Differentiation of Human Induced Pluripotent Stem Cells from Patients with Severe COPD into Functional Airway Epithelium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patients’ Clinical Characteristics
2.2. Human Embryonic Stem Cell (ESC) and hiPSC Generation and Maintenance
2.3. HIPSC Differentiation
2.4. Statistical Analysis
3. Results
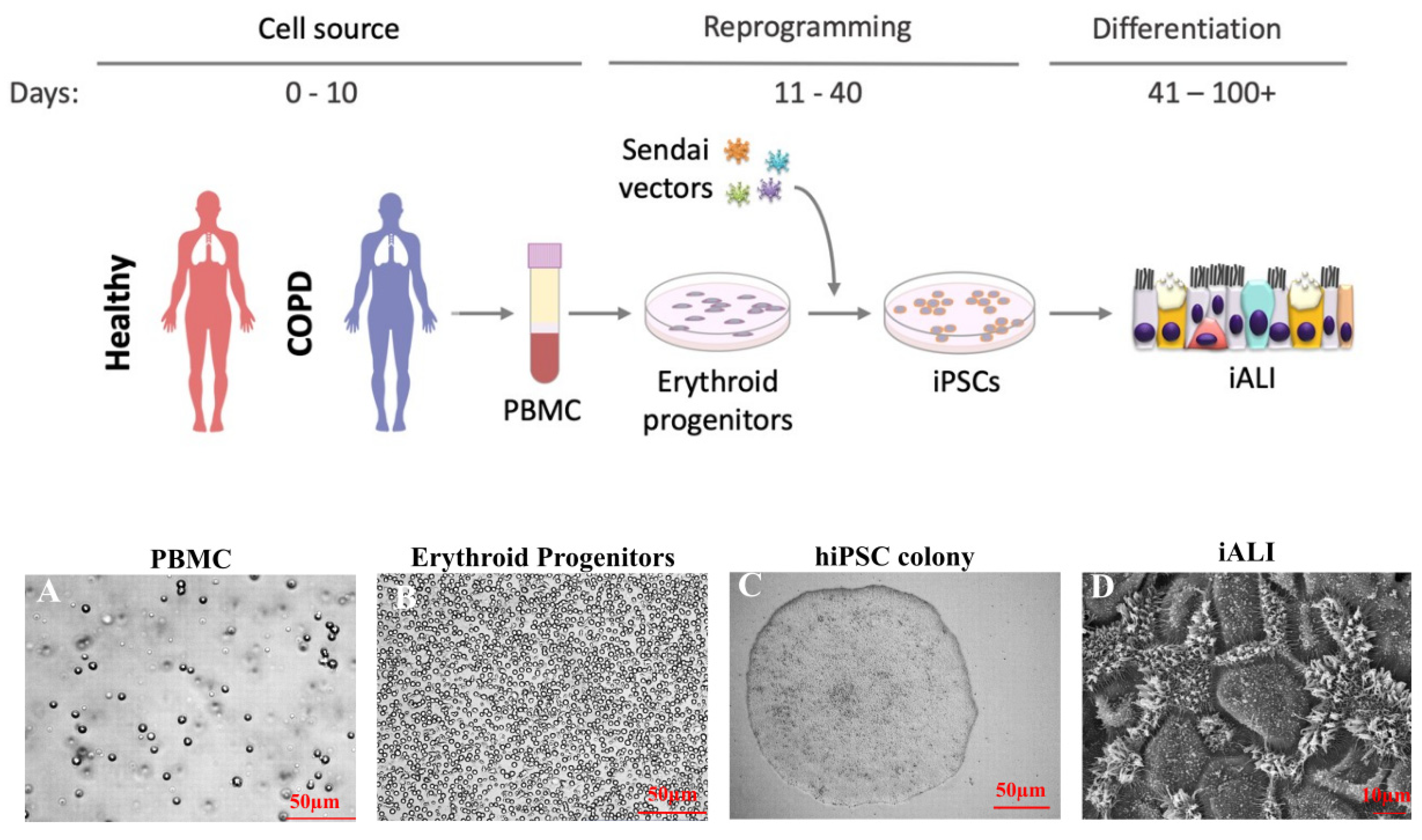
3.1. Reprogramming from Blood Samples
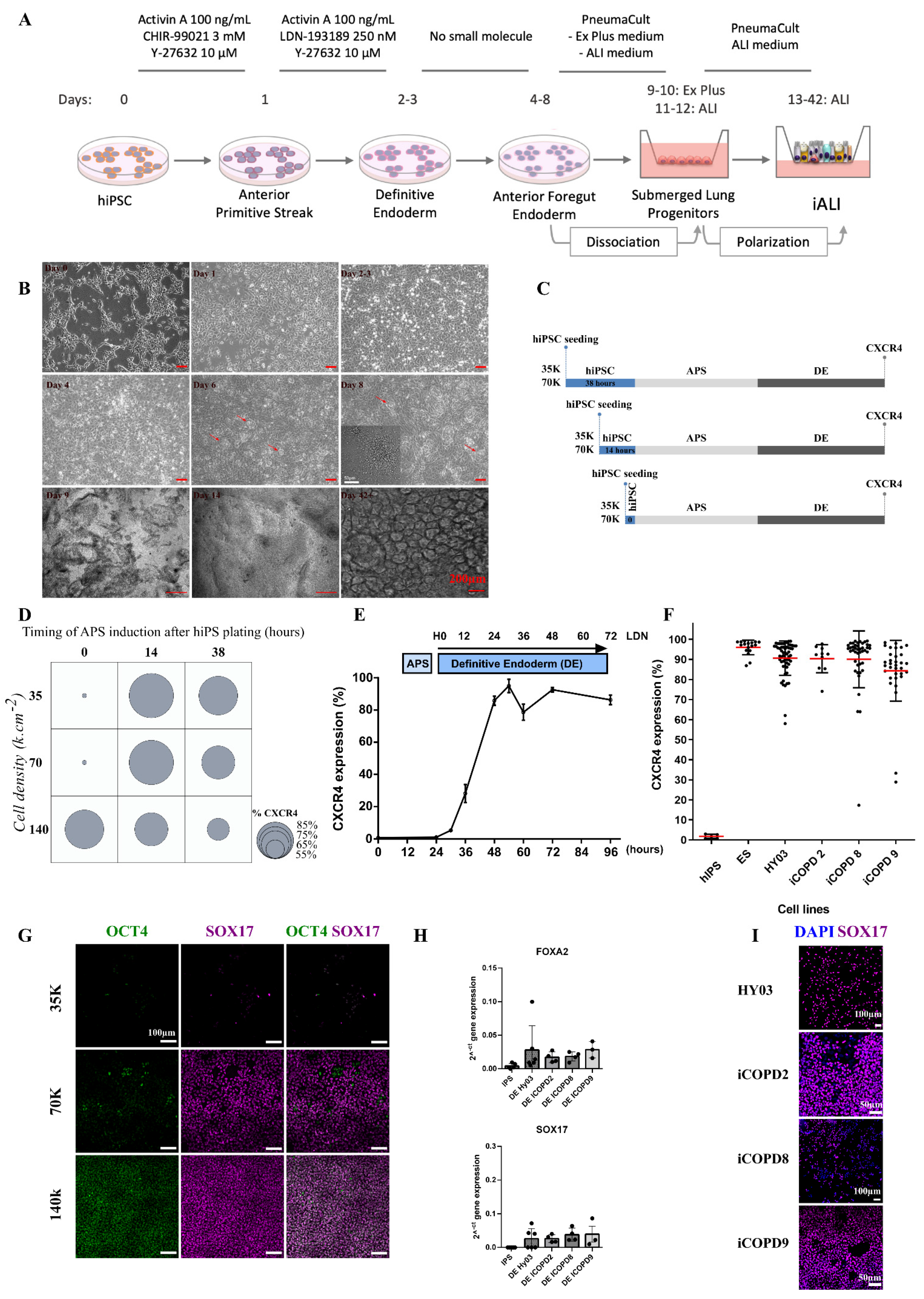
3.2. Cell Density and Induction Timing Are Critical for Successful Differentiation and Allows High Rate of Definitive Endoderm Induction
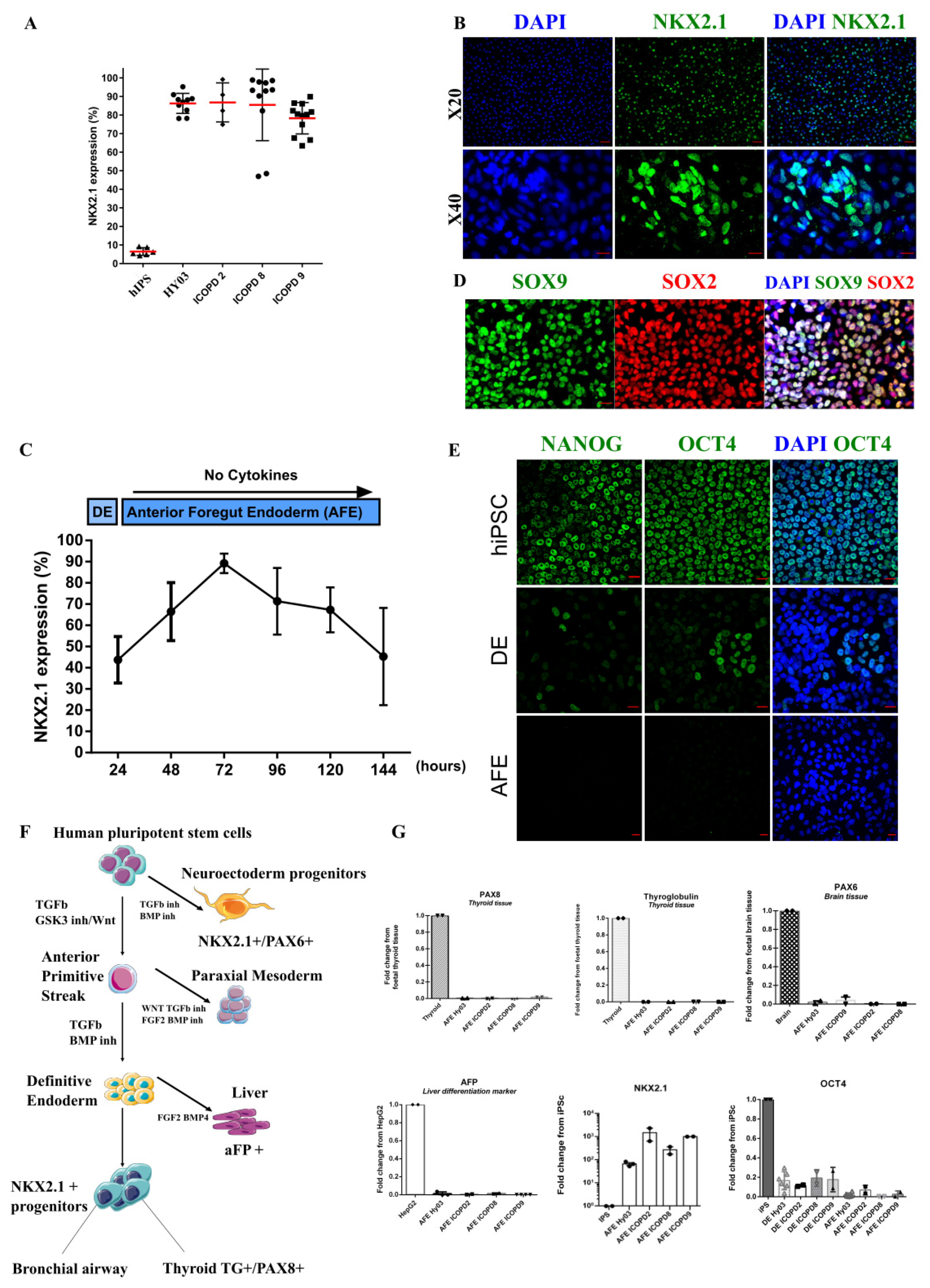
3.3. Efficient Induction of High Purity NKX2.1+ Lung Progenitors without Cell Sorting
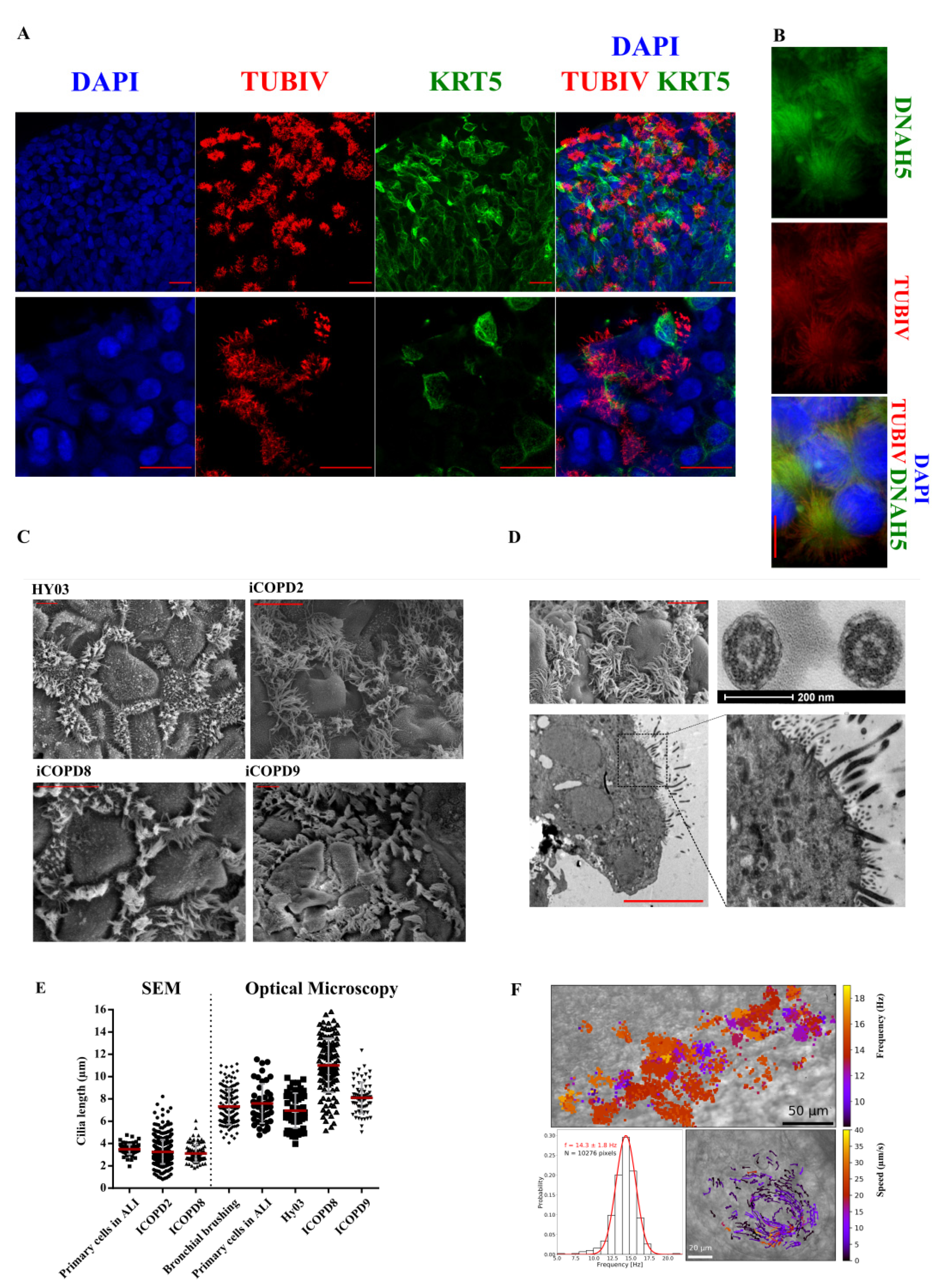
3.4. Specification of NKX2.1 Lung Progenitor Cells in 2D ALI Culture Conditions Leads to Functional, Multi-Ciliated Airway Epithelium
3.4.1. Epithelium with Barrier Function
3.4.2. IALI Bronchial Epithelium Includes Major and Rare Solitary Human Airway Epithelial Cells
3.4.3. Functional Multi-Ciliated Cell Airway Epithelium
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
COPD Patients’ Characteristics and History
References
- GBD; CODC. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980-2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1736–1788. [Google Scholar] [CrossRef] [Green Version]
- Agustí, A.; Melén, E.; DeMeo, D.L.; Breyer-Kohansal, R.; Faner, R. Pathogenesis of chronic obstructive pulmonary disease: Understanding the contributions of gene–environment interactions across the lifespan. Lancet Respir. Med. 2022, 10, 512–524. [Google Scholar] [CrossRef]
- Higham, A.; Quinn, A.M.; Cançado, J.E.D.; Singh, D. The pathology of small airways disease in COPD: Historical aspects and future directions. Respir. Res. 2019, 20, 49. [Google Scholar] [CrossRef] [Green Version]
- Hogg, J.C.; Chu, F.; Utokaparch, S.; Woods, R.; Elliott, W.M.; Buzatu, L.; Cherniack, R.M.; Rogers, R.M.; Sciurba, F.C.; Coxson, H.O.; et al. The Nature of Small-Airway Obstruction in Chronic Obstructive Pulmonary Disease. N. Engl. J. Med. 2004, 350, 2645–2653. [Google Scholar] [CrossRef]
- Xu, F.; Vasilescu, D.M.; Kinose, D.; Tanabe, N.; Ng, K.W.; Coxson, H.O.; Cooper, J.D.; Hackett, T.-L.; Verleden, S.E.; Vanaudenaerde, B.M.; et al. The molecular and cellular mechanisms associated with the destruction of terminal bronchioles in COPD. Eur. Respir. J. 2022, 59, 2101411. [Google Scholar] [CrossRef]
- Ahmed, E.; Sansac, C.; Assou, S.; Gras, D.; Petit, A.; Vachier, I.; Chanez, P.; De Vos, J.; Bourdin, A. Lung development, regeneration and plasticity: From disease physiopathology to drug design using induced pluripotent stem cells. Pharmacol. Ther. 2018, 183, 58–77. [Google Scholar] [CrossRef]
- Green, M.D.; Chen, A.; Nostro, M.-C.; D’Souza, S.L.; Schaniel, C.; Lemischka, I.R.; Gouon-Evans, V.; Keller, G.; Snoeck, H.-W. Generation of anterior foregut endoderm from human embryonic and induced pluripotent stem cells. Nat. Biotechnol. 2011, 29, 267–272. [Google Scholar] [CrossRef] [Green Version]
- Wong, A.P.; Bear, C.E.; Chin, S.; Pasceri, P.; Thompson, T.O.; Huan, L.-J.; Ratjen, F.; Ellis, J.; Rossant, J. Directed differentiation of human pluripotent stem cells into mature airway epithelia expressing functional CFTR protein. Nat. Biotechnol. 2012, 30, 876–882. [Google Scholar] [CrossRef] [Green Version]
- Mou, H.; Zhao, R.; Sherwood, R.; Ahfeldt, T.; Lapey, A.; Wain, J.; Sicilian, L.; Izvolsky, K.; Lau, F.H.; Musunuru, K.; et al. Generation of Multipotent Lung and Airway Progenitors from Mouse ESCs and Patient-Specific Cystic Fibrosis iPSCs. Cell Stem Cell 2012, 10, 385–397. [Google Scholar] [CrossRef] [Green Version]
- Huang, S.X.L.; Islam, M.N.; O’Neill, J.; Hu, Z.; Yang, Y.-G.; Chen, Y.-W.; Mumau, M.; Green, M.D.; Vunjak-Novakovic, G.; Bhattacharya, J.; et al. Efficient generation of lung and airway epithelial cells from human pluripotent stem cells. Nat. Biotechnol. 2014, 32, 84–91. [Google Scholar] [CrossRef] [Green Version]
- Firth, A.L.; Dargitz, C.T.; Qualls, S.J.; Menon, T.; Wright, R.; Singer, O.; Gage, F.H.; Khanna, A.; Verma, I.M. Generation of multiciliated cells in functional airway epithelia from human induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2014, 111, E1723–E1730. [Google Scholar] [CrossRef] [Green Version]
- Dye, B.R.; Hill, D.R.; Ferguson, M.A.; Tsai, Y.H.; Nagy, M.S.; Dyal, R.; Wells, J.M.; Mayhew, C.N.; Nattiv, R.; Klein, O.D.; et al. In vitro generation of human pluripotent stem cell derived lung organoids. eLife 2015, 4, e05098. [Google Scholar] [CrossRef]
- Konishi, S.; Gotoh, S.; Tateishi, K.; Yamamoto, Y.; Korogi, Y.; Nagasaki, T.; Matsumoto, H.; Muro, S.; Hirai, T.; Ito, I.; et al. Directed Induction of Functional Multi-ciliated Cells in Proximal Airway Epithelial Spheroids from Human Pluripotent Stem Cells. Stem Cell Rep. 2016, 6, 18–25. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.-W.; Huang, S.X.; De Carvalho, A.L.R.T.; Ho, S.-H.; Islam, M.N.; Volpi, S.; Notarangelo, L.D.; Ciancanelli, M.; Casanova, J.-L.; Bhattacharya, J.; et al. A three-dimensional model of human lung development and disease from pluripotent stem cells. Nat. Cell Biol. 2017, 19, 542–549. [Google Scholar] [CrossRef] [Green Version]
- McCauley, K.B.; Hawkins, F.; Serra, M.; Thomas, D.C.; Jacob, A.; Kotton, D.N. Efficient Derivation of Functional Human Airway Epithelium from Pluripotent Stem Cells via Temporal Regulation of Wnt Signaling. Cell Stem Cell 2017, 20, 844–857.e6. [Google Scholar] [CrossRef] [Green Version]
- Morrisey, E.E.; Hogan, B.L.M. Preparing for the First Breath: Genetic and Cellular Mechanisms in Lung Development. Dev. Cell 2010, 18, 8–23. [Google Scholar] [CrossRef] [Green Version]
- Matsuno, K.; Mae, S.-I.; Okada, C.; Nakamura, M.; Watanabe, A.; Toyoda, T.; Uchida, E.; Osafune, K. Redefining definitive endoderm subtypes by robust induction of human induced pluripotent stem cells. Differentiation 2016, 92, 281–290. [Google Scholar] [CrossRef]
- Hawkins, F.; Kramer, P.; Jacob, A.; Driver, I.; Thomas, D.C.; McCauley, K.; Skvir, N.; Crane, A.M.; Kurmann, A.A.; Hollenberg, A.N.; et al. Prospective isolation of NKX2-1–expressing human lung progenitors derived from pluripotent stem cells. J. Clin. Investig. 2017, 127, 2277–2294. [Google Scholar] [CrossRef] [Green Version]
- Gomperts, B.N. Induction of multiciliated cells from induced pluripotent stem cells. Proc. Natl. Acad. Sci. USA 2014, 111, 6120–6121. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, E.; Fieldes, M.; Mianné, J.; Bourguignon, C.; Nasri, A.; Vachier, I.; Assou, S.; Bourdin, A.; De Vos, J. Generation of four severe early-onset chronic obstructive pulmonary disease (COPD) patient-derived induced pluripotent stem cell lines from peripheral blood mononuclear cells. Stem Cell Res. 2021, 56, 102550. [Google Scholar] [CrossRef]
- Fieldes, M.; Ahmed, E.; Bourguignon, C.; Mianné, J.; Martin, M.; Arnould, C.; Vachier, I.; Assou, S.; De Vos, J.; Bourdin, A. Generation of the induced pluripotent stem cell line UHOMi002-A from peripheral blood mononuclear cells of a healthy male donor. Stem Cell Res. 2020, 49, 102037. [Google Scholar] [CrossRef]
- Ahmed, E.; Sansac, C.; Fieldes, M.; Bergougnoux, A.; Bourguignon, C.; Mianné, J.; Arnould, C.; Vachier, I.; Assou, S.; Bourdin, A.; et al. Generation of the induced pluripotent stem cell line UHOMi001-A from a patient with mutations in CCDC40 gene causing Primary Ciliary Dyskinesia (PCD). Stem Cell Res. 2018, 33, 15–19. [Google Scholar] [CrossRef] [PubMed]
- Bai, Q.; Ramirez, J.-M.; Becker, F.; Pantesco, V.; Lavabre-Bertrand, T.; Hovatta, O.; Lemaître, J.-M.; Pellestor, F.; De Vos, J. Temporal Analysis of Genome Alterations Induced by Single-Cell Passaging in Human Embryonic Stem Cells. Stem Cells Dev. 2015, 24, 653–662. [Google Scholar] [CrossRef] [Green Version]
- Assou, S.; Girault, N.; Plinet, M.; Bouckenheimer, J.; Sansac, C.; Combe, M.; Mianné, J.; Bourguignon, C.; Fieldes, M.; Ahmed, E.; et al. Recurrent Genetic Abnormalities in Human Pluripotent Stem Cells: Definition and Routine Detection in Culture Supernatant by Targeted Droplet Digital PCR. Stem Cell Rep. 2020, 14, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Miller, A.J.; Hill, D.R.; Nagy, M.S.; Aoki, Y.; Dye, B.R.; Chin, A.M.; Huang, S.; Zhu, F.; White, E.S.; Lama, V.; et al. In Vitro Induction and In Vivo Engraftment of Lung Bud Tip Progenitor Cells Derived from Human Pluripotent Stem Cells. Stem Cell Rep. 2018, 10, 101–119. [Google Scholar] [CrossRef] [Green Version]
- Gibbons, I.R.; Rowe, A.J. Dynein: A Protein with Adenosine Triphosphatase Activity from Cilia. Science 1965, 149, 424–426. [Google Scholar] [CrossRef]
- Papon, J.-F.; Bassinet, L.; Cariou-Patron, G.; Zerah-Lancner, F.; Vojtek, A.-M.; Blanchon, S.; Crestani, B.; Amselem, S.; Coste, A.; Housset, B.; et al. Quantitative analysis of ciliary beating in primary ciliary dyskinesia: A pilot study. Orphanet J. Rare Dis. 2012, 7, 78. [Google Scholar] [CrossRef]
- Hurley, K.; Ding, J.; Villacorta-Martin, C.; Herriges, M.J.; Jacob, A.; Vedaie, M.; Alysandratos, K.D.; Sun, Y.L.; Lin, C.; Werder, R.B.; et al. Reconstructed Single-Cell Fate Trajectories Define Lineage Plasticity Windows during Differentiation of Human PSC-Derived Distal Lung Progenitors. Cell Stem Cell 2020, 26, 593–608.e8. [Google Scholar] [CrossRef]
- Abo, K.M.; Sainz de Aja, J.; Lindstrom-Vautrin, J.; Alysandratos, K.D.; Richards, A.; Garcia-de-Alba, C.; Huang, J.; Hix, O.T.; Werder, R.B.; Bullitt, E.; et al. Air-liquid interface culture promotes maturation and allows environmental exposure of pluripotent stem cell-derived alveolar epithelium. JCI Insight 2022, 7, e155589. [Google Scholar] [CrossRef]
- Hawkins, F.J.; Suzuki, S.; Beermann, M.L.; Barillà, C.; Wang, R.; Villacorta-Martin, C.; Berical, A.; Jean, J.; Le Suer, J.; Matte, T.; et al. Derivation of Airway Basal Stem Cells from Human Pluripotent Stem Cells. Cell Stem Cell 2021, 28, 79–95.e8. [Google Scholar] [CrossRef]
- Gotoh, S.; Ito, I.; Nagasaki, T.; Yamamoto, Y.; Konishi, S.; Korogi, Y.; Matsumoto, H.; Muro, S.; Hirai, T.; Funato, M.; et al. Generation of Alveolar Epithelial Spheroids via Isolated Progenitor Cells from Human Pluripotent Stem Cells. Stem Cell Rep. 2014, 3, 394–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kido, T.; Koui, Y.; Suzuki, K.; Kobayashi, A.; Miura, Y.; Chern, E.Y.; Tanaka, M.; Miyajima, A. CPM Is a Useful Cell Surface Marker to Isolate Expandable Bi-Potential Liver Progenitor Cells Derived from Human iPS Cells. Stem Cell Rep. 2015, 5, 508–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lafkas, D.; Shelton, A.; Chiu, C.; Boenig, G.D.L.; Chen, Y.; Stawicki, S.S.; Siltanen, C.; Reichelt, M.; Zhou, M.; Wu, X.; et al. Therapeutic antibodies reveal Notch control of transdifferentiation in the adult lung. Nature 2015, 528, 127–131. [Google Scholar] [CrossRef] [PubMed]
- Okuda, K.; Chen, G.; Subramani, D.B.; Wolf, M.; Gilmore, R.C.; Kato, T.; Radicioni, G.; Kesimer, M.; Chua, M.; Dang, H.; et al. Localization of Secretory Mucins MUC5AC and MUC5B in Normal/Healthy Human Airways. Am. J. Respir. Crit. Care Med. 2019, 199, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Palange, P.; Testa, U.; Huertas, A.; Calabro, L.; Antonucci, R.; Petrucci, E.; Pelosi, E.; Pasquini, L.; Satta, A.; Morici, G.; et al. Circulating haemopoietic and endothelial progenitor cells are decreased in COPD. Eur. Respir. J. 2006, 27, 529–541. [Google Scholar] [CrossRef]
- Kirkham, S.; Kolsum, U.; Rousseau, K.; Singh, D.; Vestbo, J.; Thornton, D.J. MUC5B Is the Major Mucin in the Gel Phase of Sputum in Chronic Obstructive Pulmonary Disease. Am. J. Respir. Crit. Care Med. 2008, 178, 1033–1039. [Google Scholar] [CrossRef] [Green Version]
- Finicelli, M.; Squillaro, T.; Galderisi, U.; Peluso, G. Micro-RNAs: Crossroads between the Exposure to Environmental Particulate Pollution and the Obstructive Pulmonary Disease. Int. J. Mol. Sci. 2020, 21, 7221. [Google Scholar] [CrossRef]
- Verleden, S.E.; Tanabe, N.; McDonough, J.E.; Vasilescu, D.M.; Xu, F.; Wuyts, W.A.; Piloni, D.; De Sadeleer, L.; Willems, S.; Mai, C.; et al. Small airways pathology in idiopathic pulmonary fibrosis: A retrospective cohort study. Lancet Respir. Med. 2020, 8, 573–584. [Google Scholar] [CrossRef]
- Dummer, A.; Poelma, C.; DeRuiter, M.C.; Goumans, M.J.T.; Hierck, B.P. Measuring the primary cilium length: Improved method for unbiased high-throughput analysis. Cilia 2016, 5, 7. [Google Scholar] [CrossRef] [Green Version]
- Kesimer, M.; Ford, A.A.; Ceppe, A.; Radicioni, G.; Cao, R.; Davis, C.W.; Doerschuk, C.M.; Alexis, N.E.; Anderson, W.H.; Henderson, A.G.; et al. Airway Mucin Concentration as a Marker of Chronic Bronchitis. N. Engl. J. Med. 2017, 377, 911–922. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahmed, E.; Fieldes, M.; Bourguignon, C.; Mianné, J.; Petit, A.; Jory, M.; Cazevieille, C.; Boukhaddaoui, H.; Garnett, J.P.; Hirtz, C.; et al. Differentiation of Human Induced Pluripotent Stem Cells from Patients with Severe COPD into Functional Airway Epithelium. Cells 2022, 11, 2422. https://doi.org/10.3390/cells11152422
Ahmed E, Fieldes M, Bourguignon C, Mianné J, Petit A, Jory M, Cazevieille C, Boukhaddaoui H, Garnett JP, Hirtz C, et al. Differentiation of Human Induced Pluripotent Stem Cells from Patients with Severe COPD into Functional Airway Epithelium. Cells. 2022; 11(15):2422. https://doi.org/10.3390/cells11152422
Chicago/Turabian StyleAhmed, Engi, Mathieu Fieldes, Chloé Bourguignon, Joffrey Mianné, Aurélie Petit, Myriam Jory, Chantal Cazevieille, Hassan Boukhaddaoui, James P. Garnett, Christophe Hirtz, and et al. 2022. "Differentiation of Human Induced Pluripotent Stem Cells from Patients with Severe COPD into Functional Airway Epithelium" Cells 11, no. 15: 2422. https://doi.org/10.3390/cells11152422






