MiR-23b Promotes Porcine Preadipocyte Differentiation via SESN3 and ACSL4
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Approval and Sample Collection
2.2. Cell Culture and Transfection
2.3. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.4. Western Blot
2.5. Oil Red O Stain
2.6. Target Genes Prediction of miR-23b
2.7. Dual-Luciferase Reporter Gene Assay
2.8. RNA-Binding Protein Immunoprecipitation Assay (RIP)
2.9. miRNA Pulldown
2.10. Statistical Analysis
3. Results
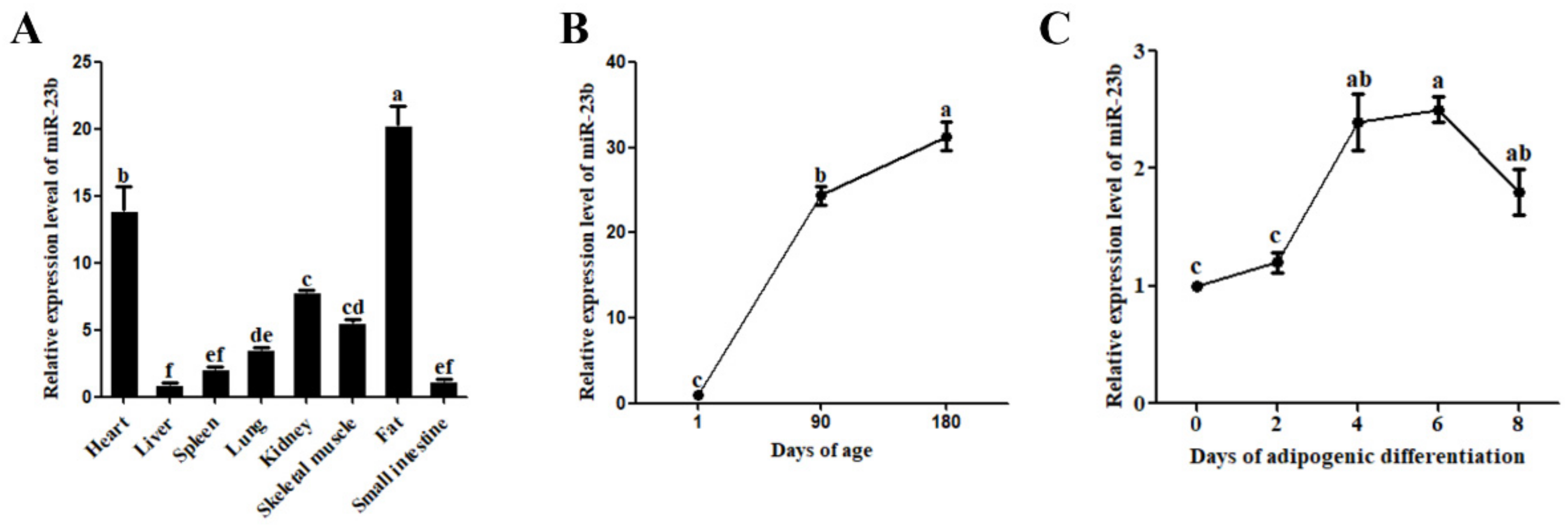
3.1. Expression Analysis of miR-23b in Pigs
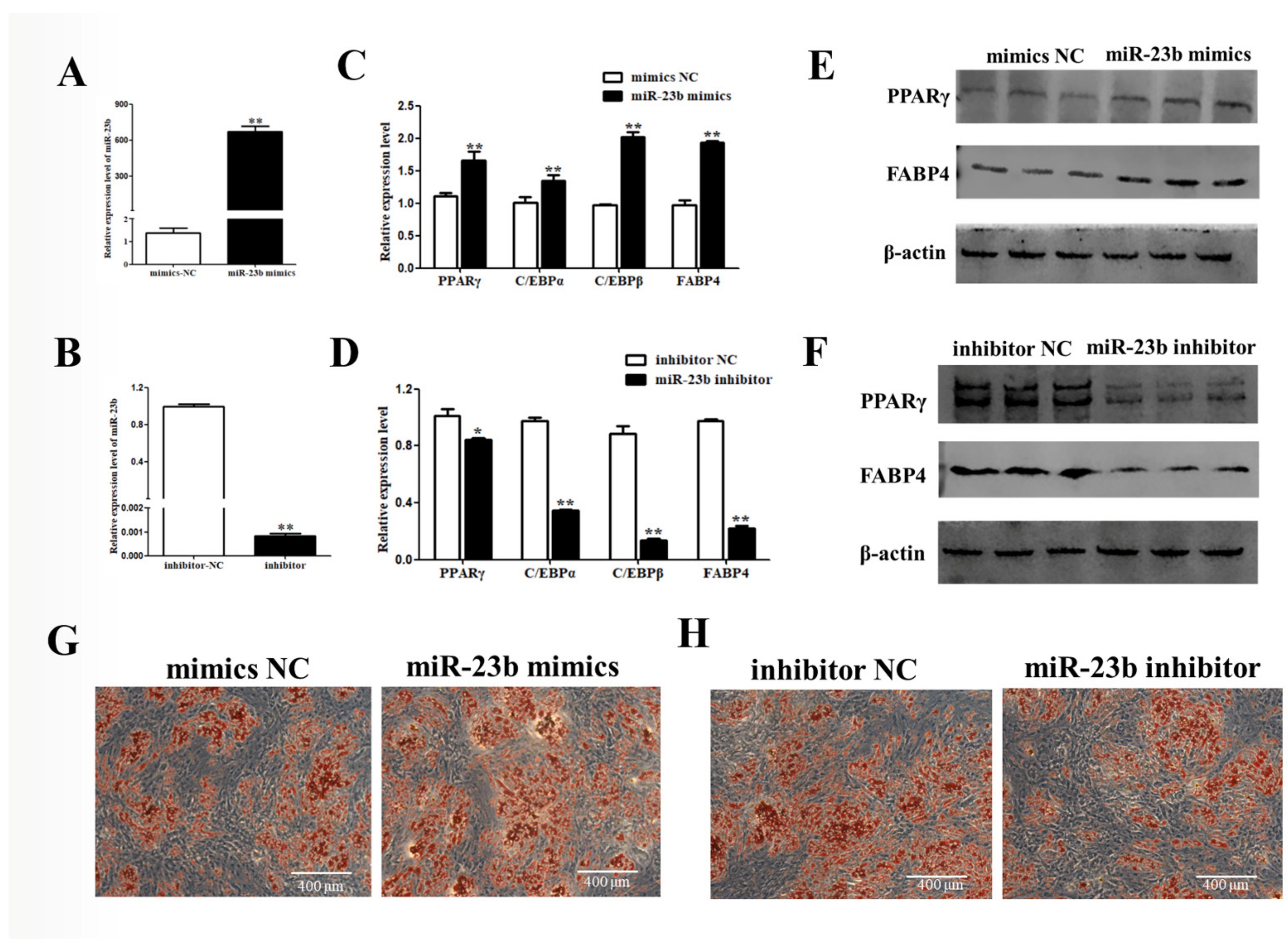
3.2. miR-23b Promotes Adipogenic Differentiation of Porcine Precursor Adipocytes
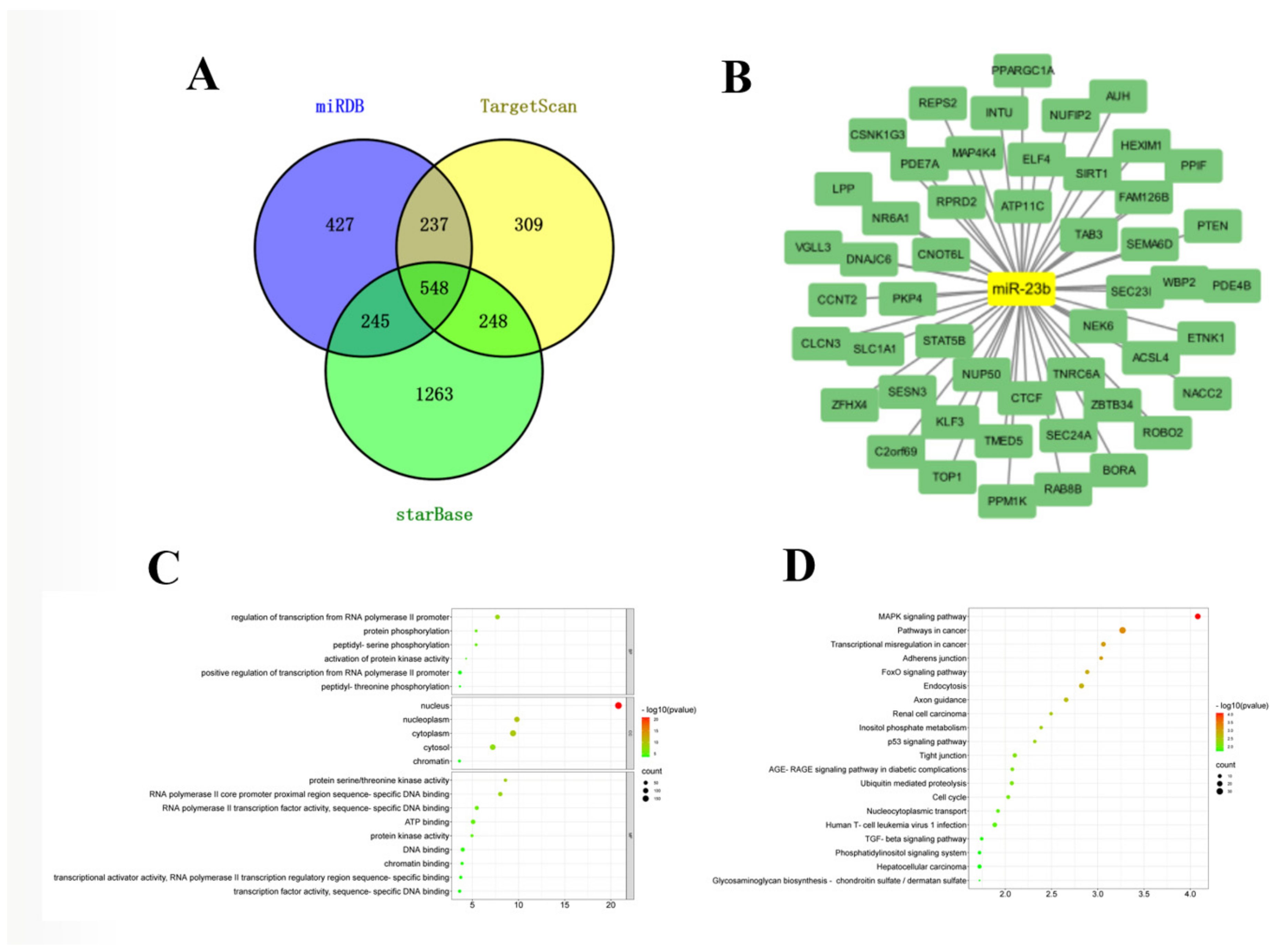
3.3. Construction of miR-23b-mRNA Network
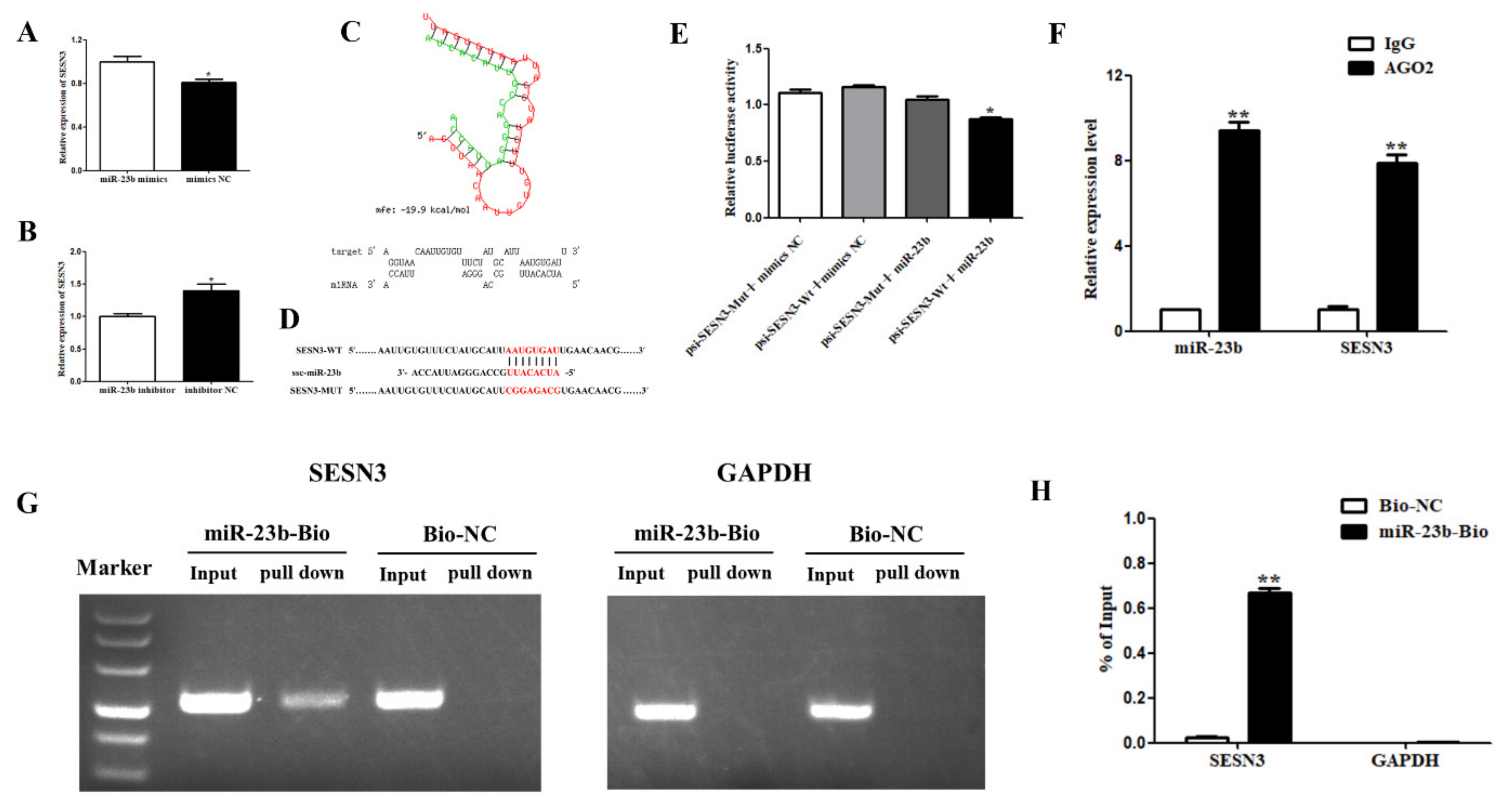
3.4. miR-23b Regulates Cells Adipogenic Differentiation by Binding SESN3
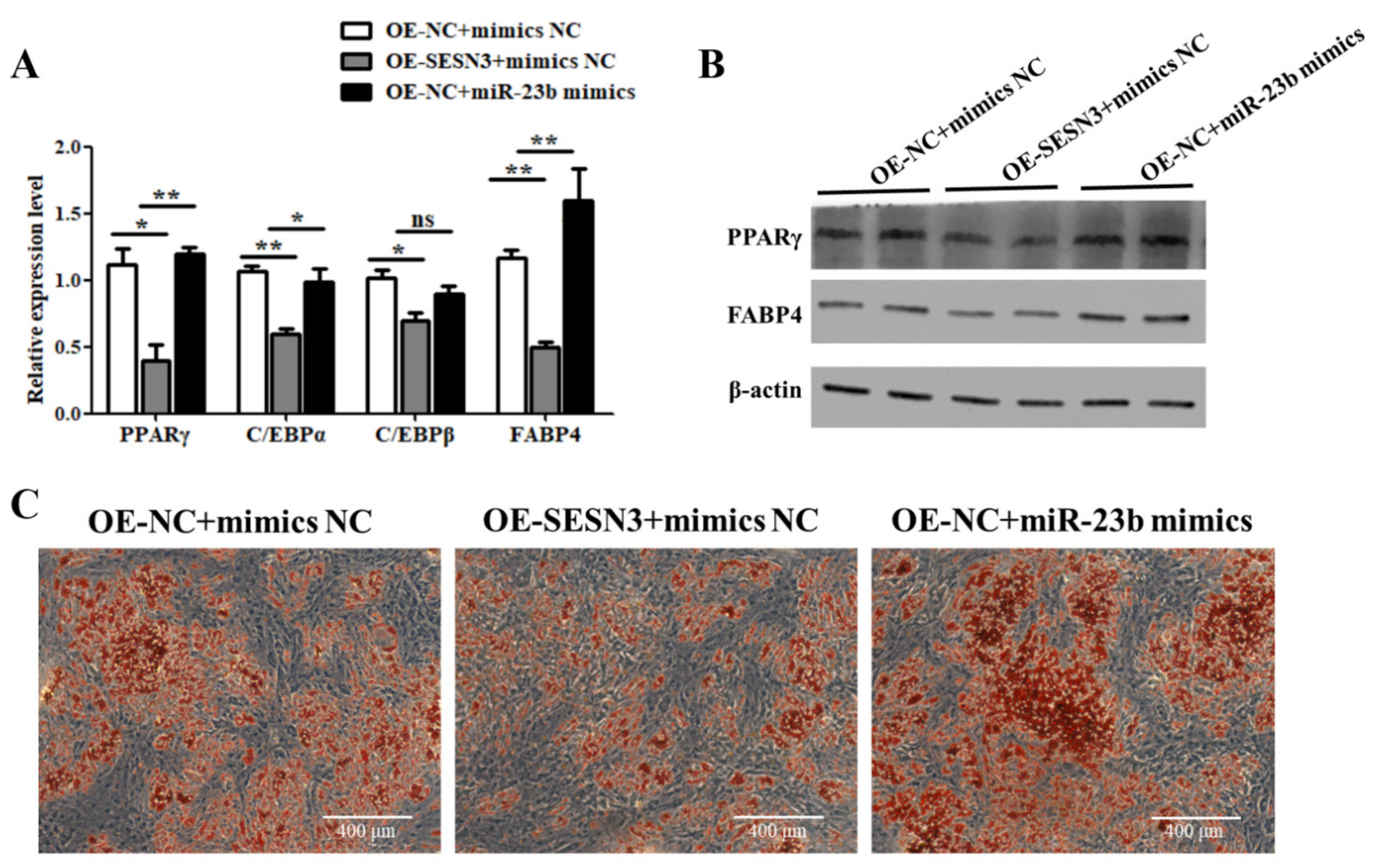
3.5. miR-23b Restores the Inhibitory Effect of SESN3 on Cells Adipogenic Differentiation
3.6. Regulatory Relationship between miR-23b and ACSL4
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bertol, T.M.; de Campos, R.M.; Ludke, J.V.; Terra, N.N.; de Figueiredo, E.A.; Coldebella, A.; dos Santos Filho, J.I.; Kawski, V.L.; Lehr, N.M. Effects of genotype and dietary oil supplementation on performance, carcass traits, pork quality and fatty acid composition of backfat and intramuscular fat. Meat Sci. 2013, 93, 507–516. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Shim, K.S.; Na, C.S.; Choe, H.S. Studies on Intramuscular Fat Percentage in Live Swine Using Real-time Ultrasound to Determine Pork Quality. Asian-Australas. J. Anim. Sci. 2015, 28, 318–322. [Google Scholar] [CrossRef] [Green Version]
- Poleti, M.D.; Regitano, L.C.A.; Souza, G.; Cesar, A.S.M.; Simas, R.C.; Silva-Vignato, B.; Oliveira, G.B.; Andrade, S.C.S.; Cameron, L.C.; Coutinho, L.L. Longissimus dorsi muscle label-free quantitative proteomic reveals biological mechanisms associated with intramuscular fat deposition. J. Proteom. 2018, 179, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Christodoulides, C.; Vidal-Puig, A. PPARs and adipocyte function. Mol. Cell. Endocrinol. 2010, 318, 61–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Yu, S.; Chen, L.; Hu, X.; Zheng, J.; Deng, X. Involvement of PPARγ/FSP27 in the pathogenic mechanism underlying insulin resistance: Tipping the balance between lipogenesis and fat storage in adult catch-up growth rats. Nutr. Metab. 2019, 16, 11. [Google Scholar] [CrossRef]
- Farmer, S.R. Transcriptional control of adipocyte formation. Cell Metab. 2006, 4, 263–273. [Google Scholar] [CrossRef] [Green Version]
- Zuo, Y.; Qiang, L.; Farmer, S.R. Activation of CCAAT/enhancer-binding protein (C/EBP) alpha expression by C/EBP beta during adipogenesis requires a peroxisome proliferator-activated receptor-gamma-associated repression of HDAC1 at the C/ebp alpha gene promoter. J. Biol. Chem. 2006, 281, 7960–7967. [Google Scholar] [CrossRef] [Green Version]
- Ertunc, M.E.; Sikkeland, J.; Fenaroli, F.; Griffiths, G.; Daniels, M.P.; Cao, H.; Saatcioglu, F.; Hotamisligil, G.S. Secretion of fatty acid binding protein aP2 from adipocytes through a nonclassical pathway in response to adipocyte lipase activity. J. Lipid Res. 2015, 56, 423–434. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.; Wang, S.; Chen, L.; Chen, Y.; Wu, M.; Zhang, Y.; Yu, K.; Huang, Z.; Qin, L.; Mo, D. MicroRNA-344 inhibits 3T3-L1 cell differentiation via targeting GSK3β of Wnt/β-catenin signaling pathway. FEBS Lett. 2014, 588, 429–435. [Google Scholar] [CrossRef] [Green Version]
- Jeong, B.C.; Kang, I.H.; Koh, J.T. MicroRNA-302a inhibits adipogenesis by suppressing peroxisome proliferator-activated receptor γ expression. FEBS Lett. 2014, 588, 3427–3434. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.Y.; Kim, A.Y.; Lee, H.W.; Son, Y.H.; Lee, G.Y.; Lee, J.W.; Lee, Y.S.; Kim, J.B. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARgamma expression. Biochem. Biophys. Res. Commun. 2010, 392, 323–328. [Google Scholar] [CrossRef]
- Chu, Y.; Yao, Y.; Li, X. MiR-370 enhances cell cycle and represses lipid accumulation in porcine adipocytes. Anim. Biotechnol. 2021, 32, 334–342. [Google Scholar] [CrossRef]
- Liu, S.; Sun, G.; Yuan, B.; Zhang, L.; Gao, Y.; Jiang, H.; Dai, L.; Zhang, J. miR-375 negatively regulates porcine preadipocyte differentiation by targeting BMPR2. FEBS Lett. 2016, 590, 1417–1427. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Gao, Y.; Xu, M.Q.; Wang, C.J.; Fu, X.H.; Liu, J.B.; Han, D.X.; Jiang, H.; Yuan, B.; Zhang, J.B. miR-181a regulate porcine preadipocyte differentiation by targeting TGFBR1. Gene 2019, 681, 45–51. [Google Scholar] [CrossRef]
- Borji, M.; Nourbakhsh, M. Down-Regulation of SIRT1 Expression by mir-23b Contributes to Lipid Accumulation in HepG2 Cells. Biochem. Genet. 2019, 57, 507–521. [Google Scholar] [CrossRef]
- Li, H.; Li, X.; Yu, S.; Hu, Y.; Xu, L.; Wang, T.; Yang, X.; Sun, X.; Zhao, B. miR-23b Ameliorates nonalcoholic steatohepatitis by targeting Acyl-CoA thioesterases 4. Exp. Cell Res. 2021, 407, 112787. [Google Scholar] [CrossRef]
- Liu, Y.; Tan, J.; Ou, S.; Chen, J.; Chen, L. Adipose-derived exosomes deliver miR-23a/b to regulate tumor growth in hepatocellular cancer by targeting the VHL/HIF axis. J. Physiol. Biochem. 2019, 75, 391–401. [Google Scholar] [CrossRef]
- Zhao, S.; Li, T.; Li, J.; Lu, Q.; Han, C.; Wang, N.; Qiu, Q.; Cao, H.; Xu, X.; Chen, H.; et al. miR-23b-3p induces the cellular metabolic memory of high glucose in diabetic retinopathy through a SIRT1-dependent signalling pathway. Diabetologia 2016, 59, 644–654. [Google Scholar] [CrossRef] [Green Version]
- Xiong, W.; Lin, Y.; Xu, L.; Tamadon, A.; Zou, S.; Tian, F.; Shao, R.; Li, X.; Feng, Y. Circulatory microRNA 23a and microRNA 23b and polycystic ovary syndrome (PCOS): The effects of body mass index and sex hormones in an Eastern Han Chinese population. J. Ovarian Res. 2017, 10, 10. [Google Scholar] [CrossRef] [Green Version]
- Ferrante, S.C.; Nadler, E.P.; Pillai, D.K.; Hubal, M.J.; Wang, Z.; Wang, J.M.; Gordish-Dressman, H.; Koeck, E.; Sevilla, S.; Wiles, A.A.; et al. Adipocyte-derived exosomal miRNAs: A novel mechanism for obesity-related disease. Pediatric Res. 2015, 77, 447–454. [Google Scholar] [CrossRef]
- Zhou, W.; Xu, J.; Wang, C.; Shi, D.; Yan, Q. miR-23b-3p regulates apoptosis and autophagy via suppressing SIRT1 in lens epithelial cells. J. Cell. Biochem. 2019, 120, 19635–19646. [Google Scholar] [CrossRef]
- Picard, F.; Kurtev, M.; Chung, N.; Topark-Ngarm, A.; Senawong, T.; Machado De Oliveira, R.; Leid, M.; McBurney, M.W.; Guarente, L. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature 2004, 429, 771–776. [Google Scholar] [CrossRef]
- Liu, X.; Zhao, H.; Jin, Q.; You, W.; Cheng, H.; Liu, Y.; Song, E.; Liu, G.; Tan, X.; Zhang, X.; et al. Resveratrol induces apoptosis and inhibits adipogenesis by stimulating the SIRT1-AMPKα-FOXO1 signalling pathway in bovine intramuscular adipocytes. Mol. Cell. Biochem. 2018, 439, 213–223. [Google Scholar] [CrossRef]
- Bai, L.; Pang, W.J.; Yang, Y.J.; Yang, G.S. Modulation of Sirt1 by resveratrol and nicotinamide alters proliferation and differentiation of pig preadipocytes. Mol. Cell. Biochem. 2008, 307, 129–140. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Budanov, A.V.; Karin, M. Sestrins orchestrate cellular metabolism to attenuate aging. Cell Metab. 2013, 18, 792–801. [Google Scholar] [CrossRef] [Green Version]
- Budanov, A.V. Stress-responsive sestrins link p53 with redox regulation and mammalian target of rapamycin signaling. Antioxid. Redox Signal. 2011, 15, 1679–1690. [Google Scholar] [CrossRef]
- Bae, S.H.; Sung, S.H.; Oh, S.Y.; Lim, J.M.; Lee, S.K.; Park, Y.N.; Lee, H.E.; Kang, D.; Rhee, S.G. Sestrins activate Nrf2 by promoting p62-dependent autophagic degradation of Keap1 and prevent oxidative liver damage. Cell Metab. 2013, 17, 73–84. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Budanov, A.V.; Talukdar, S.; Park, E.J.; Park, H.L.; Park, H.W.; Bandyopadhyay, G.; Li, N.; Aghajan, M.; Jang, I.; et al. Maintenance of metabolic homeostasis by Sestrin2 and Sestrin3. Cell Metab. 2012, 16, 311–321. [Google Scholar] [CrossRef] [Green Version]
- Park, H.W.; Park, H.; Ro, S.H.; Jang, I.; Semple, I.A.; Kim, D.N.; Kim, M.; Nam, M.; Zhang, D.; Yin, L.; et al. Hepatoprotective role of Sestrin2 against chronic ER stress. Nat. Commun. 2014, 5, 4233. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Liu, S.; Yuan, H.; Niu, Y.; Fu, L. Sestrin 2 induces autophagy and attenuates insulin resistance by regulating AMPK signaling in C2C12 myotubes. Exp. Cell Res. 2017, 354, 18–24. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Hu, W.; Niu, Y.; Liu, S.; Fu, L. Exercise improves lipid metabolism disorders induced by high-fat diet in a SESN2/JNK-independent manner. Appl. Physiol Nutr. Metab. Physiol. Appl. Nutr. Et Metab. 2021, 46, 1322–1330. [Google Scholar] [CrossRef] [PubMed]
- Kang, X.; Petyaykina, K.; Tao, R.; Xiong, X.; Dong, X.C.; Liangpunsakul, S. The inhibitory effect of ethanol on Sestrin3 in the pathogenesis of ethanol-induced liver injury. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 307, G58–G65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mashek, D.G.; Li, L.O.; Coleman, R.A. Long-chain acyl-CoA synthetases and fatty acid channeling. Future Lipidol. 2007, 2, 465–476. [Google Scholar] [CrossRef] [Green Version]
- Soupene, E.; Kuypers, F.A. Mammalian long-chain acyl-CoA synthetases. Exp. Biol. Med. 2008, 233, 507–521. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Ding, C.; Chen, Y.; Hu, W.; Yu, C.; Peng, C.; Feng, X.; Cheng, Q.; Wu, W.; Lu, Y.; et al. ACSL4 reprograms fatty acid metabolism in hepatocellular carcinoma via c-Myc/SREBP1 pathway. Cancer Lett. 2021, 502, 154–165. [Google Scholar] [CrossRef]
- Xia, H.; Lee, K.W.; Chen, J.; Kong, S.N.; Sekar, K.; Deivasigamani, A.; Seshachalam, V.P.; Goh, B.K.P.; Ooi, L.L.; Hui, K.M. Simultaneous silencing of ACSL4 and induction of GADD45B in hepatocellular carcinoma cells amplifies the synergistic therapeutic effect of aspirin and sorafenib. Cell Death Discov. 2017, 3, 17058. [Google Scholar] [CrossRef]
- Belkaid, A.; Ouellette, R.J.; Surette, M.E. 17β-estradiol-induced ACSL4 protein expression promotes an invasive phenotype in estrogen receptor positive mammary carcinoma cells. Carcinogenesis 2017, 38, 402–410. [Google Scholar] [CrossRef] [Green Version]
- Kuwata, H.; Nakatani, E.; Shimbara-Matsubayashi, S.; Ishikawa, F.; Shibanuma, M.; Sasaki, Y.; Yoda, E.; Nakatani, Y.; Hara, S. Long-chain acyl-CoA synthetase 4 participates in the formation of highly unsaturated fatty acid-containing phospholipids in murine macrophages. Biochim. Et Biophys. Acta. Mol. Cell Biol. Lipids 2019, 1864, 1606–1618. [Google Scholar] [CrossRef]
- Chen, J.N.; Jiang, Y.Z.; Cen, W.M.; Xing, S.H.; Zhu, L.; Tang, G.Q.; Li, M.Z.; Jiang, A.A.; Lou, P.E.; Wen, A.X.; et al. Distribution of H-FABP and ACSL4 gene polymorphisms and their associations with intramuscular fat content and backfat thickness in different pig populations. Genet. Mol. Res. GMR 2014, 13, 6759–6772. [Google Scholar] [CrossRef]
- Mercadé, A.; Estellé, J.; Pérez-Enciso, M.; Varona, L.; Silió, L.; Noguera, J.L.; Sánchez, A.; Folch, J.M. Characterization of the porcine acyl-CoA synthetase long-chain 4 gene and its association with growth and meat quality traits. Anim. Genet. 2006, 37, 219–224. [Google Scholar] [CrossRef]
- Ruść, A.; Sieczkowska, H.; Krzęcio, E.; Antosik, K.; Zybert, A.; Koćwin-Podsiadła, M.; Kamiński, S. The association between acyl-CoA synthetase (ACSL4) polymorphism and intramuscular fat content in (Landrace × Yorkshire) × Duroc pigs. Meat Sci. 2011, 89, 440–443. [Google Scholar] [CrossRef]
- Corominas, J.; Ramayo-Caldas, Y.; Castelló, A.; Muñoz, M.; Ibáñez-Escriche, N.; Folch, J.M.; Ballester, M. Evaluation of the porcine ACSL4 gene as a candidate gene for meat quality traits in pigs. Anim. Genet. 2012, 43, 714–720. [Google Scholar] [CrossRef]
- Wang, W.; Li, X.; Ding, N.; Teng, J.; Zhang, S.; Zhang, Q.; Tang, H. miR-34a regulates adipogenesis in porcine intramuscular adipocytes by targeting ACSL4. BMC Genet. 2020, 21, 33. [Google Scholar] [CrossRef]
- Fan, Y.; Han, Z.; Lu, X. Identification of Milk Fat Metabolism-Related Pathways of the Bovine Mammary Gland during Mid and Late Lactation and Functional Verification of the ACSL4 Gene. Genes 2020, 11, 1357. [Google Scholar] [CrossRef]
- Askari, B.; Kanter, J.E.; Sherrid, A.M.; Golej, D.L.; Bender, A.T.; Liu, J.; Hsueh, W.A.; Beavo, J.A.; Coleman, R.A.; Bornfeldt, K.E. Rosiglitazone inhibits acyl-CoA synthetase activity and fatty acid partitioning to diacylglycerol and triacylglycerol via a peroxisome proliferator-activated receptor-gamma-independent mechanism in human arterial smooth muscle cells and macrophages. Diabetes 2007, 56, 1143–1152. [Google Scholar] [CrossRef] [Green Version]
- Ren, H.; Zhang, H. ACSL4 Directs Intramuscular Adipogenesis and Fatty Acid Composition in Pigs. Anim. Open Access J. MDPI 2022, 12, 119. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [Green Version]
- Liang, Y.; Lu, Q.; Li, W.; Zhang, D.; Zhang, F.; Zou, Q.; Chen, L.; Tong, Y.; Liu, M.; Wang, S.; et al. Reactivation of tumour suppressor in breast cancer by enhancer switching through NamiRNA network. Nucleic Acids Res. 2021, 49, 8556–8572. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Zhang, N.; Li, J.; Zhang, W.; Hei, W.; Ji, M.; Yang, Y.; Cao, G.; Guo, X.; Li, B. MiR-23b Promotes Porcine Preadipocyte Differentiation via SESN3 and ACSL4. Cells 2022, 11, 2339. https://doi.org/10.3390/cells11152339
Li M, Zhang N, Li J, Zhang W, Hei W, Ji M, Yang Y, Cao G, Guo X, Li B. MiR-23b Promotes Porcine Preadipocyte Differentiation via SESN3 and ACSL4. Cells. 2022; 11(15):2339. https://doi.org/10.3390/cells11152339
Chicago/Turabian StyleLi, Meng, Na Zhang, Jiao Li, Wanfeng Zhang, Wei Hei, Mengting Ji, Yang Yang, Guoqing Cao, Xiaohong Guo, and Bugao Li. 2022. "MiR-23b Promotes Porcine Preadipocyte Differentiation via SESN3 and ACSL4" Cells 11, no. 15: 2339. https://doi.org/10.3390/cells11152339
APA StyleLi, M., Zhang, N., Li, J., Zhang, W., Hei, W., Ji, M., Yang, Y., Cao, G., Guo, X., & Li, B. (2022). MiR-23b Promotes Porcine Preadipocyte Differentiation via SESN3 and ACSL4. Cells, 11(15), 2339. https://doi.org/10.3390/cells11152339





