Golden Syrian Hamster Models for Cancer Research
Abstract
:1. History of Hamsters as Animal Models in Cancer Research
2. Is There a Need for More Genetically Engineered Rodent Models of Cancer?
3. Genetic Engineering in the Golden Syrian Hamster
4. KCNQ1, TP53, and IL2RG Genetically Engineered Hamster Cancer Models
4.1. KCNQ1 Knockout Hamster Model
4.2. TP53 Knockout Hamster Model
4.3. IL2RG KO Hamster Model
5. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, Z.; Xiong, C.; Mo, S.; Tian, H.; Yu, M.; Mao, T.; Chen, Q.; Luo, H.; Li, Q.; Lu, J.; et al. Comprehensive transcriptome analyses of the fructose-fed Syrian Golden hamster liver provides novel insights into lipid metabolism. PLoS ONE 2016, 11, e0162402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ebihara, H.; Zivcec, M.; Gardner, D.; Falzarano, D.; LaCasse, R.; Rosenke, R.; Long, D.; Haddock, E.; Fischer, E.; Kawaoka, Y.; et al. A Syrian golden hamster model recapitulating ebola hemorrhagic fever. J. Infect. Dis. 2013, 207, 306–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guillaume, V.; Wong, K.T.; Looi, R.Y.; Georges-Courbot, M.C.; Barrot, L.; Buckland, R.; Wild, T.F.; Horvat, B. Acute Hendra virus infection: Analysis of the pathogenesis and passive antibody protection in the hamster model. Virology 2009, 387, 459–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jove, M.; Ayala, V.; Ramirez-Nunez, O.; Serrano, J.C.; Cassanye, A.; Arola, L.; Caimari, A.; Del Bas, J.M.; Crescenti, A.; Pamplona, R.; et al. Lipidomic and metabolomic analyses reveal potential plasma biomarkers of early atheromatous plaque formation in hamsters. Cardiovasc. Res. 2013, 97, 642–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paciello, O.; Wojcik, S.; Gradoni, L.; Oliva, G.; Trapani, F.; Iovane, V.; Politano, L.; Papparella, S. Syrian hamster infected with Leishmania infantum: A new experimental model for inflammatory myopathies. Muscle Nerve 2010, 41, 355–361. [Google Scholar] [CrossRef] [PubMed]
- Best, E.L.; Freeman, J.; Wilcox, M.H. Models for the study of Clostridium difficile infection. Gut Microbes 2012, 3, 145–167. [Google Scholar] [CrossRef] [Green Version]
- Wahl-Jensen, V.; Bollinger, L.; Safronetz, D.; de Kok-Mercado, F.; Scott, D.P.; Ebihara, H. Use of the Syrian hamster as a new model of ebola virus disease and other viral hemorrhagic fevers. Viruses 2012, 4, 3754–3784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safronetz, D.; Ebihara, H.; Feldmann, H.; Hooper, J.W. The Syrian hamster model of hantavirus pulmonary syndrome. Antivir. Res. 2012, 95, 282–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weisburger, J.H. Colon carcinogens: Their metabolism and mode of action. Cancer 1971, 28, 60–70. [Google Scholar] [CrossRef]
- Della Porta, G. Induction of intestinal, mammary, and ovarian tumors in hamsters with oral administration of 20-Methylcholanthrene. Cancer Res. 1961, 21, 575–578. [Google Scholar] [PubMed]
- Vairaktaris, E.; Spyridonidou, S.; Papakosta, V.; Vylliotis, A.; Lazaris, A.; Perrea, D.; Yapijakis, C.; Patsouris, E. The hamster model of sequential oral oncogenesis. Oral Oncol. 2008, 44, 315–324. [Google Scholar] [CrossRef]
- Takahashi, M.; Hori, M.; Mutoh, M.; Wakabayashi, K.; Nakagama, H. Experimental animal models of pancreatic carcinogenesis for prevention studies and their relevance to human disease. Cancers 2011, 3, 582–602. [Google Scholar] [CrossRef] [PubMed]
- Wold, W.S.; Toth, K. Chapter three—Syrian hamster as an animal model to study oncolytic adenoviruses and to evaluate the efficacy of antiviral compounds. Adv. Cancer Res. 2012, 115, 69–92. [Google Scholar] [CrossRef] [PubMed]
- Ying, B.; Toth, K.; Spencer, J.F.; Aurora, R.; Wold, W.S.M. Transcriptome sequencing and development of an expression microarray platform for liver infection in adenovirus type 5-infected Syrian Golden hamsters. Virology 2015, 485, 305–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, J.; Guo, L.; VonTungeln, L.; Vanlandingham, M.; Cerniglia, C.E.; Chen, H. Smokeless tobacco impacts oral microbiota in a Syrian Golden hamster cheek pouch carcinogenesis model. Anaerobe 2018, 52, 29–42. [Google Scholar] [CrossRef]
- Terasaki, M.; Nishizaka, Y.; Murase, W.; Kubota, A.; Kojima, H.; Kojoma, M.; Tanaka, T.; Maeda, H.; Miyashita, K.; Mutoh, M.; et al. Effect of Fucoxanthinol on Pancreatic Ductal Adenocarcinoma Cells from an N-Nitrosobis(2-oxopropyl)amine-initiated Syrian Golden Hamster Pancreatic Carcinogenesis Model. Cancer Genom. Proteom. 2021, 18 (Suppl. 3), 407–423. [Google Scholar] [CrossRef]
- Wang, W.C.; Huang, M.Y.; Chen, Y.K.; Lan, W.C.; Shieh, T.M.; Shih, Y.H. Salivary Exosome Proteomics and Bioinformatics Analysis in 7,12-Dimethylbenz[a]anthracene-Induced Oral Cancer with Radiation Therapy-A Syrian Golden Hamster Model. Diagnostics 2021, 12, 65. [Google Scholar] [CrossRef] [PubMed]
- Kreimann, E.L.; Itoiz, M.E.; Dagrosa, A.; Garavaglia, R.; Farías, S.; Batistoni, D.; Schwint, A.E. The hamster cheek pouch as a model of oral cancer for boron neutron capture therapy studies: Selective delivery of boron by boronophenylalanine. Cancer Res. 2001, 61, 8775–8781. [Google Scholar] [PubMed]
- Santa Cruz, I.S.; Garabalino, M.A.; Trivillin, V.A.; Itoiz, M.E.; Pozzi, E.C.C.; Thorp, S.; Curotto, P.; Guidobono, J.S.; Heber, E.M.; Nigg, D.W.; et al. Optimization of the classical oral cancerization protocol in hamster to study oral cancer therapy. Oral Dis. 2020, 26, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Yapijakis, C.; Kalogera, S.; Papakosta, V.; Vassiliou, S. The Hamster Model of Sequential Oral Carcinogenesis: An Update. In Vivo 2019, 33, 1751–1755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tchitchek, N.; Safronetz, D.; Rasmussen, A.L.; Martens, C.; Virtaneva, K.; Porcella, S.F.; Feldmann, H.; Ebihara, H.; Katze, M.G. Sequencing, annotation and analysis of the Syrian hamster (Mesocricetus auratus) transcriptome. PLoS ONE 2014, 9, e112617. [Google Scholar] [CrossRef] [Green Version]
- Fan, Z.; Li, W.; Lee, S.R.; Meng, Q.; Shi, B.; Bunch, T.D.; White, K.L.; Kong, I.K.; Wang, Z. Efficient gene targeting in golden Syrian hamsters by the CRISPR/Cas9 system. PLoS ONE 2014, 9, e109755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, R.; Miao, J.; Fan, Z.; Song, S.; Kong, I.K.; Wang, Y.; Wang, Z. Production of Genetically Engineered Golden Syrian Hamsters by Pronuclear Injection of the CRISPR/Cas9 Complex. J. Vis. Exp. 2018, 131, e56263. [Google Scholar] [CrossRef] [PubMed]
- Sanmamed, M.F.; Chester, C.; Melero, I.; Kohrt, H. Defining the optimal murine models to investigate immune checkpoint blockers and their combination with other immunotherapies. Ann. Oncol. 2016, 27, 1190–1198. [Google Scholar] [CrossRef] [PubMed]
- Yip, H.; Haupt, C.; Maresh, G.; Zhang, X.; Li, L. Humanized mice for immune checkpoint blockade in human solid tumors. Am. J. Clin. Exp. Urol. 2019, 7, 313–320. [Google Scholar] [PubMed]
- Doty, D.T.; Schueler, J.; Mott, V.L.; Bryan, C.M.; Moore, N.F.; Ho, J.C.; Borenstein, J.T. Modeling Immune Checkpoint Inhibitor Efficacy in Syngeneic Mouse Tumors in an Ex Vivo Immuno-Oncology Dynamic Environment. Int. J. Mol. Sci. 2020, 21, 6478. [Google Scholar] [CrossRef] [PubMed]
- Adam, K.; Iuga, A.; Tocheva, A.S.; Mor, A. A novel mouse model for checkpoint inhibitor-induced adverse events. PLoS ONE 2021, 16, e0246168. [Google Scholar] [CrossRef] [PubMed]
- Kersten, K.; de Visser, K.E.; van Miltenburg, M.H.; Jonkers, J. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol. Med. 2017, 9, 137–153. [Google Scholar] [CrossRef] [PubMed]
- Contreras, X.; Amberg, N.; Davaatseren, A.; Hansen, A.H.; Sonntag, J.; Andersen, L.; Bernthaler, T.; Streicher, C.; Heger, A.; Johnson, R.L.; et al. A genome-wide library of MADM mice for single-cell genetic mosaic analysis. Cell Rep. 2021, 35, 109274. [Google Scholar] [CrossRef]
- Cheon, D.J.; Orsulic, S. Mouse models of cancer. Annu. Rev. Pathol. 2011, 6, 95–119. [Google Scholar] [CrossRef]
- Ireson, C.R.; Alavijeh, M.S.; Palmer, A.M.; Fowler, E.R.; Jones, H.J. The role of mouse tumour models in the discovery and development of anticancer drugs. Br. J. Cancer 2019, 121, 101–108. [Google Scholar] [CrossRef] [PubMed]
- de Jong, M.; Maina, T. Of mice and humans: Are they the same?—Implications in cancer translational research. J. Nucl. Med. 2010, 51, 501–504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kulke, M.H.; Bergsland, E.K.; Ryan, D.P.; Enzinger, P.C.; Lynch, T.J.; Zhu, A.X.; Meyerhardt, J.A.; Heymach, J.V.; Fogler, W.E.; Sidor, C.; et al. Phase II study of recombinant human endostatin in patients with advanced neuroendocrine tumors. J. Clin. Oncol. 2006, 24, 3555–3561. [Google Scholar] [CrossRef]
- Macpherson, D. Insights from mouse models into human retinoblastoma. Cell Div. 2008, 3, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacks, T.; Remington, L.; Williams, B.O.; Schmitt, E.M.; Halachmi, S.; Bronson, R.T.; Weinberg, R.A. Tumor spectrum analysis in p53-mutant mice. Curr. Biol 1994, 4, 1–7. [Google Scholar] [CrossRef]
- Su, L.K.; Kinzler, K.W.; Vogelstein, B.; Preisinger, A.C.; Moser, A.R.; Luongo, C.; Gould, K.A.; Dove, W.F. Multiple intestinal neoplasia caused by a mutation in the murine homolog of the APC gene. Science 1992, 256, 668–670. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Li, X.; Wang, J.; Gao, D.; Li, Y.; Li, H.; Chu, Y.; Zhang, Z.; Liu, H.; Jiang, G.; et al. Re-designing Interleukin-12 to enhance its safety and potential as an anti-tumor immunotherapeutic agent. Nat. Commun. 2017, 8, 1395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, J.X.; Wang, J.Y.; Li, H.Z.; Guo, H.R.; Dunmall, L.S.C.; Zhang, Z.X.; Cheng, Z.G.; Gao, D.L.; Dong, J.Z.; Wang, Z.D.; et al. Promising xenograft animal model recapitulating the features of human pancreatic cancer. World J. Gastroenterol. 2020, 26, 4802–4816. [Google Scholar] [CrossRef]
- Jandrig, B.; Krause, H.; Zimmermann, W.; Vasiliunaite, E.; Gedvilaite, A.; Ulrich, R.G. Hamster Polyomavirus Research: Past, Present, and Future. Viruses 2021, 13, 907. [Google Scholar] [CrossRef]
- Golden, J.W.; Li, R.; Cline, C.R.; Zeng, X.; Mucker, E.M.; Fuentes-Lao, A.J.; Spik, K.W.; Williams, J.A.; Twenhafel, N.; Davis, N.; et al. Hamsters Expressing Human Angiotensin-Converting Enzyme 2 Develop Severe Disease following Exposure to SARS-CoV-2. mBio 2022, 13, e0290621. [Google Scholar] [CrossRef]
- Halfmann, P.J.; Kuroda, M.; Maemura, T.S.; Armbrust, T.; Wright, R.; Balaram, A.; Florek, K.R.; Bateman, A.C.; Kawaoka, Y. Efficacy of vaccination and previous infection against the Omicron BA.1 variant in Syrian hamsters. Cell Rep. 2022, 39, 110688. [Google Scholar] [CrossRef] [PubMed]
- Uraki, R.; Kiso, M.; Iida, S.; Imai, M.; Takashita, E.; Kuroda, M.; Halfmann, P.J.; Loeber, S.; Maemura, T.; Yamayoshi, S.; et al. Characterization and antiviral susceptibility of SARS-CoV-2 Omicron BA.2. Nature 2022, 607, 119–127. [Google Scholar] [CrossRef] [PubMed]
- den Uil, S.H.; Coupé, V.M.; Linnekamp, J.F.; van den Broek, E.; Goos, J.A.; Delis-van, D.P.M.; Belt, E.J.; van Grieken, N.C.; Scott, P.M.; Vermeulen, L.; et al. Loss of KCNQ1 expression in stage II and stage III colon cancer is a strong prognostic factor for disease recurrence. Br. J. Cancer 2016, 115, 1565–1574. [Google Scholar] [CrossRef] [Green Version]
- Than, B.L.; Goos, J.A.; Sarver, A.L.; O’Sullivan, M.G.; Rod, A.; Starr, T.K.; Fijneman, R.J.; Meijer, G.A.; Zhao, L.; Zhang, Y.; et al. The role of KCNQ1 in mouse and human gastrointestinal cancers. Oncogene 2014, 33, 3861–3868. [Google Scholar] [CrossRef] [Green Version]
- Uil, S.H.; Coupé, V.M.H.; Bril, H.; Meijer, G.A.; Fijneman, R.J.A.; Stockmann, H.B.A.C. KCNQ1 and lymphovascular invasion are key features in a prognostic classifier for stage II and III colon cancer. BMC Cancer 2022, 22, 372. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Miao, J.; Tabaran, A.F.; O’Sullivan, M.G.; Anderson, K.J.; Scott, P.M.; Wang, Z.; Cormier, R.T. A novel cancer syndrome caused by KCNQ1-deficiency in the golden Syrian hamster. J. Carcinog. 2018, 17, 6. [Google Scholar] [CrossRef]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aubrey, B.J.; Strasser, A. Tumor-Suppressor Functions of the TP53 Pathway. Cold Spring Harb. Perspect Med. 2016, 6, a026062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miao, J.; Li, R.; Wettere, A.J.V.; Guo, H.; Tabaran, A.F.; Carlson, T.; Scott, P.M.; Chen, K.; Cormier, R.T. Cancer spectrum in TP53-deficient golden Syrian hamsters: A new model for Li-Fraumeni syndrome. J. Carcinog. 2021, 20, 18. [Google Scholar] [CrossRef] [PubMed]
- Molica, M.; Mazzone, C.; Niscola, P.; de Fabritiis, P. TP53 Mutations in Acute Myeloid Leukemia: Still a Daunting Challenge? Front. Oncol. 2021, 10, 610820. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.X.; Leonard, W.J. The Common Cytokine Receptor γ Chain Family of Cytokines. Cold Spring Harb. Perspect Biol. 2018, 10, a028449. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Ying, B.; Liu, Y.; Spencer, J.F.; Miao, J.; Tollefson, A.E.; Brien, J.D.; Wang, Y.; Wold, W.S.M.; Wang, Z.; et al. Generation and characterization of an IL2rg knockout Syrian hamster model for XSCID and HAdV-C6 infection in immunocompromised patients. Dis. Model Mech. 2020, 13, dmm044602. [Google Scholar] [CrossRef] [PubMed]
- Lan, T.; Xue, X.; Dunmall, L.C.; Miao, J.; Wang, Y. Patient-derived xenograft: A developing tool for screening biomarkers and potential therapeutic targets for human esophageal cancers. Aging 2021, 13, 12273–12293. [Google Scholar] [CrossRef]
- Dobrolecki, L.E.; Airhart, S.D.; Alferez, D.G.; Aparicio, S.; Behbod, F.; Bentires-Alj, M.; Brisken, C.; Bult, C.J. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev. 2016, 35, 547–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, N.C.; Kenney, L.L.; Jangalwe, S.; Aryee, K.E.; Greiner, D.L.; Brehm, M.A.; Shultz, L.D. Humanized Mouse Models of Clinical Disease. Annu. Rev. Pathol. 2017, 12, 187–215. [Google Scholar] [CrossRef] [Green Version]
- Akkina, R.; Allam, A.; Balazs, A.B.; Blankson, J.N.; Burnett, J.C.; Casares, S.; Garcia, J.V.; Hasenkrug, K.J.; Kashanchi, F.; Kitchen, S.G.; et al. Improvements and Limitations of Humanized Mouse Models for HIV Research: NIH/NIAID “Meet the Experts” 2015 Workshop Summary. AIDS Res. Hum. Retrovir. 2016, 32, 109–119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, T.M.; Brehm, M.A.; Bridges, S.; Ferguson, S.; Kumar, P.; Mirochnitchenko, O.; Palucka, K.; Pelanda, R.; Sanders-Beer, B.; Shultz, L.D.; et al. Humanized immune system mouse models: Progress, challenges and opportunities. Nat. Immunol. 2019, 20, 770–774. [Google Scholar] [CrossRef] [PubMed]
- Shultz, L.D.; Lyons, B.L.; Burzenski, L.M.; Gott, B.; Chen, X.; Chaleff, S.; Kotb, M.; Gillies, S.D.; King, M.; Mangada, J.; et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J. Immunol. 2005, 174, 6477–6489. [Google Scholar] [CrossRef] [Green Version]
- Theocharides, A.P.; Rongvaux, A.; Fritsch, K.; Flavell, R.A.; Manz, M.G. Humanized hemato-lymphoid system mice. Haematologica 2016, 101, 5–19. [Google Scholar] [CrossRef]
- Willinger, T.; Rongvaux, A.; Takizawa, H.; Yancopoulos, G.D.; Valenzuela, D.M.; Murphy, A.J.; Auerbach, W.; Eynon, E.E.; Stevens, S.; Manz, M.G.; et al. Human IL-3/GM-CSF knock-in mice support human alveolar macrophage development and human immune responses in the lung. Proc. Natl. Acad. Sci. USA 2011, 108, 2390–2395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rongvaux, A.; Willinger, T.; Martinek, J.; Strowig, T.; Gearty, S.V.; Teichmann, L.L.; Saito, Y.; Marches, F.; Halene, S.; Palucka, A.K.; et al. Development and function of human innate immune cells in a humanized mouse model. Nat. Biotechnol. 2014, 32, 364–372. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Rongvaux, A.; Taylor, A.; Jiang, T.; Tebaldi, T.; Balasubramanian, K.; Bagale, A.; Terzi, Y.K.; Gbyli, R.; Wang, X.; et al. A highly efficient and faithful MDS patient-derived xenotransplantation model for pre-clinical studies. Nat. Commun. 2019, 10, 366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cho, S.A.; Park, J.H.; Seok, S.H.; Juhn, J.H.; Kim, S.J.; Ji, H.J.; Choo, Y.S.; Park, J.H. Effect of granulocyte macrophage-colony stimulating factor (GM-CSF) on 5-FU-induced ulcerative mucositis in hamster buccal pouches. Exp. Toxicol. Pathol. 2006, 57, 321–328. [Google Scholar] [CrossRef] [PubMed]
- Bortolanza, S.; Bunuales, M.; Otano, I.; Gonzalez-Aseguinolaza, G.; Ortiz-de-Solorzano, C.; Perez, D.; Prieto, J.; Hernandez-Alcoceba, R. Treatment of pancreatic cancer with an oncolytic adenovirus expressing interleukin-12 in Syrian hamsters. Mol. Ther. 2009, 17, 614–622. [Google Scholar] [CrossRef] [PubMed]
- Yunis, A.A.; Arimura, G.K.; Russin, D.J. Human pancreatic carcinoma (MIA PaCa-2) in continuous culture: Sensitivity to asparaginase. Int. J. Cancer 1977, 19, 128–135. [Google Scholar] [CrossRef] [PubMed]


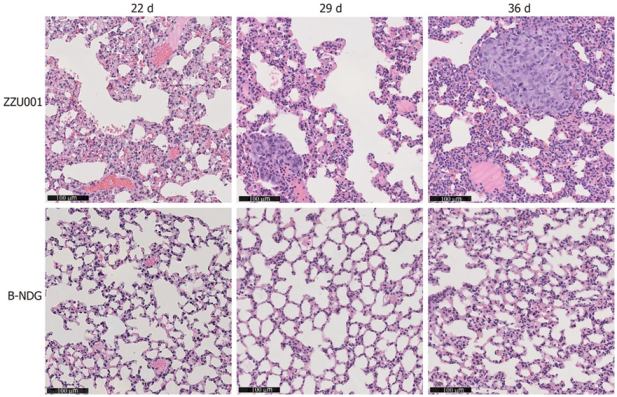
| MIAPaCa-2 (SC) | MIAPaCa-2 (OrT) | |||
|---|---|---|---|---|
| B-NDG | ZZU001 | B-NDG | ZZU001 | |
| Distant metastasis | ||||
| Liver | - | - | 3/5 (60) | 5/5 (100) |
| Lung | - | 5/5 (100) | - | 5/5 (100) |
| Retroperitoneum | - | - | 3/5 (60) | 5/5 (100) |
| Mesentery | - | - | 3/5 (60) | 5/5 (100) |
| Diaphragm | - | - | 2/5 (40) | 2/5 (40) |
| Spleen | - | - | 2/5 (40) | - |
| Stomach | - | - | - | 1/5 (20) |
| Kidney | - | 2/5 (40) | - | 5/5 (100) |
| Adrenal gland | - | 1/5 (20) | - | 2/5 (40) |
| Local infiltration | ||||
| Spleen | - | - | 3/5(60) | 4/5 (80) |
| Stomach | - | - | 1/5 (20) | 3/5 (60) |
| Liver (hilus) | - | - | 3/5 (60) | 5/5 (100) |
| Kidney (hilus) | - | - | - | 1/5 (20) |
| Retroperitoneum | - | - | 3/5 (60) | 5/5 (100) |
| Bowel | - | - | 3/5 (60) | 5/5 (100) |
| Mesentery (adjacent to pancreas) | - | 4/5 (80) | 5/5 (100) | |
| Signs of tumor burden | ||||
| Ascites | - | - | 3/5 (60) | 3/5 (60) |
| Jaundice | - | - | 2/5 (40) | 3/5 (60) |
| Ileus | - | - | - | 2/5 (40) |
| Cachexia | - | - | 1/5 (20) | 3/5 (60) |
| Panc-1 (SC) | SUIT-2 (SC) | Patu8988T (SC) | Capan-1 (SC) | |
|---|---|---|---|---|
| ZZU001 | ZZU001 | ZZU001 | ZZU001 | |
| Distant metastasis | ||||
| Liver | - | 1/3 (33) | 3/5 (60) | - |
| Lung | 5/5 (100) | 3/3 (100) | - | 5/5 (100) |
| Retroperitoneum | - | - | - | - |
| Mesentery | - | - | - | - |
| Diaphragm | - | - | - | - |
| Spleen | - | - | - | - |
| Stomach | - | - | - | - |
| Kidney | - | 1/3 (33) | 1/5 (20) | - |
| Adrenal gland | - | - | - | - |
| Head and Neck Cancer | 2008 Review of the Hamster Model of Sequential Oral Oncogenesis [11] |
| Results of the effect of smokeless tobacco on oral microbiota in the hamster cheek pouch carcinogenesis model [15] | |
| Salivary exosome proteomics and bioinformatics analysis of DMBA-induced oral cancer with radiation therapy in the hamster oral carcinogenesis model [17] | |
| Hamster cheek pouch model of oral cancer for boron neutron capture therapy studies: Selective delivery of boron by boronphenylalanine [18] | |
| A 2020 study describing optimization of the oral cancerization model in hamsters to study oral cancer therapy [19] | |
| A 2019 review of the hamster model of sequential oral carcinogenesis [20] | |
| Pancreatic cancer | A 2011 review of the use of hamsters for chemically induced pancreatic cancer, use in prevention, treatment and relevance to the human disease [12] |
| Effect of use of Fucoxanthinol on BOP-treated pancreatic ductal adenocarcinoma cells in a hamster pancreatic cancer model [16] | |
| IL2RG knockout created by CRISPR/cas9 technology for creation of PDX metastatic pancreatic cancer models—described in detail in this review [38] | |
| Other chemicallyinduced cancers | Very early (1961) study employing 20-Methylcholanthrene oral administration induction of intestinal, mammary and ovarian cancers [10] |
| Oncolytic adenoviruses | Chapter review by Wold and Toth, pioneers in this field, from 2012 that summarizes the use of the hamster as an animal model to study oncolytic adenoviruses and to evaluate the efficiacy of antiviral compounds [13] |
| Describes the use of an oncolytic adenoviral vector to express IL-12 to treat chemically induced pancreatic cancer in the hamster [64] | |
| Hamster polyoma virus | A very recent review of HaPV research, including the prevalence of HaPV in hamster colonies worldwide and the risk of lymphomas in HaPV positive hamsters [39] |
| KCNQ1 Knockout | Knockout generated by CRISPR/cas9 technology, develop a wide range of cancers, with the top four cancers being T-cell lymphomas, plasma cell tumors, hemangiosarcomas and myeloproliferative disorders, discussed in detail in this review [46] |
| TP53 Knockout | Knockout generated by CRISPR/cas9 technology, develop a wide range of cancers, with the top three cancers being lymphomas, AML and hemangiosarcomas, discussed in detail in this review [49] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Cormier, R.T. Golden Syrian Hamster Models for Cancer Research. Cells 2022, 11, 2395. https://doi.org/10.3390/cells11152395
Wang Z, Cormier RT. Golden Syrian Hamster Models for Cancer Research. Cells. 2022; 11(15):2395. https://doi.org/10.3390/cells11152395
Chicago/Turabian StyleWang, Zhongde, and Robert T. Cormier. 2022. "Golden Syrian Hamster Models for Cancer Research" Cells 11, no. 15: 2395. https://doi.org/10.3390/cells11152395
APA StyleWang, Z., & Cormier, R. T. (2022). Golden Syrian Hamster Models for Cancer Research. Cells, 11(15), 2395. https://doi.org/10.3390/cells11152395







