Nuclear Receptor Atlases of Choroidal Tissues Reveal Candidate Receptors Associated with Age-Related Macular Degeneration
Abstract
:1. Introduction
2. Materials and Methods
2.1. Model Systems
2.2. Tube Formation Assay
2.3. RNA Isolation and cDNA Preparation
2.4. AMD-Specific Genotyping
2.5. Primer Validation, qPCR Assay
2.6. Venn Diagrams and Pathway Analysis
2.7. Experimental Laser Induced Choroidal Injury in Mice
2.8. In Vivo Imaging
2.9. RPE-Choroid Flat Mount Preparation
2.10. RT2 Profiler PCR Array for Mouse Nuclear Receptors and Coregulators
2.11. Statistical Analyses and Rigor
2.12. Institutional Review Board Statement
3. Results
3.1. Rigor of Human Choroidal Endothelial Cell and Human Choroid Tissue Sample Preparation
3.2. Nuclear Receptor Atlas of Primary Choroidal Endothelial Cells and Freshly Isolated Choroid
3.3. Pathway Analysis of Nuclear Receptors Expressed in Choroidal Endothelial Cells and Freshly Isolated Choroid
3.4. Comparison of Choroidal Endothelial Cell and RPE Atlases
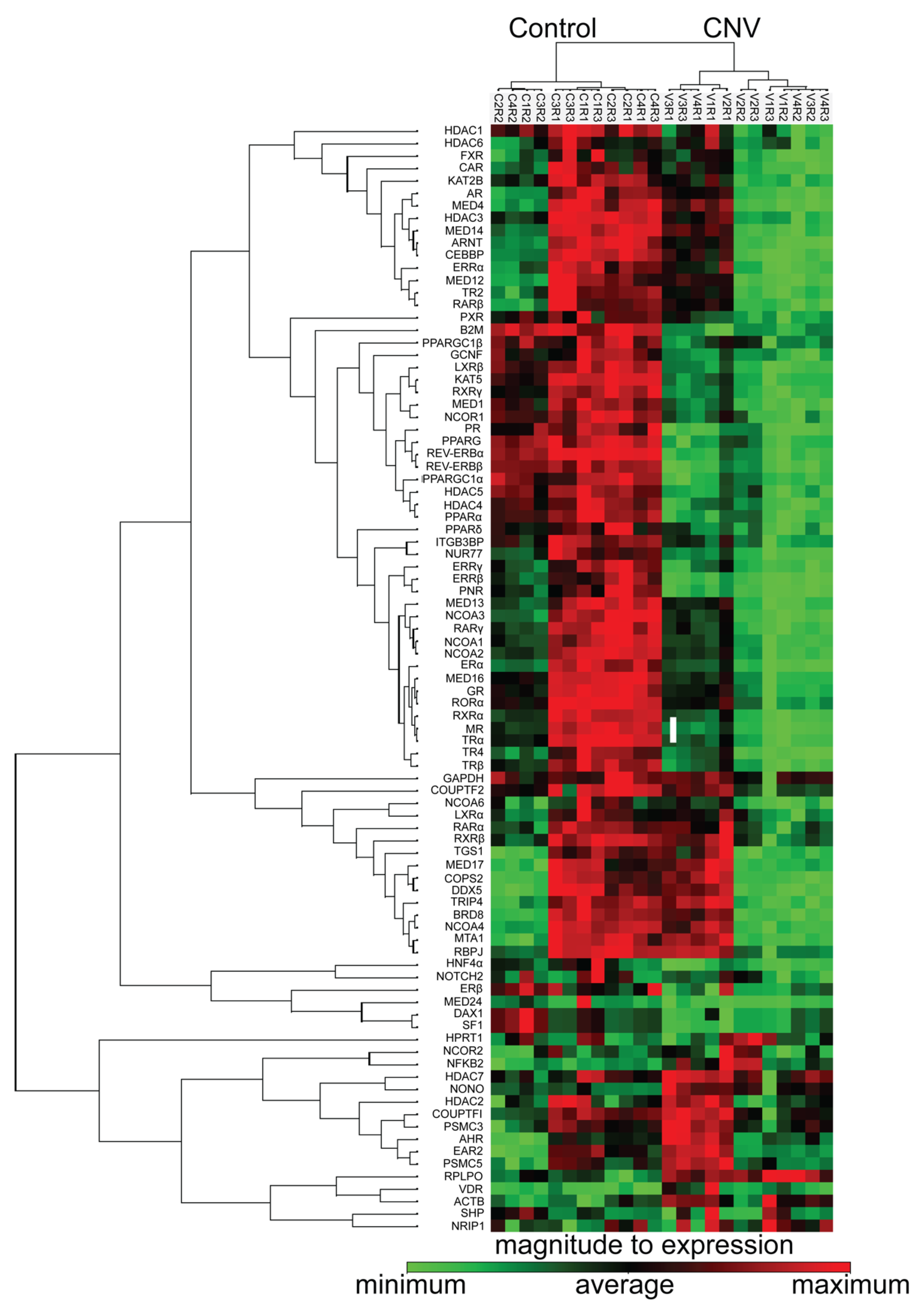
3.5. Nuclear Receptors Are Differentially Regulated Following RPE/Choroidal Injury
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Nickla, D.L.; Wallman, J. The multifunctional choroid. Prog. Retin. Eye Res. 2010, 29, 144–168. [Google Scholar] [CrossRef]
- Hogan, M.J.; Alvarado, J.A.; Weddell, J.E. Histology of the Human Eye; An Atlas and Textbook; Saunders: Philadelphia, PA, USA, 1971. [Google Scholar]
- Zhao, J.; Wang, Y.X.; Zhang, Q.; Wei, W.B.; Xu, L.; Jonas, J.B. Macular Choroidal Small-Vessel Layer, Sattler’s Layer and Haller’s Layer Thicknesses: The Beijing Eye Study. Sci. Rep. 2018, 8, 4411. [Google Scholar] [CrossRef] [PubMed]
- Campbell, M.; Humphries, P. The blood-retina barrier: Tight junctions and barrier modulation. Adv. Exp. Med. Biol. 2012, 763, 70–84. [Google Scholar] [PubMed]
- Streilein, J.W.; Ma, N.; Wenkel, H.; Ng, T.F.; Zamiri, P. Immunobiology and privilege of neuronal retina and pigment epithelium transplants. Vis. Res. 2002, 42, 487–495. [Google Scholar] [CrossRef]
- Farazdaghi, M.K.; Ebrahimi, K.B. Role of the Choroid in Age-related Macular Degeneration: A Current Review. J. Ophthalmic Vis. Res. 2019, 14, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Tao, Y. Choroidal structural changes correlate with severity of diabetic retinopathy in diabetes mellitus. BMC Ophthalmol. 2019, 19, 186. [Google Scholar] [CrossRef] [PubMed]
- Chirco, K.R.; Sohn, E.H.; Stone, E.M.; Tucker, B.A.; Mullins, R.F. Structural and molecular changes in the aging choroid: Implications for age-related macular degeneration. Eye 2017, 31, 10–25. [Google Scholar] [CrossRef]
- Wong, W.L.; Su, X.; Li, X.; Cheung, C.M.; Klein, R.; Cheng, C.Y.; Wong, T.Y. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: A systematic review and meta-analysis. Lancet Glob. Health 2014, 2, e106–e116. [Google Scholar] [CrossRef]
- Cheloni, R.; Gandolfi, S.A.; Signorelli, C.; Odone, A. Global prevalence of diabetic retinopathy: Protocol for a systematic review and meta-analysis. BMJ Open 2019, 9, e022188. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, P.; Smith, W.; Attebo, K.; Wang, J.J. Prevalence of age-related maculopathy in Australia: The Blue Mountains Eye Study. Ophthalmology 1995, 102, 1450–1460. [Google Scholar] [CrossRef]
- Klein, R.; Klein, B.E.; Knudtson, M.D.; Meuer, S.M.; Swift, M.; Gangnon, R.E. Fifteen-year cumulative incidence of age-related macular degeneration: The Beaver Dam Eye Study. Ophthalmology 2007, 114, 253–262. [Google Scholar] [CrossRef]
- Chakravarthy, U.; Wong, T.Y.; Fletcher, A.; Piault, E.; Evans, C.; Zlateva, G.; Buggage, R.; Pleil, A.; Mitchell, P. Clinical risk factors for age-related macular degeneration: A systematic review and meta-analysis. BMC Ophthalmol. 2010, 10, 31. [Google Scholar] [CrossRef] [PubMed]
- Fritsche, L.G.; Fariss, R.N.; Stambolian, D.; Abecasis, G.R.; Curcio, C.A.; Swaroop, A. Age-related macular degeneration: Genetics and biology coming together. Annu. Rev. Genom. Hum. Genet. 2014, 15, 151–171. [Google Scholar] [CrossRef]
- Das, U.N. Diabetic macular edema, retinopathy and age-related macular degeneration as inflammatory conditions. Arch. Med. Sci. 2016, 12, 1142–1157. [Google Scholar] [CrossRef]
- Jun, S.; Datta, S.; Wang, L.; Pegany, R.; Cano, M.; Handa, J.T. The impact of lipids, lipid oxidation, and inflammation on AMD, and the potential role of miRNAs on lipid metabolism in the RPE. Exp. Eye Res. 2019, 181, 346–355. [Google Scholar] [CrossRef]
- Hu, P.; Herrmann, R.; Bednar, A.; Saloupis, P.; Dwyer, M.A.; Yang, P.; Qi, X.; Thomas, R.S.; Jaffe, G.J.; Boulton, M.E.; et al. Aryl hydrocarbon receptor deficiency causes dysregulated cellular matrix metabolism and age-related macular degeneration-like pathology. Proc. Natl. Acad. Sci. USA 2013, 110, E4069–E4078. [Google Scholar] [CrossRef]
- Nita, M.; Strzalka-Mrozik, B.; Grzybowski, A.; Mazurek, U.; Romaniuk, W. Age-related macular degeneration and changes in the extracellular matrix. Med. Sci. Monit. 2014, 20, 1003–1016. [Google Scholar] [CrossRef] [PubMed]
- Safi, S.Z.; Qvist, R.; Kumar, S.; Batumalaie, K.; Ismail, I.S. Molecular mechanisms of diabetic retinopathy, general preventive strategies, and novel therapeutic targets. Biomed Res. Int. 2014, 2014, 801269. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Miao, X.; Li, F.; Wang, S.; Liu, Q.; Wang, Y.; Sun, J. Oxidative Stress-Related Mechanisms and Antioxidant Therapy in Diabetic Retinopathy. Oxid. Med. Cell. Longev. 2017, 2017, 9702820. [Google Scholar] [CrossRef] [PubMed]
- Fisher, C.R.; Ferrington, D.A. Perspective on AMD Pathobiology: A Bioenergetic Crisis in the RPE. Investig. Ophthalmol. Vis. Sci. 2018, 59, AMD41–AMD47. [Google Scholar] [CrossRef]
- Tan, W.; Zou, J.; Yoshida, S.; Jiang, B.; Zhou, Y. The Role of Inflammation in Age-Related Macular Degeneration. Int. J. Biol. Sci. 2020, 16, 2989–3001. [Google Scholar] [CrossRef]
- Whitmore, S.S.; Sohn, E.H.; Chirco, K.R.; Drack, A.V.; Stone, E.M.; Tucker, B.A.; Mullins, R.F. Complement activation and choriocapillaris loss in early AMD: Implications for pathophysiology and therapy. Prog. Retin. Eye Res. 2015, 45, 1–29. [Google Scholar] [CrossRef]
- Seddon, J.M.; McLeod, D.S.; Bhutto, I.A.; Villalonga, M.B.; Silver, R.E.; Wenick, A.S.; Edwards, M.M.; Lutty, G.A. Histopathological Insights Into Choroidal Vascular Loss in Clinically Documented Cases of Age-Related Macular Degeneration. JAMA Ophthalmol. 2016, 134, 1272–1280. [Google Scholar] [CrossRef] [PubMed]
- Ambati, J.; Fowler, B.J. Mechanisms of age-related macular degeneration. Neuron 2012, 75, 26–39. [Google Scholar] [CrossRef] [PubMed]
- Yao, P.L.; Peavey, J.; Malek, G. Leveraging Nuclear Receptors as Targets for Pathological Ocular Vascular Diseases. Int. J. Mol. Sci 2020, 21, 2889. [Google Scholar] [CrossRef]
- Santos, R.; Ursu, O.; Gaulton, A.; Bento, A.P.; Donadi, R.S.; Bologa, C.G.; Karlsson, A.; Al-Lazikani, B.; Hersey, A.; Oprea, T.I.; et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017, 16, 19–34. [Google Scholar] [CrossRef]
- Overington, J.P.; Al-Lazikani, B.; Hopkins, A.L. How many drug targets are there? Nat. Rev. Drug Discov. 2006, 5, 993–996. [Google Scholar] [CrossRef]
- Zhao, L.; Zhou, S.; Gustafsson, J.A. Nuclear Receptors: Recent Drug Discovery for Cancer Therapies. Endocr. Rev. 2019, 40, 1207–1249. [Google Scholar] [CrossRef]
- Landreth, G. Therapeutic use of agonists of the nuclear receptor PPARγ in Alzheimer’s disease. Curr. Alzheimer. Res. 2007, 4, 159–164. [Google Scholar] [CrossRef]
- Dwyer, M.A.; Kazmin, D.; Hu, P.; McDonnell, D.P.; Malek, G. Research resource: Nuclear receptor atlas of human retinal pigment epithelial cells: Potential relevance to age-related macular degeneration. Mol. Endocrinol. 2011, 25, 360–372. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, M.; Ding, J.D.; Qi, X.; Boulton, M.E.; Yao, P.L.; Peters, J.M.; Malek, G. PPARβ/δ selectively regulates phenotypic features of age-related macular degeneration. Aging 2016, 8, 1952–1978. [Google Scholar] [CrossRef]
- Choudhary, M.; Malek, G. The Aryl Hydrocarbon Receptor: A Mediator and Potential Therapeutic Target for Ocular and Non-Ocular Neurodegenerative Diseases. Int. J. Mol. Sci. 2020, 21, 6777. [Google Scholar] [CrossRef]
- Choudhary, M.; Safe, S.; Malek, G. Suppression of aberrant choroidal neovascularization through activation of the aryl hydrocarbon receptor. Biochim. Biophys. Acta Mol. Basis. Dis. 2018, 1864, 1583–1595. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, M.; Ismail, E.N.; Yao, P.L.; Tayyari, F.; Radu, R.A.; Nusinowitz, S.; Boulton, M.E.; Apte, R.S.; Ruberti, J.W.; Handa, J.T.; et al. LXRs regulate features of age-related macular degeneration and may be a potential therapeutic target. JCI Insight 2020, 5, e131928. [Google Scholar] [CrossRef] [PubMed]
- Choudhary, M.; Kazmin, D.; Hu, P.; Thomas, R.S.; McDonnell, D.P.; Malek, G. Aryl hydrocarbon receptor knock-out exacerbates choroidal neovascularization via multiple pathogenic pathways. J. Pathol. 2015, 235, 101–112. [Google Scholar] [CrossRef]
- Li, T.; Diao, H.; Zhao, L.; Xing, Y.; Zhang, J.; Liu, N.; Yan, Y.; Tian, X.; Sun, W.; Liu, B. Identification of suitable reference genes for real-time quantitative PCR analysis of hydrogen peroxide-treated human umbilical vein endothelial cells. BMC Mol. Biol. 2017, 18, 10. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Gillespie, M.; Jassal, B.; Stephan, R.; Milacic, M.; Rothfels, K.; Senff-Ribeiro, A.; Griss, J.; Sevilla, C.; Matthews, L.; Gong, C.; et al. The reactome pathway knowledgebase 2022. Nucleic Acids Res. 2022, 50, D687–D692. [Google Scholar] [CrossRef]
- Espinosa-Heidmann, D.G.; Suner, I.; Hernandez, E.P.; Frazier, W.D.; Csaky, K.G.; Cousins, S.W. Age as an independent risk factor for severity of experimental choroidal neovascularization. Investig. Ophthalmol. Vis. Sci. 2002, 43, 1567–1573. [Google Scholar]
- Zhu, Z.; Chen, J.; Lin, Y.; Zhang, C.; Li, W.; Qiao, H.; Fu, M.; Dang, E.; Wang, G. Aryl Hydrocarbon Receptor in Cutaneous Vascular Endothelial Cells Restricts Psoriasis Development by Negatively Regulating Neutrophil Recruitment. J. Investig. Dermatol. 2020, 140, 1233–1243.e9. [Google Scholar] [CrossRef]
- Ikeda, R.; Tsuchiya, Y.; Koike, N.; Umemura, Y.; Inokawa, H.; Ono, R.; Inoue, M.; Sasawaki, Y.; Grieten, T.; Okubo, N.; et al. REV-ERBα and REV-ERBβ function as key factors regulating Mammalian Circadian Output. Sci. Rep. 2019, 9, 10171. [Google Scholar] [CrossRef] [PubMed]
- Strauss, O. The retinal pigment epithelium in visual function. Physiol. Rev. 2005, 85, 845–881. [Google Scholar] [CrossRef]
- Lutty, G.A.; McLeod, D.S.; Bhutto, I.A.; Edwards, M.M.; Seddon, J.M. Choriocapillaris dropout in early age-related macular degeneration. Exp. Eye Res. 2020, 192, 107939. [Google Scholar] [CrossRef]
- Biesemeier, A.; Taubitz, T.; Julien, S.; Yoeruek, E.; Schraermeyer, U. Choriocapillaris breakdown precedes retinal degeneration in age-related macular degeneration. Neurobiol. Aging 2014, 35, 2562–2573. [Google Scholar] [CrossRef]
- Wang, B.; Tontonoz, P. Liver X receptors in lipid signalling and membrane homeostasis. Nat. Rev. Endocrinol. 2018, 14, 452–463. [Google Scholar] [CrossRef]
- Zechel, C. The germ cell nuclear factor (GCNF). Mol. Reprod. Dev. 2005, 72, 550–556. [Google Scholar] [CrossRef] [PubMed]
- Le Maire, A.; Teyssier, C.; Balaguer, P.; Bourguet, W.; Germain, P. Regulation of RXR-RAR Heterodimers by RXR- and RAR-Specific Ligands and Their Combinations. Cells 2019, 8, 1392. [Google Scholar] [CrossRef] [PubMed]
- Dawson, M.I.; Xia, Z. The retinoid X receptors and their ligands. Biochim. Biophys. Acta (BBA)-Mol. Cell Biol. Lipids 2012, 1821, 21–56. [Google Scholar] [CrossRef]
- Qiu, F.; Matlock, G.; Chen, Q.; Zhou, K.; Du, Y.; Wang, X.; Ma, J.X. Therapeutic Effects of PPARα Agonist on Ocular Neovascularization in Models Recapitulating Neovascular Age-Related Macular Degeneration. Investig. Ophthalmol. Vis. Sci. 2017, 58, 5065–5075. [Google Scholar] [CrossRef]
- Cruz, N.M.; Yuan, Y.; Leehy, B.D.; Baid, R.; Kompella, U.; DeAngelis, M.M.; Escher, P.; Haider, N.B. Modifier genes as therapeutics: The nuclear hormone receptor Rev Erb alpha (Nr1d1) rescues Nr2e3 associated retinal disease. PLoS ONE 2014, 9, e87942. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Datta, S.; Brabbit, E.; Love, Z.; Woytowicz, V.; Flattery, K.; Capri, J.; Yao, K.; Wu, S.; Imboden, M.; et al. Nr2e3 is a genetic modifier that rescues retinal degeneration and promotes homeostasis in multiple models of retinitis pigmentosa. Gene Ther. 2021, 28, 223–241. [Google Scholar] [CrossRef]
- Mollema, N.J.; Yuan, Y.; Jelcick, A.S.; Sachs, A.J.; von Alpen, D.; Schorderet, D.; Escher, P.; Haider, N.B. Nuclear receptor Rev-erb alpha (Nr1d1) functions in concert with Nr2e3 to regulate transcriptional networks in the retina. PLoS ONE 2011, 6, e17494. [Google Scholar] [CrossRef]
- Park, Y.Y.; Kim, K.; Kim, S.B.; Hennessy, B.T.; Kim, S.M.; Park, E.S.; Lim, J.Y.; Li, J.; Lu, Y.; Gonzalez-Angulo, A.M.; et al. Reconstruction of nuclear receptor network reveals that NR2E3 is a novel upstream regulator of ESR1 in breast cancer. EMBO Mol. Med. 2012, 4, 52–67. [Google Scholar] [CrossRef]
- Khanal, T.; Leung, Y.K.; Jiang, W.; Timchenko, N.; Ho, S.M.; Kim, K. NR2E3 is a key component in p53 activation by regulating a long noncoding RNA DINO in acute liver injuries. FASEB J. 2019, 33, 8335–8348. [Google Scholar] [CrossRef]
- Minneman, K.P. Postsynaptic induction of serotonin N-acetyltransferase activity and the control of cyclic nucleotide metabolism in organ cultures of the rat pineal. Mol. Pharmacol. 1977, 13, 735–745. [Google Scholar]
- Peters, J.M.; Shah, Y.M.; Gonzalez, F.J. The role of peroxisome proliferator-activated receptors in carcinogenesis and chemoprevention. Nat. Rev. Cancer 2012, 12, 181–195. [Google Scholar] [CrossRef] [PubMed]
- Herzlich, A.A.; Ding, X.; Shen, D.; Ross, R.J.; Tuo, J.; Chan, C.C. Peroxisome Proliferator-Activated Receptor Expression in Murine Models and Humans with Age-related Macular Degeneration. Open Biol. J. 2009, 2, 141–148. [Google Scholar] [CrossRef]
- Sulaiman, R.S.; Kadmiel, M.; Cidlowski, J.A. Glucocorticoid receptor signaling in the eye. Steroids 2018, 133, 60–66. [Google Scholar] [CrossRef] [PubMed]
- Arimura, S.; Takamura, Y.; Miyake, S.; Gozawa, M.; Iwasaki, K.; Tomomatsu, T.; Matsumura, T.; Inatani, M. The effect of triamcinolone acetonide or bevacizumab on the levels of proinflammatory cytokines after retinal laser photocoagulation in pigmented rabbits. Exp. Eye Res. 2016, 149, 1–7. [Google Scholar] [CrossRef] [PubMed]
- McCurley, A.; Jaffe, I.Z. Mineralocorticoid receptors in vascular function and disease. Mol. Cell. Endocrinol. 2012, 350, 256–265. [Google Scholar] [CrossRef] [PubMed]
- Nguyen Dinh Cat, A.; Griol-Charhbili, V.; Loufrani, L.; Labat, C.; Benjamin, L.; Farman, N.; Lacolley, P.; Henrion, D.; Jaisser, F. The endothelial mineralocorticoid receptor regulates vasoconstrictor tone and blood pressure. FASEB J. 2010, 24, 2454–2463. [Google Scholar] [CrossRef] [PubMed]
- Zhao, M.; Celerier, I.; Bousquet, E.; Jeanny, J.C.; Jonet, L.; Savoldelli, M.; Offret, O.; Curan, A.; Farman, N.; Jaisser, F.; et al. Mineralocorticoid receptor is involved in rat and human ocular chorioretinopathy. J. Clin. Investig. 2012, 122, 2672–2679. [Google Scholar] [CrossRef] [PubMed]
- Jamali, N.; Wang, S.; Darjatmoko, S.R.; Sorenson, C.M.; Sheibani, N. Vitamin D receptor expression is essential during retinal vascular development and attenuation of neovascularization by 1, 25(OH)2D3. PLoS ONE 2017, 12, e0190131. [Google Scholar] [CrossRef] [PubMed]
- Jamali, N.; Sorenson, C.M.; Sheibani, N. Vitamin D and regulation of vascular cell function. Am. J. Physiol. Heart Circ. Physiol. 2018, 314, H753–H765. [Google Scholar] [CrossRef]
- Layana, A.G.; Minnella, A.M.; Garhöfer, G.; Aslam, T.; Holz, F.G.; Leys, A.; Silva, R.; Delcourt, C.; Souied, E.; Seddon, J.M. Vitamin D and Age-Related Macular Degeneration. Nutrients 2017, 9, 1120. [Google Scholar] [CrossRef]
- Perez Serena, A.; Martinez Betancourt, D.P.; Gonzalez Del Valle, F.; Ruiz-Moreno, J.M. Serum 25-hydroxy vitamin D levels in age-related macular degeneration. Int. J. Retin. Vitr. 2022, 8, 17. [Google Scholar] [CrossRef] [PubMed]
- Audet-Walsh, E.; Giguére, V. The multiple universes of estrogen-related receptor α and γ in metabolic control and related diseases. Acta Pharmacol. Sin. 2015, 36, 51–61. [Google Scholar] [CrossRef]
- Son, Y.O.; Park, S.; Kwak, J.S.; Won, Y.; Choi, W.S.; Rhee, J.; Chun, C.H.; Ryu, J.H.; Kim, D.K.; Choi, H.S.; et al. Estrogen-related receptor γ causes osteoarthritis by upregulating extracellular matrix-degrading enzymes. Nat. Commun. 2017, 8, 2133. [Google Scholar] [CrossRef]
- Percharde, M.; Lavial, F.; Ng, J.H.; Kumar, V.; Tomaz, R.A.; Martin, N.; Yeo, J.C.; Gil, J.; Prabhakar, S.; Ng, H.H.; et al. Ncoa3 functions as an essential Esrrb coactivator to sustain embryonic stem cell self-renewal and reprogramming. Genes Dev. 2012, 26, 2286–2298. [Google Scholar] [CrossRef]
- Razzaque, M.A.; Masuda, N.; Maeda, Y.; Endo, Y.; Tsukamoto, T.; Osumi, T. Estrogen receptor-related receptor γ has an exceptionally broad specificity of DNA sequence recognition. Gene 2004, 340, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Huppunen, J.; Aarnisalo, P. Dimerization modulates the activity of the orphan nuclear receptor ERRγ. Biochem. Biophys. Res. Commun. 2004, 314, 964–970. [Google Scholar] [CrossRef] [PubMed]
- Harding, H.P.; Lazar, M.A. The orphan receptor Rev-ErbA alpha activates transcription via a novel response element. Mol. Cell. Biol. 1993, 13, 3113–3121. [Google Scholar] [CrossRef] [PubMed]
- Wilson, T.E.; Paulsen, R.E.; Padgett, K.A.; Milbrandt, J. Participation of non-zinc finger residues in DNA binding by two nuclear orphan receptors. Science 1992, 256, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Festuccia, N.; Owens, N.; Chervova, A.; Dubois, A.; Navarro, P. The combined action of Esrrb and Nr5a2 is essential for murine naive pluripotency. Development 2021, 148, dev199604. [Google Scholar] [CrossRef]
- Xu, J.; Zhu, D.; Sonoda, S.; He, S.; Spee, C.; Ryan, S.J.; Hinton, D.R. Over-expression of BMP4 inhibits experimental choroidal neovascularization by modulating VEGF and MMP-9. Angiogenesis 2012, 15, 213–227. [Google Scholar] [CrossRef]
- Marin-Castano, M.E.; Striker, G.E.; Alcazar, O.; Catanuto, P.; Espinosa-Heidmann, D.G.; Cousins, S.W. Repetitive nonlethal oxidant injury to retinal pigment epithelium decreased extracellular matrix turnover in vitro and induced sub-RPE deposits in vivo. Investig. Ophthalmol. Vis. Sci. 2006, 47, 4098–4112. [Google Scholar] [CrossRef] [PubMed]
- Lambert, V.; Wielockx, B.; Munaut, C.; Galopin, C.; Jost, M.; Itoh, T.; Werb, Z.; Baker, A.; Libert, C.; Krell, H.W.; et al. MMP-2 and MMP-9 synergize in promoting choroidal neovascularization. FASEB J. 2003, 17, 2290–2292. [Google Scholar] [CrossRef]
- Puistola, U.; Westerlund, A.; Kauppila, A.; Turpeenniemi-Hujanen, T. Regulation of 72-kd type IV collagenase-matrix metalloproteinase-2 by estradiol and gonadotropin-releasing hormone agonist in human granulosa-lutein cells. Fertil. Steril. 1995, 64, 81–87. [Google Scholar] [CrossRef]
- Marin-Castano, M.E.; Elliot, S.J.; Potier, M.; Karl, M.; Striker, L.J.; Striker, G.E.; Csaky, K.G.; Cousins, S.W. Regulation of estrogen receptors and MMP-2 expression by estrogens in human retinal pigment epithelium. Investig. Ophthalmol. Vis. Sci. 2003, 44, 50–59. [Google Scholar] [CrossRef]
- Cousins, S.W.; Marin-Castano, M.E.; Espinosa-Heidmann, D.G.; Alexandridou, A.; Striker, L.; Elliot, S. Female gender, estrogen loss, and Sub-RPE deposit formation in aged mice. Investig. Ophthalmol. Vis. Sci. 2003, 44, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Pare, G.; Krust, A.; Karas, R.H.; Dupont, S.; Aronovitz, M.; Chambon, P.; Mendelsohn, M.E. Estrogen receptor-α mediates the protective effects of estrogen against vascular injury. Circ. Res. 2002, 90, 1087–1092. [Google Scholar] [CrossRef] [PubMed]
- Marcantoni, E.; Di Francesco, L.; Totani, L.; Piccoli, A.; Evangelista, V.; Tacconelli, S.; Patrignani, P. Effects of estrogen on endothelial prostanoid production and cyclooxygenase-2 and heme oxygenase-1 expression. Prostaglandins Other Lipid Mediat. 2012, 98, 122–128. [Google Scholar] [CrossRef]
- Bolego, C.; Rossoni, G.; Fadini, G.P.; Vegeto, E.; Pinna, C.; Albiero, M.; Boscaro, E.; Agostini, C.; Avogaro, A.; Gaion, R.M.; et al. Selective estrogen receptor-alpha agonist provides widespread heart and vascular protection with enhanced endothelial progenitor cell mobilization in the absence of uterotrophic action. FASEB J. 2010, 24, 2262–2272. [Google Scholar] [CrossRef] [PubMed]
- Harris, H.A.; Katzenellenbogen, J.A.; Katzenellenbogen, B.S. Characterization of the biological roles of the estrogen receptors, ERα and ERβ, in estrogen target tissues in vivo through the use of an ERalpha-selective ligand. Endocrinology 2002, 143, 4172–4177. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, D.P.; Norris, J.D. Analysis of the molecular pharmacology of estrogen receptor agonists and antagonists provides insights into the mechanism of action of estrogen in bone. Osteoporos. Int. 1997, 7 (Suppl. S1), S29–S34. [Google Scholar] [CrossRef] [PubMed]
- DuSell, C.D.; Nelson, E.R.; Wang, X.; Abdo, J.; Modder, U.I.; Umetani, M.; Gesty-Palmer, D.; Javitt, N.B.; Khosla, S.; McDonnell, D.P. The endogenous selective estrogen receptor modulator 27-hydroxycholesterol is a negative regulator of bone homeostasis. Endocrinology 2010, 151, 3675–3685. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.M.; Flamini, M.I.; Zullino, S.; Gopal, S.; Genazzani, A.R.; Simoncini, T. Estrogen receptor-α promotes endothelial cell motility through focal adhesion kinase. Mol. Hum. Reprod. 2011, 17, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Johns, A.; Freay, A.D.; Fraser, W.; Korach, K.S.; Rubanyi, G.M. Disruption of estrogen receptor gene prevents 17 beta estradiol-induced angiogenesis in transgenic mice. Endocrinology 1996, 137, 4511–4513. [Google Scholar] [CrossRef] [PubMed]
- Haran, E.F.; Maretzek, A.F.; Goldberg, I.; Horowitz, A.; Degani, H. Tamoxifen enhances cell death in implanted MCF7 breast cancer by inhibiting endothelium growth. Cancer Res. 1994, 54, 5511–5514. [Google Scholar] [PubMed]
- Aranda, A.; Pascual, A. Nuclear hormone receptors and gene expression. Physiol. Rev. 2001, 81, 1269–1304. [Google Scholar] [CrossRef]
- Furrer, R.; Eisele, P.S.; Schmidt, A.; Beer, M.; Handschin, C. Paracrine cross-talk between skeletal muscle and macrophages in exercise by PGC-1α-controlled BNP. Sci. Rep. 2017, 7, 40789. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Bai, P.; Xiao, L.; Shen, M.; Yu, Q.; Lei, Y.; Huang, W.; Lin, X.; Zheng, X.; Wei, T.; et al. Mediator complex subunit 16 is down-regulated in papillary thyroid cancer, leading to increased transforming growth factor-β signaling and radioiodine resistance. J. Biol. Chem. 2020, 295, 10726–10740. [Google Scholar] [CrossRef]
- Wang, X.; Ma, W.; Han, S.; Meng, Z.; Zhao, L.; Yin, Y.; Wang, Y.; Li, J. TGF-β participates choroid neovascularization through Smad2/3-VEGF/TNF-α signaling in mice with Laser-induced wet age-related macular degeneration. Sci. Rep. 2017, 7, 9672. [Google Scholar] [CrossRef] [PubMed]
- Malek, G.; Li, C.M.; Guidry, C.; Medeiros, N.E.; Curcio, C.A. Apolipoprotein B in cholesterol-containing drusen and basal deposits of human eyes with age-related maculopathy. Am. J. Pathol. 2003, 162, 413–425. [Google Scholar] [CrossRef]
- Voigt, A.P.; Mulfaul, K.; Mullin, N.K.; Flamme-Wiese, M.J.; Giacalone, J.C.; Stone, E.M.; Tucker, B.A.; Scheetz, T.E.; Mullins, R.F. Single-cell transcriptomics of the human retinal pigment epithelium and choroid in health and macular degeneration. Proc. Natl. Acad. Sci. USA 2019, 116, 24100–24107. [Google Scholar] [CrossRef]






| Gene | 1°CEC | fCh |
|---|---|---|
| AR | (++) | (+) |
| ARNT | (++) | (+++) |
| COUP-TF2 | (+++) | (+) |
| EAR2 | (+++) | (++) |
| ERRα | (++) | (+) |
| ERRβ | (−) | (+) |
| ERRγ | (−) | (+) |
| ERβ | (+) | (−) |
| FXR | (−) | (+) |
| GR | (+) | (++) |
| HNF4α | (−) | (+) |
| HNF4γ | (−) | (+) |
| NUR77 | (−) | (+) |
| NOR1 | (+) | (++) |
| NURR1 | (+) | (++) |
| PNR | (++) | (−) |
| PPAR-β/δ2 | (++) | (+++) |
| PR | (−) | (+) |
| REV-ERBα | (++) | (+) |
| RORα | (++) | (+++) |
| RORβ | (+) | (+++) |
| RXRβ | (−) | (++) |
| RORγ | (−) | (+++) |
| TLX | (−) | (+) |
| TR2 | (+) | (−) |
| TRβ | (+) | (++) |
| Sr. No. | Gene | Alias | Fold Regulation | p-Value |
|---|---|---|---|---|
| 1. | ERα | Estrogen Receptor 1 | −2.04 | 0.000066 |
| 2. | ERRβ | Estrogen Related Receptor Beta | −6.56 | 0 |
| 3. | ERRγ | Estrogen Related Receptor Gamma | −3 | 0.000009 |
| 4. | HNF4α | Hepatocyte Nuclear Factor 4 Alpha | −2.46 | 0.000059 |
| 5. | MED16 | Mediator Complex Subunit 16 | −2.14 | 0.000001 |
| 6. | REV-ERBα | V-ErbA-Related Protein 1 | −2.71 | 0 |
| 7. | REV-ERBβ | V-ErbA-Related Protein 1-Related | −2.22 | 0 |
| 8. | PNR | Photoreceptor-Specific Nuclear Receptor | −5.55 | 0 |
| 9. | MR | Mineralocorticoid Receptor | −2.58 | 0 |
| 10. | NUR77 | Nuclear Hormone Receptor NUR/77 | −2.03 | 0.000002 |
| 11. | PR | Progesterone Receptor | −2.93 | 0 |
| 12. | PPARα | Peroxisome Proliferator Activated Receptor Alpha | −2.14 | 0 |
| 13. | PPARGC1A | PPARG Coactivator 1 Alpha | −2.13 | 0 |
| 14. | RXRα | Retinoid X Receptor Alpha | −2.03 | 0.000004 |
| 15. | RARγ | Retinoid X Receptor Gamma | −2.34 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peavey, J.; Parmar, V.M.; Malek, G. Nuclear Receptor Atlases of Choroidal Tissues Reveal Candidate Receptors Associated with Age-Related Macular Degeneration. Cells 2022, 11, 2386. https://doi.org/10.3390/cells11152386
Peavey J, Parmar VM, Malek G. Nuclear Receptor Atlases of Choroidal Tissues Reveal Candidate Receptors Associated with Age-Related Macular Degeneration. Cells. 2022; 11(15):2386. https://doi.org/10.3390/cells11152386
Chicago/Turabian StylePeavey, Jeremy, Vipul M. Parmar, and Goldis Malek. 2022. "Nuclear Receptor Atlases of Choroidal Tissues Reveal Candidate Receptors Associated with Age-Related Macular Degeneration" Cells 11, no. 15: 2386. https://doi.org/10.3390/cells11152386






