Phaseolus vulgaris Erythroagglutinin (PHA-E)-Positive Ceruloplasmin Acts as a Potential Biomarker in Pancreatic Cancer Diagnosis
Abstract
1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Lectins and Antibodies
2.3. Purification of N-Glycans from Serum for Mass Spectrometry (MS) Analysis
2.4. Lectin and Western Blotting
2.5. Lectin Pull-Down Assay
2.6. In-Gel Protein Digestion
2.7. Identification of Proteins via Nano-LC-MS/MS
2.8. Immunohistochemistry (IHC)
2.9. Expression of Cp and N-Acetylglucosaminyltransferase III (GnT III) in PC from GEO Datasets
2.10. Statistical Analysis
3. Results
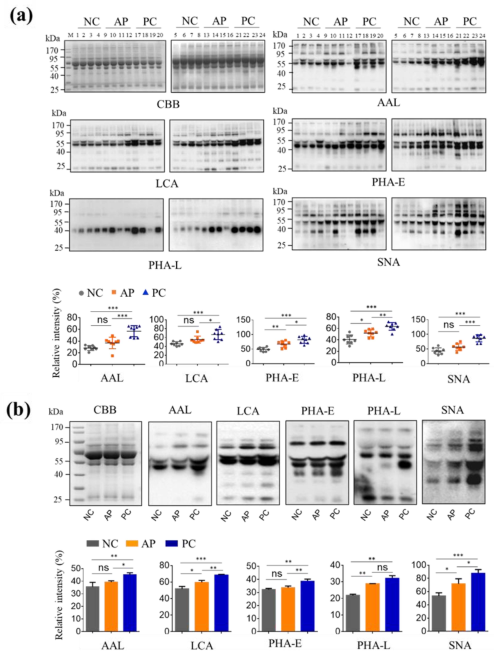
3.1. Altered Glycan Patterns of Glycoproteins in PC Patients’ Sera
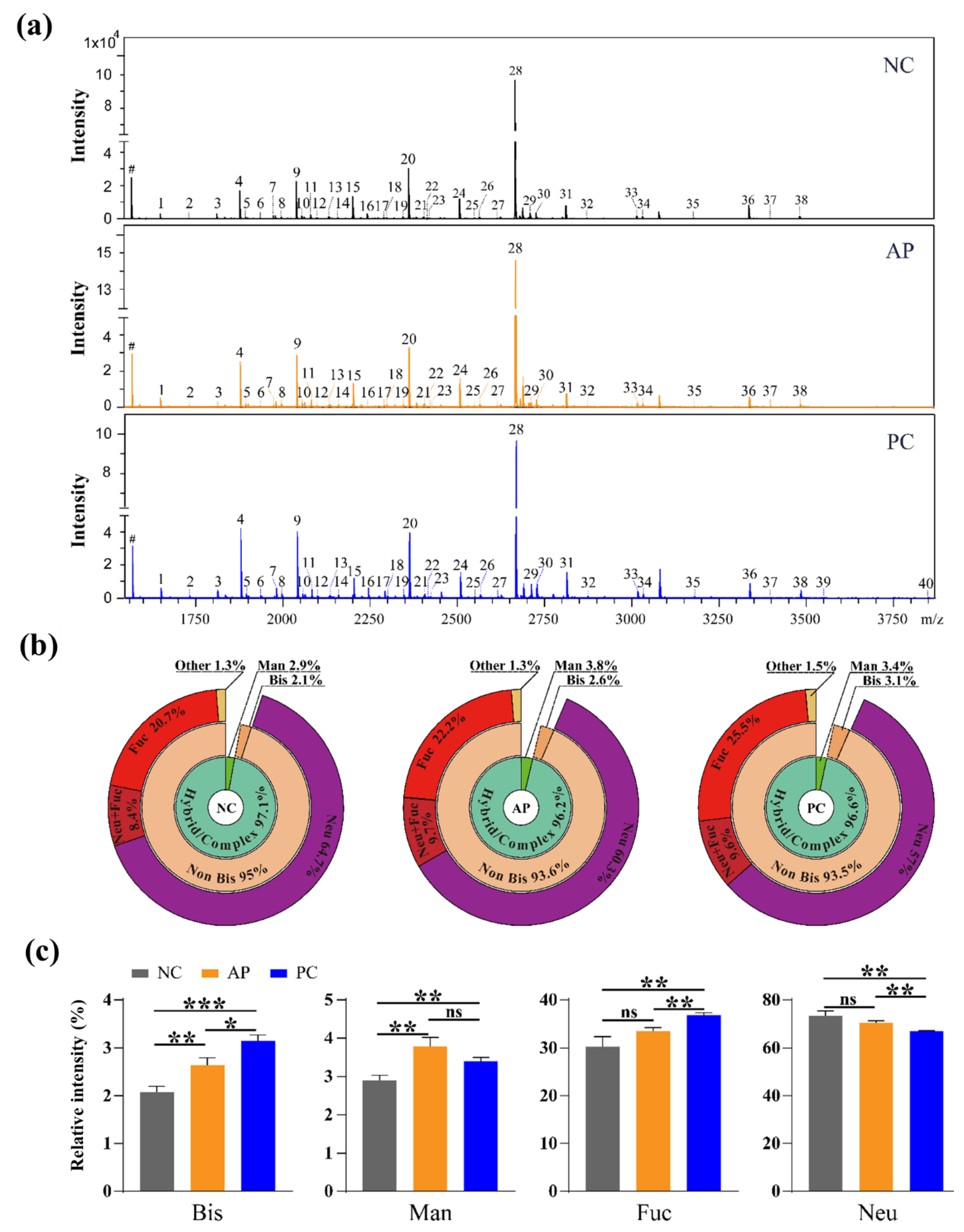
3.2. Altered N-Glycan Types in PC patients’ Sera
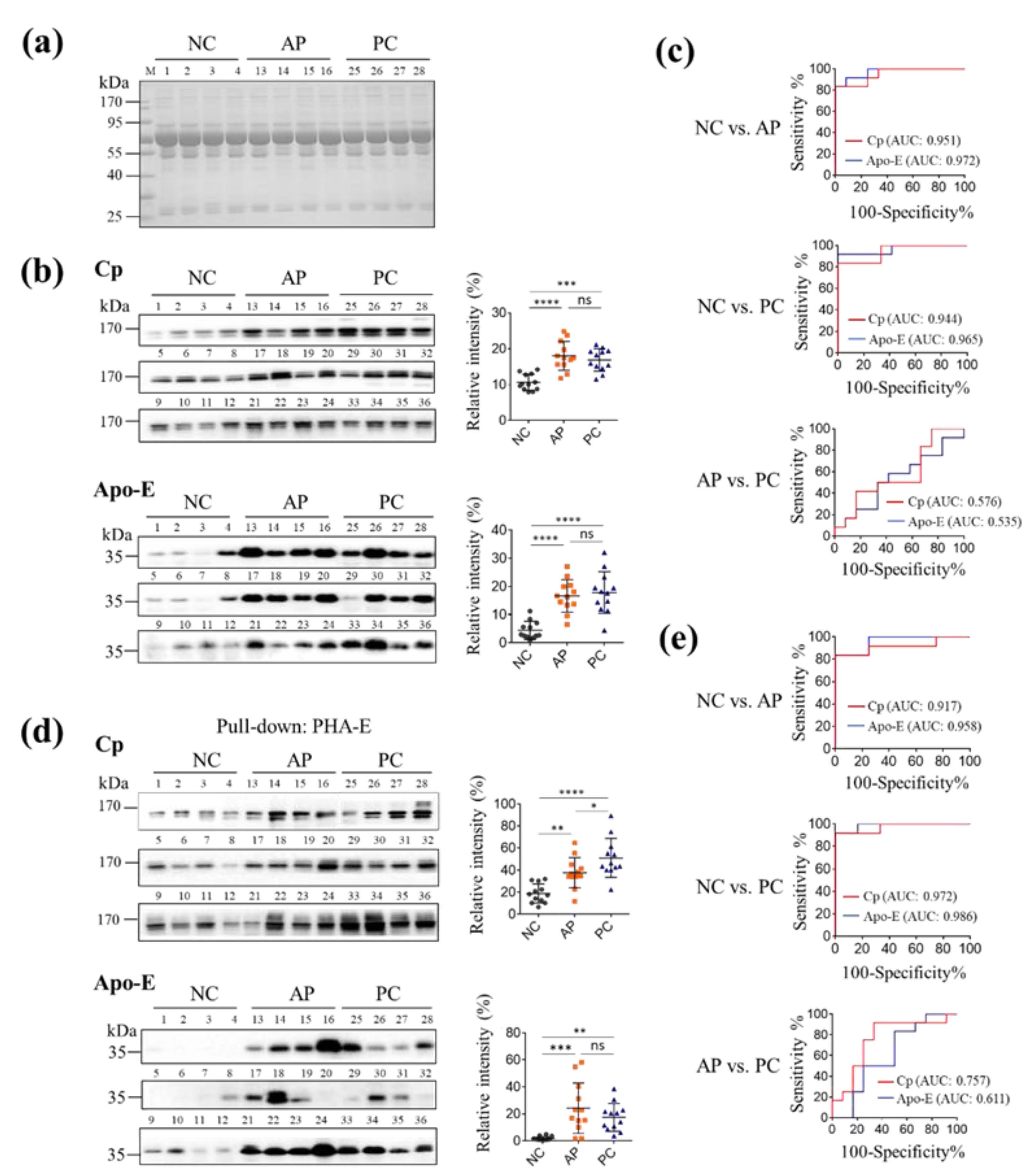
3.3. Pull-Down Glycoproteins with PHA-E Lectin
3.4. The Levels of PHA-E-Positive Cp and Apo-E in PC Patients’ Sera
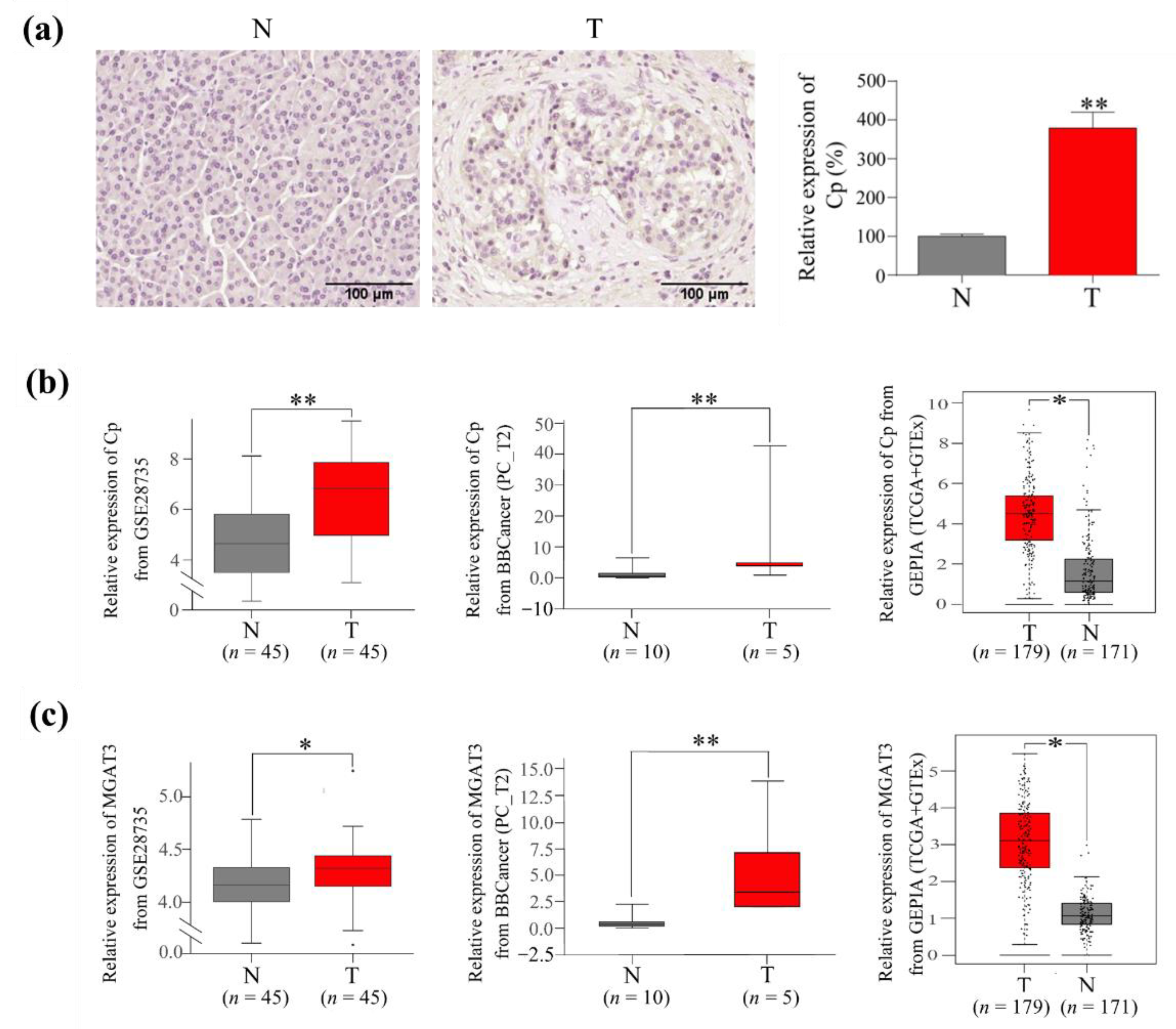
3.5. The Expression of Cp and GnT III in PC Tumor Tissues
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Goel, G.; Sun, W. Novel approaches in the management of pancreatic ductal adenocarcinoma: Potential promises for the future. J. Hematol. Oncol. 2015, 8, 44. [Google Scholar] [CrossRef]
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic cancer. Lancet 2016, 388, 73–85. [Google Scholar] [CrossRef]
- Zhang, L.; Sanagapalli, S.; Stoita, A. Challenges in diagnosis of pancreatic cancer. World J. Gastroenterol. 2018, 24, 2047–2060. [Google Scholar] [CrossRef]
- Ni, X.G.; Bai, X.F.; Mao, Y.L.; Shao, Y.F.; Wu, J.X.; Shan, Y.; Wang, C.F.; Wang, J.; Tian, Y.T.; Liu, Q.; et al. The clinical value of serum CEA, CA19-9, and CA242 in the diagnosis and prognosis of pancreatic cancer. Eur. J. Surg. Oncol. 2005, 31, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Goonetilleke, K.S.; Siriwardena, A.K. Systematic review of carbohydrate antigen (CA 19-9) as a biochemical marker in the diagnosis of pancreatic cancer. Eur. J. Surg. Oncol. 2007, 33, 266–270. [Google Scholar] [CrossRef]
- Bagaria, B.; Sood, S.; Sharma, R.; Lalwani, S. Comparative study of CEA and CA19-9 in esophageal, gastric and colon cancers individually and in combination (ROC curve analysis). Cancer Biol. Med. 2013, 10, 148–157. [Google Scholar] [PubMed]
- Waanders, E.; van Keimpema, L.; Brouwer, J.T.; van Oijen, M.G.; Aerts, R.; Sweep, F.C.; Nevens, F.; Drenth, J.P. Carbohydrate antigen 19-9 is extremely elevated in polycystic liver disease. Liver Int. 2009, 29, 1389–1395. [Google Scholar] [CrossRef]
- Luo, G.; Liu, C.; Guo, M.; Cheng, H.; Lu, Y.; Jin, K.; Liu, L.; Long, J.; Xu, J.; Lu, R.; et al. Potential biomarkers in Lewis negative patients with pancreatic cancer. Ann. Surg. 2017, 265, 800–805. [Google Scholar] [CrossRef]
- Stroszczynski, C.; Grützmann, R.; Kittner, T. CT and MR imaging of pancreatic cancer. Recent Results Cancer Res. 2008, 177, 5–14. [Google Scholar] [PubMed]
- Vijayakumar, A.; Vijayakumar, A. Imaging of focal autoimmune pancreatitis and differentiating it from pancreatic cancer. ISRN Radiol. 2013, 2013, 569489. [Google Scholar] [CrossRef] [PubMed]
- Narimatsu, H. Strategy for development of clinically useful glyco-biomarkers. Glycoconj J. 2014, 31, 403–407. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Taniguchi, N. Potential of N-glycan in cell adhesion and migration as either a positive or negative regulator. Cell Adhes. Migr. 2008, 2, 243–245. [Google Scholar] [CrossRef] [PubMed]
- Kailemia, M.J.; Park, D.; Lebrilla, C.B. Glycans and glycoproteins as specific biomarkers for cancer. Anal. Bioanal. Chem. 2017, 409, 395–410. [Google Scholar] [CrossRef]
- Guo, J.M.; Zhang, X.Y.; Chen, H.L.; Wang, G.M.; Zhang, Y.K. Structural alterations of sugar chains in urine fibronectin from bladder cancer patients and its enzymatic mechanism. J. Cancer Res. Clin. Oncol. 2001, 127, 512–519. [Google Scholar] [CrossRef]
- Tan, Z.; Cao, L.; Wu, Y.; Wang, B.; Song, Z.; Yang, J.; Cheng, L.; Yang, X.; Zhou, X.; Dai, Z.; et al. Bisecting GlcNAc modification diminishes the pro-metastatic functions of small extracellular vesicles from breast cancer cells. J. Extracell. Vesicles 2020, 10, e12005. [Google Scholar] [CrossRef]
- Cheng, L.; Cao, L.; Wu, Y.; Xie, W.; Li, J.; Guan, F.; Tan, Z. Bisecting N-Acetylglucosamine on EGFR Inhibits Malignant Phenotype of Breast Cancer via Down-Regulation of EGFR/Erk Signaling. Front. Oncol. 2020, 10, 929. [Google Scholar] [CrossRef]
- Li, D.; Mallory, T.; Satomura, S. AFP-L3: A new generation of tumor marker for hepatocellular carcinoma. Clin. Chim. Acta 2001, 313, 15–19. [Google Scholar] [CrossRef]
- Tang, S.; Wei, L.; Sun, Y.; Zhou, F.; Zhu, S.; Yang, R.; Huang, Y.; Zhang, H.; Xu, H.; Yang, J. CA153 in breast secretions as a potential molecular marker for diagnosing breast cancer: A Meta Analysis. PLoS ONE 2016, 11, e0163030. [Google Scholar] [CrossRef]
- Matsuda, A.; Kuno, A.; Kawamoto, T.; Matsuzaki, H.; Irimura, T.; Ikehara, Y.; Zen, Y.; Nakanuma, Y.; Yamamoto, M.; Ohkohchi, N.; et al. Wisteria floribunda agglutinin-positive mucin 1 is a sensitive biliary marker for human cholangiocarcinoma. Hepatology 2010, 52, 174–182. [Google Scholar] [CrossRef]
- Kirwan, A.; Utratna, M.; O’Dwyer, M.E.; Joshi, L.; Kilcoyne, M. Glycosylation-Based Serum Biomarkers for Cancer Diagnostics and Prognostics. Biomed. Res. Int. 2015, 2015, 490531. [Google Scholar] [CrossRef] [PubMed]
- Zowczak, M.; Iskra, M.; Paszkowski, J.; Mańczak, M.; Torliński, L.; Wysocka, E. Oxidase activity of ceruloplasmin and concentrations of copper and zinc in serum of cancer patients. J. Trace Elem. Med. Biol. 2001, 15, 193–196. [Google Scholar] [CrossRef]
- Hanas, J.S.; Hocker, J.R.; Cheung, J.Y.; Larabee, J.L.; Lerner, M.R.; Lightfoot, S.A.; Morgan, D.L.; Denson, K.D.; Prejeant, K.C.; Gusev, Y.; et al. Biomarker identification in human pancreatic cancer sera. Pancreas 2008, 36, 61–69. [Google Scholar] [CrossRef]
- Balmaña, M.; Sarrats, A.; Llop, E.; Barrabés, S.; Saldova, R.; Ferri, M.J.; Figueras, J.; Fort, E.; de Llorens, R.; Rudd, P.M.; et al. Identification of potential pancreatic cancer serum markers: Increased sialyl-Lewis X on ceruloplasmin. Clin. Chim. Acta 2015, 442, 56–62. [Google Scholar] [CrossRef]
- Xu, J.; Sun, H.; Huang, G.; Liu, G.; Li, Z.; Yang, H.; Jin, L.; Cui, X.; Shi, L.; Ma, T.; et al. A fixation method for the optimisation of western blotting. Sci. Rep. 2019, 9, 6649. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.Y.; Lin, S.Y.; Yeh, Y.Y.; Hsiao, H.H.; Wu, C.Y.; Chen, S.T.; Wang, A.H. A modified protein precipitation procedure for efficient removal of albumin from serum. Electrophoresis 2005, 26, 2117–2127. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zheng, W.; Wang, W.; Shen, H.; Liu, L.; Lou, W.; Wang, X.; Yang, P. A new panel of pancreatic cancer biomarkers discovered using a mass spectrometry-based pipeline. Br. J. Cancer 2017, 117, 1846–1854. [Google Scholar] [CrossRef]
- Satoh, J.; Darley-Usmar, V.M.; Kashimura, H.; Fukutomi, H.; Anan, K.; Ohsuga, T. Analysis of pure pancreatic juice proteins by two-dimensional gel electrophoresis in cases of pancreatic cancer. Gastroenterol. Jpn. 1986, 21, 623–629. [Google Scholar] [CrossRef]
- Ihara, Y.; Nishikawa, A.; Tohma, T.; Soejima, H.; Niikawa, N.; Taniguchi, N. cDNA cloning, expression, and chromosomal localization of human N-acetylglucosaminyltransferase III (GnT-III). J. Biochem. 1993, 113, 692–698. [Google Scholar] [CrossRef] [PubMed]
- Maisonneuve, P.; Lowenfels, A.B. Epidemiology of pancreatic cancer: An update. Dig. Dis. 2010, 28, 645–656. [Google Scholar] [CrossRef]
- Chambers, A.F.; Groom, A.C.; MacDonald, I.C. Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer 2002, 2, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Chan, A.; Diamandis, E.P.; Blasutig, I.M. Strategies for discovering novel pancreatic cancer biomarkers. J. Proteomics. 2013, 81, 126–134. [Google Scholar] [CrossRef]
- Kirkegård, J.; Cronin-Fenton, D.; Heide-Jørgensen, U.; Mortensen, F.V. Acute Pancreatitis and Pancreatic Cancer Risk: A Nationwide Matched-Cohort Study in Denmark. Gastroenterology 2018, 154, 1729–1736. [Google Scholar] [CrossRef]
- Zhang, X.; An, R.; Tian, H.; Zhao, J. Increased risk of pancreatic cancer after acute pancreatitis: A meta-analysis of prospective cohort studies. Clin. Res. Hepatol. Gastroenterol. 2019, 43, e39–e41. [Google Scholar] [CrossRef] [PubMed]
- Sadr-Azodi, O.; Oskarsson, V.; Discacciati, A.; Videhult, P.; Askling, J.; Ekbom, A. Pancreatic cancer following acute pancreatitis: A population-based matched cohort study. Am. J. Gastroenterol. 2018, 113, 1711–1719. [Google Scholar] [CrossRef] [PubMed]
- Lankisch, P.G.; Apte, M.; Banks, P.A. Acute pancreatitis. Lancet 2015, 386, 85–96. [Google Scholar] [CrossRef]
- Meher, S.; Mishra, T.S.; Sasmal, P.K.; Rath, S.; Sharma, R.; Rout, B.; Sahu, M.K. Role of Biomarkers in Diagnosis and Prognostic Evaluation of Acute Pancreatitis. J. Biomark. 2015, 2015, 519534. [Google Scholar] [CrossRef]
- Falconer, J.S.; Fearon, K.C.; Ross, J.A.; Elton, R.; Wigmore, S.J.; Garden, O.J.; Carter, D.C. Acute-phase protein response and survival duration of patients with pancreatic cancer. Cancer 1995, 75, 2077–2082. [Google Scholar] [CrossRef]
- Kontro, H.; Joenväärä, S.; Haglund, C.; Renkonen, R. Comparison of sialylated N-glycopeptide levels in serum of pancreatic cancer patients, acute pancreatitis patients, and healthy controls. Proteomics 2014, 14, 1713–1723. [Google Scholar] [CrossRef]
- Terao, N.; Takamatsu, S.; Minehira, T.; Sobajima, T.; Nakayama, K.; Kamada, Y.; Miyoshi, E. Fucosylation is a common glycosylation type in pancreatic cancer stem cell-like phenotypes. World J. Gastroenterol. 2015, 21, 3876–3887. [Google Scholar] [CrossRef]
- Zhao, J.; Simeone, D.M.; Heidt, D.; Anderson, M.A.; Lubman, D.M. Comparative serum glycoproteomics using lectin selected sialic acid glycoproteins with mass spectrometric analysis: Application to pancreatic cancer serum. J. Proteome. Res. 2006, 5, 1792–1802. [Google Scholar] [CrossRef]
- Zhao, J.; Patwa, T.H.; Qiu, W.; Shedden, K.; Hinderer, R.; Misek, D.E.; Anderson, M.A.; Simeone, D.M.; Lubman, D.M. Glycoprotein microarrays with multi-lectin detection: Unique lectin binding patterns as a tool for classifying normal, chronic pancreatitis and pancreatic cancer sera. J. Proteome. Res. 2007, 6, 1864–1874. [Google Scholar] [CrossRef] [PubMed]
- Nan, B.C.; Shao, D.M.; Chen, H.L.; Huang, Y.; Gu, J.X.; Zhang, Y.B.; Wu, Z.G. Alteration of N-acetylglucosaminyltransferases in pancreatic carcinoma. Glycoconj. J. 1998, 15, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, N.; Ohkawa, Y.; Maeda, K.; Harada, Y.; Nagae, M.; Kizuka, Y.; Ihara, H.; Ikeda, Y. True significance of N-acetylglucosaminyltransferases GnT-III, V and α1,6 fucosyltransferase in epithelial-mesenchymal transition and cancer. Mol. Aspects. Med. 2021, 79, 100905. [Google Scholar] [CrossRef] [PubMed]
- Allam, H.; Johnson, B.P.; Zhang, M.; Lu, Z.; Cannon, M.J.; Abbott, K.L. The glycosyltransferase GnT-III activates Notch signaling and drives stem cell expansion to promote the growth and invasion of ovarian cancer. J. Biol. Chem. 2017, 292, 16351–16359. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Tan, Z.; Guan, F.; Ren, Y. The Essential Functions and Detection of Bisecting GlcNAc in Cell Biology. Front. Chem. 2020, 8, 511. [Google Scholar] [CrossRef]
- Nouso, K.; Amano, M.; Ito, Y.M.; Miyahara, K.; Morimoto, Y.; Kato, H.; Tsutsumi, K.; Tomoda, T.; Yamamoto, N.; Nakamura, S.; et al. Clinical utility of high-throughput glycome analysis in patients with pancreatic cancer. J. Gastroenterol. 2013, 48, 1171–1179. [Google Scholar] [CrossRef]
- Yan, L.; Tonack, S.; Smith, R.; Dodd, S.; Jenkins, R.E.; Kitteringham, N.; Greenhalf, W.; Ghaneh, P.; Neoptolemos, J.P.; Costello, E. Confounding effect of obstructive jaundice in the interpretation of proteomic plasma profiling data for pancreatic cancer. J. Proteome. Res. 2009, 8, 142–148. [Google Scholar] [CrossRef]
- Zhou, B.; Fang, B.; Yan, S.; Wang, W. Comparison of tumor markers for predicting disease-free survival in surgically resected pancreatic neuroendocrine tumors. Clin. Lab. 2017, 63, 1251–1259. [Google Scholar] [CrossRef]
- Jeong, D.H.; Kim, H.K.; Prince, A.E.; Lee, D.S.; Kim, Y.N.; Han, J.; Kim, K.T. Plasma proteomic analysis of patients with squamous cell carcinoma of the uterine cervix. J. Gynecol. Oncol. 2008, 19, 173–180. [Google Scholar] [CrossRef]
- Chen, S.Y.; Chang, T.C.; Lin, C.Y.; Lai, C.H.; Wu, R.C.; Yang, L.Y.; Chang, W.Y.; Lee, Y.S.; Yang, W.V.; Chao, A. Serum levels of alpha1-antitrypsin isoforms in patients with ovarian clear cell carcinoma: An exploratory study. J. Chin. Med. Assoc. 2021, 84, 1048–1053. [Google Scholar] [CrossRef]
- Zhao, W.; Yang, Z.; Liu, X.; Tian, Q.; Lv, Y.; Liang, Y.; Li, C.; Gao, X.; Chen, L. Identification of α1-antitrypsin as a potential prognostic biomarker for advanced nonsmall cell lung cancer treated with epidermal growth factor receptor tyrosine kinase inhibitors by proteomic analysis. J. Int. Med. Res. 2013, 41, 573–583. [Google Scholar] [CrossRef] [PubMed]
- Drabik, A. Glycopeptides as a Tool for Early Detection of Cancer. Proteomics Clin. Appl. 2018, 12, e1800108. [Google Scholar] [CrossRef] [PubMed]
- Krzyminiewski, R.; Dobosz, B.; Kubiak, T. The influence of radiotherapy on ceruloplasmin and transferrin in whole blood of breast cancer patients. Radiat. Environ. Biophys. 2017, 56, 345–352. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Han, I.W.; Jang, J.Y.; Kwon, W.; Park, T.; Kim, Y.; Lee, K.B.; Kim, S.W. Ceruloplasmin as a prognostic marker in patients with bile duct cancer. Oncotarget 2017, 8, 29028–29037. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Liu, F. Diagnostic value of serum carbohydrate antigen 19-9 in pancreatic cancer: A meta-analysis. Tumour. Biol. 2014, 35, 7459–7465. [Google Scholar] [CrossRef]
- Li, A.; Yu, J.; Kim, H.; Wolfgang, C.L.; Canto, M.I.; Hruban, R.H.; Goggins, M. MicroRNA array analysis finds elevated serum miR-1290 accurately distinguishes patients with low-stage pancreatic cancer from healthy and disease controls. Clin. Cancer Res. 2013, 19, 3600–3610. [Google Scholar] [CrossRef]

| Peak | Calculated m/z (a) | Chemical Composition (b) | Relative Intensity (%) | Structure | ||
|---|---|---|---|---|---|---|
| No. | NC | AP | PC | |||
| 1 | 1649.67 | (Hex)2 + (Man)3(GlcNAc)2 | 1.18 | 1.62 | 1.44 | 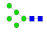 |
| 2 | 1731.72 | (HexNAc)2 + (Man)3(GlcNAc)2 | 0.13 | 0.21 | 0.28 | 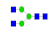 |
| 3 | 1811.72 | (Hex)3 + (Man)3(GlcNAc)2 | 1.08 | 1.31 | 1.17 | 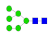 |
| 4 | 1877.78 | (HexNAc)2(Fuc)1 + (Man)3(GlcNAc)2 | 6.31 | 8.05 | 10.97 | 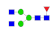 |
| 5 | 1893.77 | (Hex)1(HexNAc)2 + (Man)3(GlcNAc)2 | 0.37 | 0.43 | 0.49 |  |
| 6 | 1934.80 | (HexNAc)3 + (Man)3(GlcNAc)2 | 0.13 | 0.17 | 0.38 | 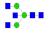 |
| 7 | 1973.77 | (Hex)4 + (Man)3(GlcNAc)2 | 0.18 | 0.26 | 0.25 | 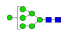 |
| 8 | 1995.78 | (Hex)1 (HexNAc)1 (NeuAc)1 + (Man)3(GlcNAc)2 | 0.41 | 0.47 | 0.53 | 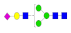 |
| 9 | 2039.83 | (Hex)1(HexNAc)2(Fuc)1 + (Man)3(GlcNAc)2 | 8.45 | 9.19 | 10.15 |  |
| 10 | 2055.83 | (Hex)2(HexNAc)2 + (Man)3(GlcNAc)2 | 0.69 | 0.57 | 0.51 |  |
| 11 | 2080.86 | (HexNAc)3(Fuc)1 + (Man)3(GlcNAc)2 | 0.74 | 1.07 | 1.21 | 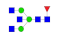 |
| 12 | 2096.85 | (Hex)1(HexNAc)3 + (Man)3(GlcNAc)2 | 0.14 | 0.19 | 0.26 | 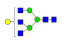 |
| 13 | 2135.82 | (Hex)5 + (Man)3(GlcNAc)2 | 0.23 | 0.34 | 0.27 | 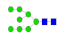 |
| 14 | 2157.83 | (Hex)2(HexNAc)1(NeuAc)1 + (Man)3(GlcNAc)2 | 0.25 | 0.42 | 0.35 | 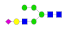 |
| 15 | 2201.88 | (Hex)2(HexNAc)2(Fuc)1 + (Man)3(GlcNAc)2 | 5.00 | 4.00 | 3.01 |  |
| 16 | 2242.91 | (Hex)1(HexNAc)3(Fuc)1 + (Man)3(GlcNAc)2 | 0.90 | 1.01 | 1.32 | 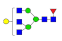 |
| 17 | 2297.88 | (Hex)6 + (Man)3(GlcNAc)2 | 0.21 | 0.26 | 0.27 |  |
| 18 | 2319.89 | (Hex)3(HexNAc)1(NeuAc)1 + (Man)3(GlcNAc)2 | 0.16 | 0.31 | 0.26 | 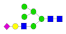 |
| 19 | 2344.92 | (Hex)1(HexNAc)2(Fuc)1(NeuAc)1 + (Man)3(GlcNAc)2 | 0.29 | 0.3 | 0.38 | 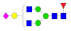 |
| 20 | 2360.91 | (Hex)2(HexNAc)2(NeuAc)1 + (Man)3(GlcNAc)2 | 10.93 | 10.50 | 8.60 | 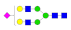 |
| 21 | 2401.94 | (Hex)1(HexNAc)3(NeuAc)1 + (Man)3(GlcNAc)2 | 0.14 | 0.20 | 0.25 | 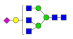 |
| 22 | 2404.96 | (Hex)2(HexNAc)3(Fuc)1 + (Man)3(GlcNAc)2 | 0.43 | 0.50 | 0.52 | 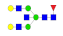 |
| 23 | 2420.96 | (Hex)3(HexNAc)3 + (Man)3(GlcNAc)2 | 0.12 | 0.13 | 0.18 |  |
| 24 | 2506.97 | (Hex)2(HexNAc)2(Fuc)1(NeuAc)1 + (Man)3(GlcNAc)2 | 3.90 | 4.03 | 3.10 |  |
| 25 | 2548.00 | (Hex)1(HexNAc)3(Fuc)1(NeuAc)1 + (Man)3(GlcNAc)2 | 0.08 | 0.14 | 0.20 | 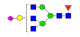 |
| 26 | 2563.99 | (Hex)2(HexNAc)3(NeuAc)1 + (Man)3(GlcNAc)2 | 0.31 | 0.38 | 0.38 | 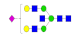 |
| 27 | 2624.04 | (Hex)3(HexNAc)4 + (Man)3(GlcNAc)2 | 0.32 | 0.33 | 0.39 | 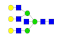 |
| 28 | 2666.01 | (Hex)2(HexNAc)2(NeuAc)2 + (Man)3(GlcNAc)2 | 49.77 | 45.42 | 44.14 |  |
| 29 | 2710.05 | (Hex)2(HexNAc)3(Fuc)1(NeuAc)1 + (Man)3(GlcNAc)2 | 1.36 | 1.73 | 1.60 |  |
| 30 | 2726.05 | (Hex)3(HexNAc)3(NeuAc)1 + (Man)3(GlcNAc)2 | 1.02 | 0.96 | 1.34 | 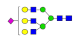 |
| 31 | 2812.07 | (Hex)2(HexNAc)2(Fuc)1(NeuAc)2 + (Man)3(GlcNAc)2 | 2.04 | 2.55 | 2.67 | 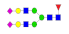 |
| 32 | 2872.10 | (Hex)3(HexNAc)3(Fuc)1(NeuAc)1 + (Man)3(GlcNAc)2 | 0.11 | 0.13 | 0.19 |  |
| 33 | 3015.15 | (Hex)2(HexNAc)3(Fuc)1(NeuAc)2 + (Man)3(GlcNAc)2 | 0.32 | 0.48 | 0.65 | 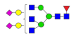 |
| 34 | 3031.14 | (Hex)3(HexNAc)3(NeuAc)2 + (Man)3(GlcNAc)2 | 0.37 | 0.32 | 0.36 | 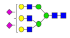 |
| 35 | 3177.20 | (Hex)3(HexNAc)3(Fuc)1(NeuAc)2 + (Man)3(GlcNAc)2 | 0.06 | 0.06 | 0.13 |  |
| 36 | 3336.24 | (Hex)3(HexNAc)3(NeuAc)3 + (Man)3(GlcNAc)2 | 1.55 | 1.65 | 1.06 | 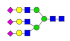 |
| 37 | 3396.27 | (Hex)4(HexNAc)4 (NeuAc)2 + (Man)3(GlcNAc)2 | 0.05 | 0.05 | 0.07 |  |
| 38 | 3482.29 | (Hex)3(HexNAc)3(Fuc)1(NeuAc)3 + (Man)3(GlcNAc)2 | 0.23 | 0.26 | 0.58 | 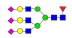 |
| 39 | 3542.33 | (Hex)4(HexNAc)4 (Fuc)1 (NeuAc)2 + (Man)3(GlcNAc)2 | — | 0.00 | 0.05 | 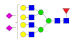 |
| 40 | 3847.43 | (Hex)4(HexNAc)4 (Fuc)1 (NeuAc)3 + (Man)3(GlcNAc)2 | — | 0.00 | 0.04 | 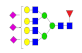 |
| Protein Name | Accession | Unique Peptides | Score | Coverage | mol wt (kDa) | PI |
|---|---|---|---|---|---|---|
| chain B, Human Complement Component C3 | gi|78101268 | 30 | 1131.09 | 31.5% | 112.9 | 5.46 |
| keratin 1 | gi|11935049 | 11 | 1057.63 | 29.7% | 66 | 8.83 |
| keratin 10 | gi|21961605 | 9 | 1034.71 | 38.5% | 58.8 | 4.95 |
| complement C3 precursor | gi|115298678 | 27 | 1003.96 | 18.2% | 187 | 5.98 |
| keratin, type II cytoskeletal 6A | gi|5031839 | 21 | 885.6 | 34.9% | 60 | 8.89 |
| type I keratin 16 | gi|1195531 | 17 | 875.29 | 36.4% | 51.2 | 4.84 |
| keratin type II | gi|908790 | 3 | 847.97 | 34.0% | 60 | 8.89 |
| keratin 1 | gi|7331218 | 11 | 666.54 | 20.3% | 66 | 8.83 |
| immunoglobulin light chain | gi|149673887 | 7 | 650.28 | 63.3% | 23.4 | 7.62 |
| chain A, Alpha1-Antitrypsin | gi|157831596 | 13 | 642.67 | 43.9% | 44.2 | 5.27 |
| human apolipoprotein A-I | gi|90108664 | 12 | 615.34 | 51.9% | 28.1 | 5.15 |
| alpha-1-antichymotrypsin precursor, partial | gi|177933 | 14 | 598.47 | 36.1% | 45.5 | 5.21 |
| immunoglobulin kappa light chain VLJ region | gi|21669395 | 9 | 582.35 | 41.5% | 30.5 | 9.34 |
| keratin K5 | gi|18999435 | 13 | 539.06 | 21.7% | 62.3 | 5.5 |
| cytokeratin 9 | gi|435476 | 11 | 490.09 | 23.0% | 62.1 | 5.06 |
| serum transferrin | gi|110590597 | 12 | 459.46 | 18.2% | 74.7 | 6.61 |
| unnamed protein product | gi|34527290 | 8 | 450.68 | 23.2% | 53.2 | 5.46 |
| plasma protease (C1) inhibitor precursor | gi|179619 | 5 | 399.48 | 19.6% | 55.1 | 5.09 |
| complement factor B | gi|291922 | 10 | 375.61 | 14.7% | 85.6 | 6.59 |
| unnamed protein product | gi|28317 | 8 | 371.36 | 15.7% | 59.5 | 5.04 |
| ALB | gi|37222202 | 8 | 367.55 | 57.5% | 19.3 | 5.14 |
| haptoglobin precursor | gi|306882 | 7 | 362.64 | 15.8% | 45.8 | 6.26 |
| immunoglobulin alpha-2 heavy chain | gi|184761 | 6 | 342.37 | 19.2% | 36.4 | 5.68 |
| epidermal cytokeratin 2 | gi|181402 | 8 | 341.02 | 14.0% | 65.8 | 8.85 |
| ceruloplasmin | gi|1620909 | 8 | 339.04 | 9.6% | 115.4 | 5.35 |
| immunoglobulin heavy chain constant region | gi|10799664 | 8 | 312.54 | 19.6% | 35.9 | 8.8 |
| Ig A1 Bur | gi|229585 | 5 | 310.32 | 7.4% | 73.3 | 10.09 |
| Ig A L | gi|229536 | 5 | 307.89 | 24.5% | 22.8 | 9.79 |
| haptoglobin Hp2 | gi|223976 | 5 | 284.97 | 10.1% | 41.7 | 6.25 |
| immunoglobulin heavy chain | gi|10334587 | 6 | 277.133 | 15.6% | 41.3 | 9.5 |
| hypothetical protein | gi|51476390 | 6 | 271.58 | 10.2% | 69.4 | 5.84 |
| alpha-2-macroglobulin precursor | gi|177870 | 6 | 254.79 | 5.2% | 163.2 | 5.98 |
| Ig G1 H Nie | gi|229601 | 4 | 240.54 | 25.4% | 49.2 | 9.59 |
| serum albumin | gi|28592 | 32 | 239.86 | 10.5% | 69.3 | 6.04 |
| apolipoprotein E | gi|178849 | 5 | 235.07 | 19.6% | 36.2 | 5.53 |
| complement component C5, partial | gi|179692 | 5 | 233.34 | 4.2% | 141.7 | 6.51 |
| apo-B100 precursor | gi|28780 | 12 | 222.07 | 1.7% | 514.9 | 6.68 |
| Ig heavy chain V-III region (ART) | gi|106482 | 4 | 208.47 | 24.8% | 23.7 | 5.04 |
| alpha1 antitrypsin | gi|225768 | 5 | 182.04 | 14.9% | 45.7 | 6.1 |
| immunoglobulin heavy chain constant region | gi|10799664 | 5 | 169.05 | 16.0% | 35.9 | 8.8 |
| alpha-2-macroglobulin precursor | gi|177870 | 3 | 128.07 | 2.7% | 163.2 | 5.98 |
| cytokeratin 9 | gi|435476 | 2 | 127.28 | 4.7% | 62.1 | 5.06 |
| hemoglobin beta | gi|229149 | 2 | 70.83 | 18.5% | 15.9 | 5.14 |
| immunoglobulin heavy chain variable region | gi|114147353 | 2 | 66.97 | 16.1% | 12.8 | 10.17 |
| complement component C3 | gi|179665 | 5 | 60.7 | 2.5% | 187 | 5.46 |
| alpha-1-B-glycoprotein | gi|69990 | 2 | 50.72 | 6.3% | 51.9 | 5.6 |
| alpha-1 antitrypsin | gi|28637 | 2 | 41.58 | 8.6% | 22.8 | 6.1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sha, S.; Wang, Y.; Liu, M.; Liu, G.; Fan, N.; Li, Z.; Dong, W. Phaseolus vulgaris Erythroagglutinin (PHA-E)-Positive Ceruloplasmin Acts as a Potential Biomarker in Pancreatic Cancer Diagnosis. Cells 2022, 11, 2453. https://doi.org/10.3390/cells11152453
Sha S, Wang Y, Liu M, Liu G, Fan N, Li Z, Dong W. Phaseolus vulgaris Erythroagglutinin (PHA-E)-Positive Ceruloplasmin Acts as a Potential Biomarker in Pancreatic Cancer Diagnosis. Cells. 2022; 11(15):2453. https://doi.org/10.3390/cells11152453
Chicago/Turabian StyleSha, Shanshan, Yating Wang, Menglu Liu, Gang Liu, Ning Fan, Zhi Li, and Weijie Dong. 2022. "Phaseolus vulgaris Erythroagglutinin (PHA-E)-Positive Ceruloplasmin Acts as a Potential Biomarker in Pancreatic Cancer Diagnosis" Cells 11, no. 15: 2453. https://doi.org/10.3390/cells11152453
APA StyleSha, S., Wang, Y., Liu, M., Liu, G., Fan, N., Li, Z., & Dong, W. (2022). Phaseolus vulgaris Erythroagglutinin (PHA-E)-Positive Ceruloplasmin Acts as a Potential Biomarker in Pancreatic Cancer Diagnosis. Cells, 11(15), 2453. https://doi.org/10.3390/cells11152453






