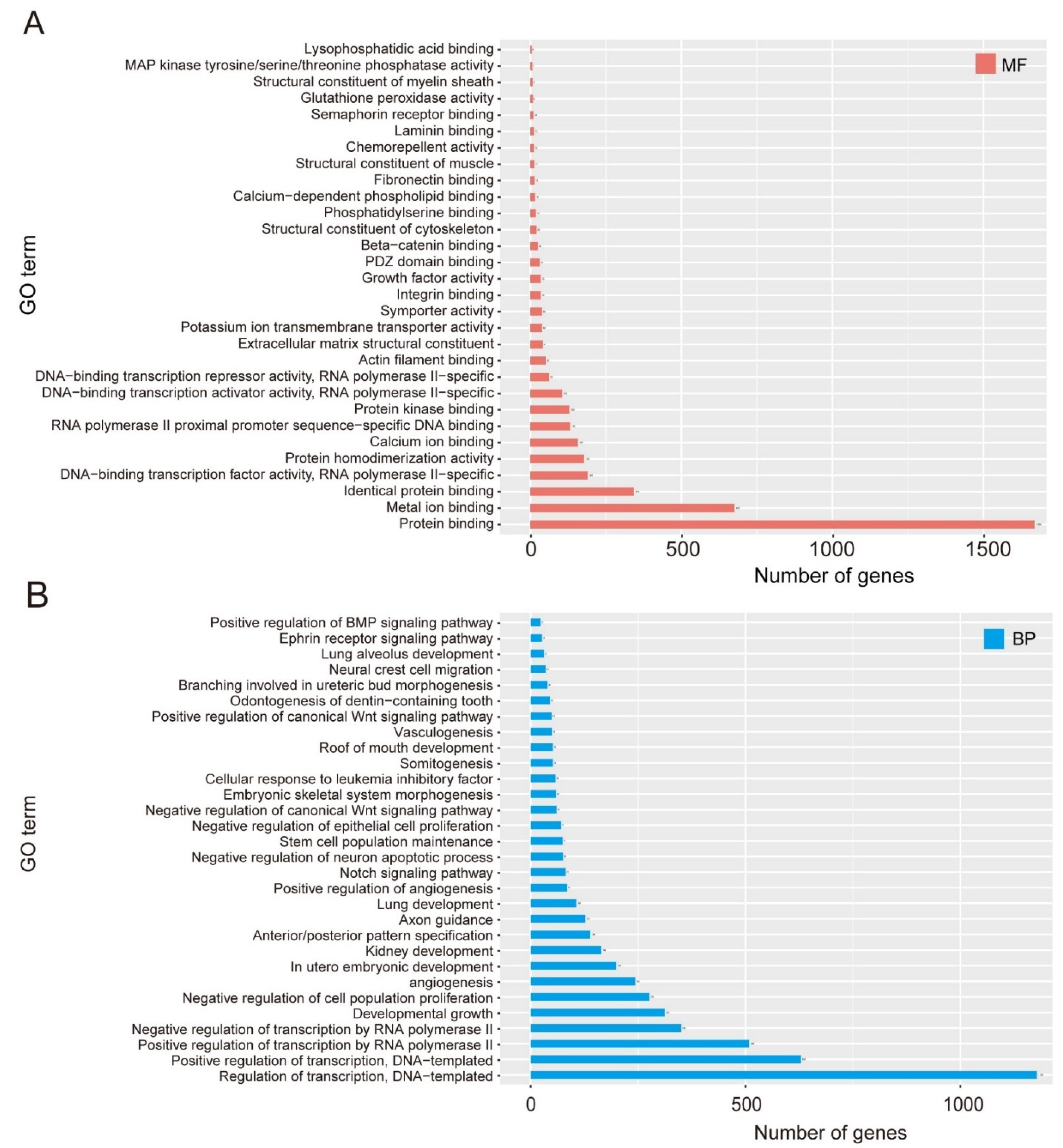
2.7. GO Enrichment and KEGG Analysis
2.8. Flow Cytometry
2.9. 8-Cell Embryo Chimerism Experiment
3. Results
3.1. CRISPR Library Screening of NANOG-GFP mESCs in a Differentiation Medium
3.2. Candidate Genes in the Screen and Their Function in Exit from Pluripotency
3.3. Zfp771 Function Is Required for Exit from Pluripotency
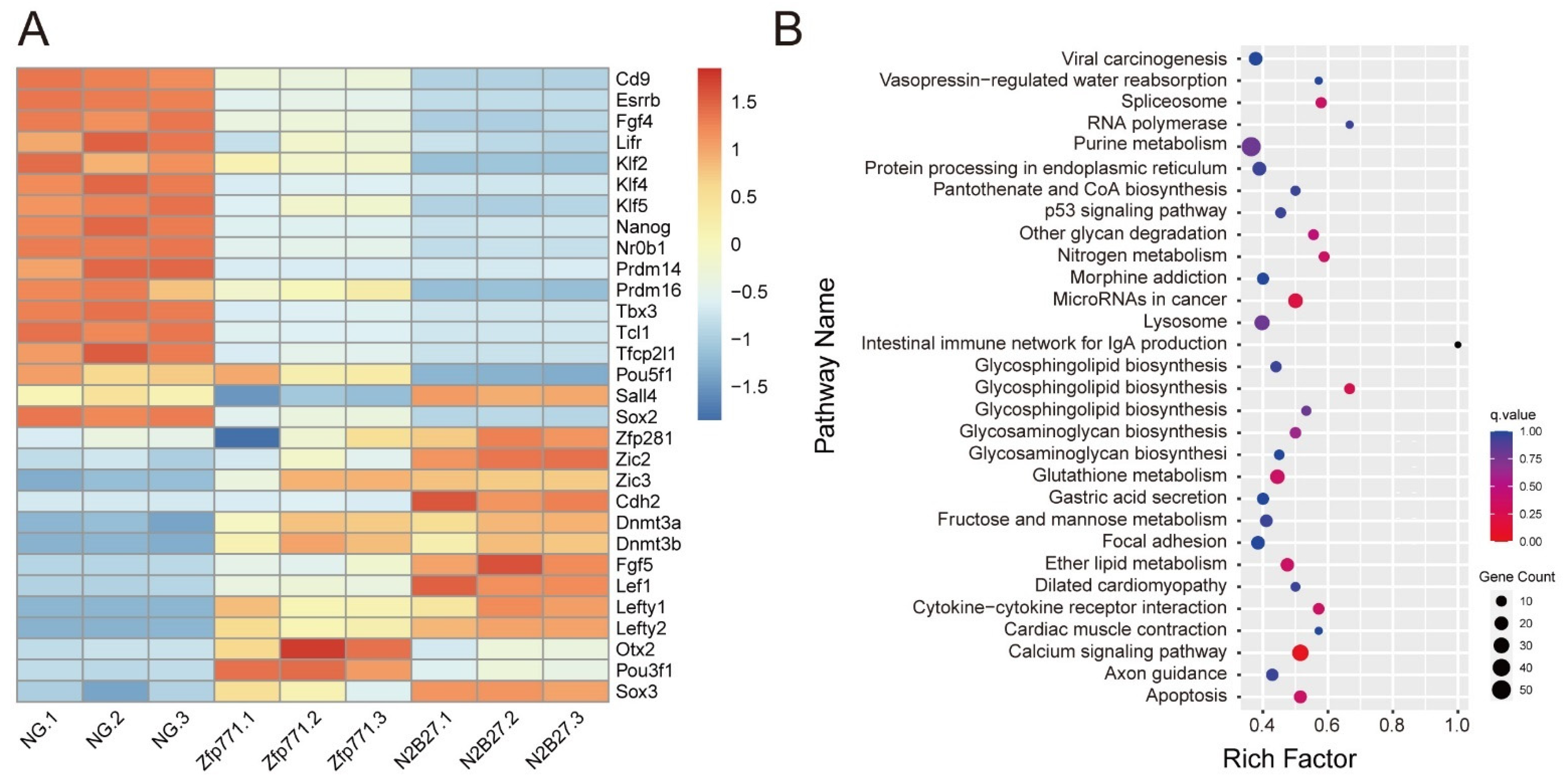
3.4. Zfp771-KO Cell Transcriptomes Reveal Differences in Pluripotency Gene Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ESCs | Embryonic stem cells |
| mESCs | Mouse embryonic stem cells |
| PB | piggyBac |
| GFP | Green fluorescent protein |
| LIF | Leukemia inhibitory factor |
| 2i | 2 inhibitors (CHIR99021, PD0325901) |
| KO | Knockout |
| cDNA | Complementary DNA |
| qPCR | Quantitative RT-PCR |
| GFP+ | GFP-positive |
| GO | Gene Ontology |
References
- Young, R.A. Control of the embryonic stem cell state. Cell 2011, 144, 940–954. [Google Scholar] [CrossRef] [Green Version]
- Capecchi, M.R. Gene targeting in mice: Functional analysis of the mammalian genome for the twenty-first century. Nat. Rev. Genet. 2005, 6, 507–512. [Google Scholar] [CrossRef]
- Nichols, J.; Smith, A. Pluripotency in the embryo and in culture. Cold Spring Harb. Perspect Biol. 2012, 4, a008128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hackett, J.A.; Surani, M.A. Regulatory principles of pluripotency: From the ground state up. Cell Stem. Cell 2014, 15, 416–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, S.; Liu, D.; Ying, Q.L. Signaling pathways in induced naive pluripotency. Curr. Opin. Genet. Dev. 2014, 28, 10–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Do, D.V.; Ueda, J.; Messerschmidt, D.M.; Lorthongpanich, C.; Zhou, Y.; Feng, B.; Guo, G.; Lin, P.J.; Hossain, M.Z.; Zhang, W.; et al. A genetic and developmental pathway from STAT3 to the OCT4-NANOG circuit is essential for maintenance of ICM lineages in vivo. Genes Dev. 2013, 27, 1378–1390. [Google Scholar] [CrossRef] [Green Version]
- Hirai, H.; Karian, P.; Kikyo, N. Regulation of embryonic stem cell self-renewal and pluripotency by leukaemia inhibitory factor. Biochem. J. 2011, 438, 11–23. [Google Scholar] [CrossRef] [Green Version]
- Wray, J.; Kalkan, T.; Gomez-Lopez, S.; Eckardt, D.; Cook, A.; Kemler, R.; Smith, A. Inhibition of glycogen synthase kinase-3 alleviates Tcf3 repression of the pluripotency network and increases embryonic stem cell resistance to differentiation. Nat. Cell Biol. 2011, 13, 838–845. [Google Scholar] [CrossRef] [Green Version]
- Leeb, M.; Dietmann, S.; Paramor, M.; Niwa, H.; Smith, A. Genetic exploration of the exit from self-renewal using haploid embryonic stem cells. Cell Stem. Cell 2014, 14, 385–393. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Yu, J.; Tilgner, K.; Ong, S.H.; Koike-Yusa, H.; Yusa, K. Genome-wide CRISPR-KO Screen Uncovers mTORC1-Mediated Gsk3 Regulation in Naive Pluripotency Maintenance and Dissolution. Cell Rep. 2018, 24, 489–502. [Google Scholar] [CrossRef] [Green Version]
- Sharma, S.; Petsalaki, E. Application of CRISPR-Cas9 Based Genome-Wide Screening Approaches to Study Cellular Signalling Mechanisms. Int. J. Mol. Sci. 2018, 19, 933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrangou, R.; Doudna, J.A. Applications of CRISPR technologies in research and beyond. Nat. Biotechnol. 2016, 34, 933–941. [Google Scholar] [CrossRef] [PubMed]
- Zhan, T.; Rindtorff, N.; Betge, J.; Ebert, M.P.; Boutros, M. CRISPR/Cas9 for cancer research and therapy. Semin. Cancer Biol. 2019, 55, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.; Konermann, S.; Gootenberg, J.S.; O Abudayyeh, O.; Platt, R.J.; Brigham, M.D.; E Sanjana, N.; Zhang, F. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 2017, 12, 828–863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelsen, T.S.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef] [Green Version]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Evers, B.; Jastrzebski, K.; Heijmans, J.P.M.; Grernrum, W.; Beijersbergen, R.; Bernards, R. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat. Biotechnol. 2016, 34, 631–633. [Google Scholar] [CrossRef]
- Hu, G.; Kim, J.; Xu, Q.; Leng, Y.; Orkin, S.H.; Elledge, S.J. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009, 23, 837–848. [Google Scholar] [CrossRef] [Green Version]
- Ying, Q.-L.; Wray, J.; Nichols, J.; Batlle-Morera, L.; Doble, B.; Woodgett, J.; Cohen, P.; Smith, A. The ground state of embryonic stem cell self-renewal. Nature 2008, 453, 519–523. [Google Scholar] [CrossRef] [Green Version]
- MacDougall, M.S.; Clarke, R.; Merrill, B.J. Intracellular Ca2+ Homeostasis and Nuclear Export Mediate Exit from Naive Pluripotency. Cell Stem. Cell 2019, 25, 210–224.e6. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Kuang, J.; Wu, C.; Zhou, W.; Zhu, S.; Jiang, H.; Zhai, Z.; Wu, Y.; Peng, J.; Liu, N.; et al. Screening Genes Promoting Exit from Naive Pluripotency Based on Genome-Scale CRISPR-Cas9 Knockout. Stem. Cells Int. 2020, 2020, 8483035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Qi, X.; Du, X.; Zou, H.; Gao, F.; Feng, T.; Lu, H.; Li, S.; An, X.; Zhang, L.; et al. Piggybac mediates efficient in vivo CRISPR library screening for tumorigenesis in mice. Proc. Natl. Acad. Sci. USA 2017, 114, 722–727. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanjana, N.E.; Shalem, O.; Zhang, F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 2014, 11, 783–784. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [Green Version]
- Foissac, S.; Sammeth, M. ASTALAVISTA: Dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res. 2007, 35, W297–W299. [Google Scholar] [CrossRef] [Green Version]
- Sammeth, M.; Foissac, S.; Guigo, R. A general definition and nomenclature for alternative splicing events. PLoS Comput. Biol. 2008, 4, e1000147. [Google Scholar] [CrossRef] [Green Version]
- Mitsui, K.; Tokuzawa, Y.; Itoh, H.; Segawa, K.; Murakami, M.; Takahashi, K.; Maruyama, M.; Maeda, M.; Yamanaka, S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 2003, 113, 631–642. [Google Scholar] [CrossRef] [Green Version]
- Loh, Y.-H.; Wu, Q.; Chew, J.-L.; Vega, V.B.; Zhang, W.; Chen, X.; Bourque, G.; George, J.; Leong, B.; Liu, J.; et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 2006, 38, 431–440. [Google Scholar] [CrossRef]
- Singh, A.M.; Hamazaki, T.; Hankowski, K.E.; Terada, N. A heterogeneous expression pattern for nanog in embryonic stem cells. Stem. Cells 2007, 25, 2534–2542. [Google Scholar] [CrossRef]
- Ying, Q.-L.; Nichols, J.; Evans, E.P.; Smith, A. Changing potency by spontaneous fusion. Nature 2002, 416, 545–548. [Google Scholar] [CrossRef] [PubMed]
- Chambers, I.; Colby, D.; Robertson, M.; Nichols, J.; Lee, S.; Tweedie, S.; Smith, A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 2003, 113, 643–655. [Google Scholar] [CrossRef] [Green Version]
- Ying, Q.L.; Smith, A.G. Defined conditions for neural commitment and differentiation. Methods Enzym. 2003, 365, 327–341. [Google Scholar]
- Hamilton, W.B.; Kaji, K.; Kunath, T. ERK2 suppresses self-renewal capacity of embryonic stem cells, but is not required for multi-lineage commitment. PLoS ONE 2013, 8, e60907. [Google Scholar] [CrossRef] [Green Version]
- Chambers, I.; Silva, J.C.R.; Colby, D.; Nichols, J.; Nijmeijer-Winter, B.; Robertson, M.; Vrana, J.; Jones, K.; Grotewold, L.; Smith, A. Nanog safeguards pluripotency and mediates germline development. Nature 2007, 450, 1230–1234. [Google Scholar] [CrossRef]
- Ohtsuka, S.; Nakai-Futatsugi, Y.; Niwa, H. LIF signal in mouse embryonic stem cells. JAKSTAT 2015, 4, e1086520. [Google Scholar] [CrossRef] [Green Version]
- Lackner, A.; Sehlke, R.; Garmhausen, M.; Stirparo, G.G.; Huth, M.; Titz-Teixeira, F.; van der Lelij, P.; Ramesmayer, J.; Thomas, H.F.; Ralser, M.; et al. Cooperative genetic networks drive embryonic stem cell transition from naive to formative pluripotency. EMBO J. 2021, 40, e105776. [Google Scholar] [CrossRef]
- Babon, J.J.; Kershaw, N.; Murphy, J.; Varghese, L.N.; Laktyushin, A.; Young, S.N.; Lucet, I.S.; Norton, R.S.; Nicola, N.A. Suppression of cytokine signaling by SOCS3: Characterization of the mode of inhibition and the basis of its specificity. Immunity 2012, 36, 239–250. [Google Scholar] [CrossRef] [Green Version]
- Babon, J.J.; Nicola, N.A. The biology and mechanism of action of suppressor of cytokine signaling 3. Growth Factors 2012, 30, 207–219. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Gayen, S.; Xiong, J.; Zhou, B.; Shanmugam, A.K.; Sun, Y.; Karatas, H.; Liu, L.; Rao, R.; Wang, S.; et al. MLL1 Inhibition Reprograms Epiblast Stem Cells to Naive Pluripotency. Cell Stem. Cell 2016, 18, 481–494. [Google Scholar] [CrossRef] [Green Version]
- Wagner, E.F.; Nebreda, A.R. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat. Rev. Cancer 2009, 9, 537–549. [Google Scholar] [CrossRef] [PubMed]
- He, D.; Wu, H.; Xiang, J.; Ruan, X.; Peng, P.; Ruan, Y.; Chen, Y.-G.; Wang, Y.; Yu, Q.; Zhang, H.; et al. Gut stem cell aging is driven by mTORC1 via a p38 MAPK-p53 pathway. Nat. Commun. 2020, 11, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laplante, M.; Sabatini, D.M. mTOR Signaling in Growth Control and Disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, A.; Hochedlinger, K. The Sox Family of Transcription Factors: Versatile Regulators of Stem and Progenitor Cell Fate. Cell Stem. Cell 2013, 12, 15–30. [Google Scholar] [CrossRef] [Green Version]
- Sabour, D.; Srinivasan, S.P.; Rohani, S.; Wagh, V.; Gaspar, J.A.; Panek, D.; Ardestani, M.A.; Doss, M.X.; Riet, N.; Abken, H.; et al. STRIP2 Is Indispensable for the Onset of Embryonic Stem Cell Differentiation. Mol. Ther. Methods Clin. Dev. 2017, 5, 116–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strober, B.J.; Elorbany, R.; Rhodes, K.; Krishnan, N.; Tayeb, K.; Battle, A.; Gilad, Y. Dynamic genetic regulation of gene expression during cellular differentiation. Science 2019, 364, 1287–1290. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, C.; Qi, X.; Gao, X.; Li, J.; Qin, Y.; Yin, Y.; Gao, F.; Feng, T.; Wu, S.; Du, X. A Genome-Wide CRISPR Screen Identifies Factors Regulating Pluripotency Exit in Mouse Embryonic Stem Cells. Cells 2022, 11, 2289. https://doi.org/10.3390/cells11152289
Gao C, Qi X, Gao X, Li J, Qin Y, Yin Y, Gao F, Feng T, Wu S, Du X. A Genome-Wide CRISPR Screen Identifies Factors Regulating Pluripotency Exit in Mouse Embryonic Stem Cells. Cells. 2022; 11(15):2289. https://doi.org/10.3390/cells11152289
Chicago/Turabian StyleGao, Chen, Xiaolan Qi, Xin Gao, Jin Li, Yumin Qin, Yunjun Yin, Fei Gao, Tao Feng, Sen Wu, and Xuguang Du. 2022. "A Genome-Wide CRISPR Screen Identifies Factors Regulating Pluripotency Exit in Mouse Embryonic Stem Cells" Cells 11, no. 15: 2289. https://doi.org/10.3390/cells11152289
APA StyleGao, C., Qi, X., Gao, X., Li, J., Qin, Y., Yin, Y., Gao, F., Feng, T., Wu, S., & Du, X. (2022). A Genome-Wide CRISPR Screen Identifies Factors Regulating Pluripotency Exit in Mouse Embryonic Stem Cells. Cells, 11(15), 2289. https://doi.org/10.3390/cells11152289






