Endogenous Retroviruses and Placental Evolution, Development, and Diversity
Abstract
1. Introduction
2. Structural Diversity of Mammalian Placentas
3. Cells Gain Their Mobility through Epithelial-Mesenchymal Transition (EMT)
4. Formation of Syncytiotrophoblast Layer in the Mammalian Species
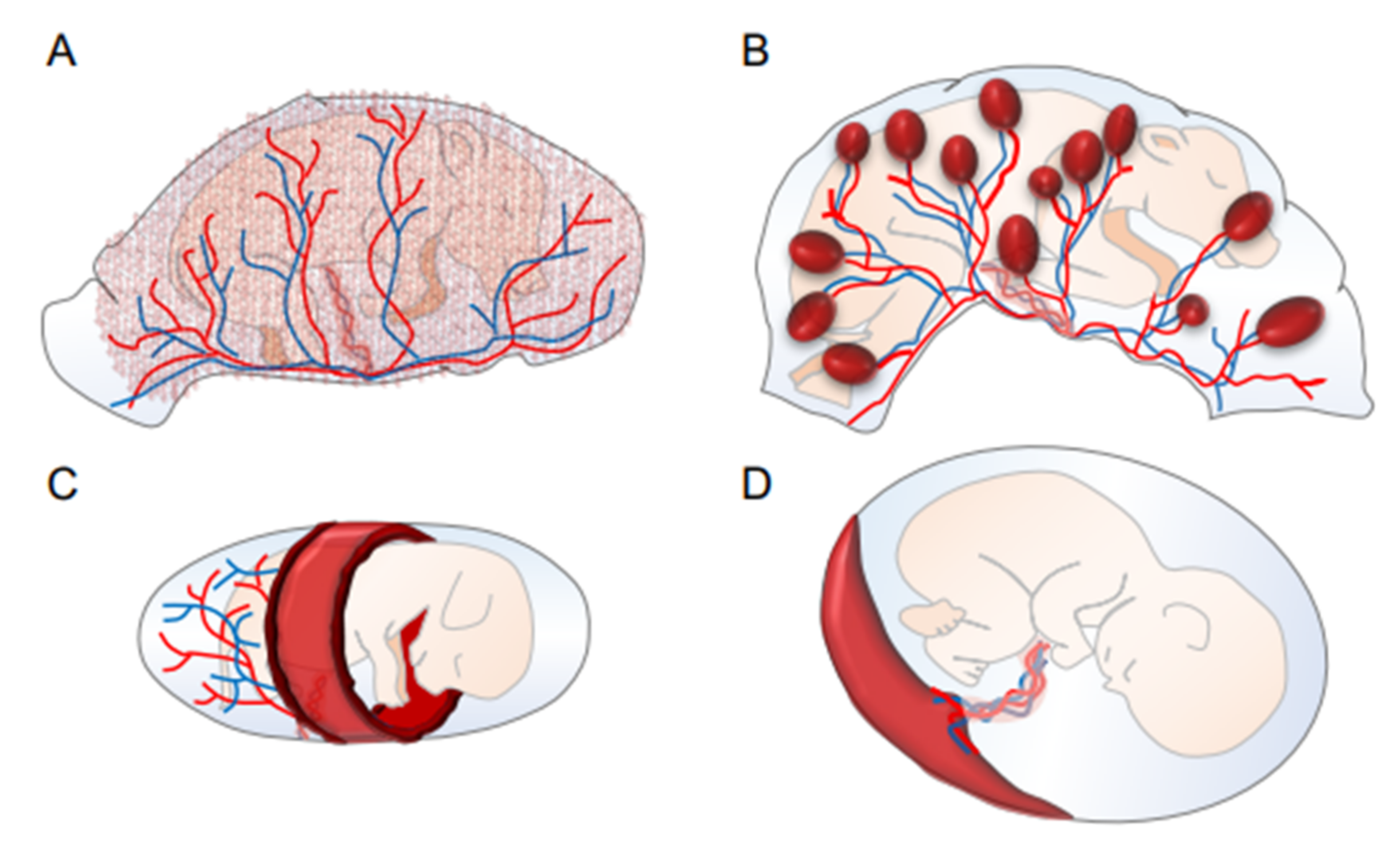
5. Requirement of ERVs for Placental Evolution
5.1. PEG10 and PEG11/RTL1 Are Required for the Acquisition of Placenta
5.2. Requirement of LDOC1/SIRH7/RTL7 for the Maintenance of Placental Function
6. Placental Diversity Requires Syncytin ERVs
7. Molecular Mechanisms Regulating the ERV Expression and ERVs Serving as Transcriptional Regulators
8. Baton Pass Hypothesis: Successive Integration of ERVs
9. New Model Explaining Placental Diversity
- (a)
- The insertion of ERVs can make functional genes of the host placenta-specific. i.e., Fematirn-1 integration into the intron 18 of pregnancy-specific FAT2 gene;
- (b)
- Its own LTR is sufficient to transcribe its own gene segments, which serve as the cis-acting element(s), resulting in the activation of a host gene. i.e., IFNG, THE1B on CRH;
- (c)
- It can make use of transcription factors utilized by the pre-existing gene, as per the baton-pass hypothesis. i.e., A transcription factor GCM1 for syncytin-1 and syncytin-2.
- (d)
- The ERV is co-opted along with its promoter/enhancer in the integrated genome; i.e., SPRE (syncytin post-transcriptional regulatory element);
- (e)
- There is cooperation with miRNAs and/or lncRNAs, yet not definitely characterized under placental/trophectodermal conditions, either alone or together with ERVs.
10. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Laird, M.K.; Thompson, M.B.; Whittington, C.M. Facultative oviparity in a viviparous skink (Saiphos equalis). Biol. Lett. 2019, 15, 20180827. [Google Scholar] [CrossRef]
- Whittington, C.M.; Van Dyke, J.U.; Liang, S.Q.T.; Edwards, S.V.; Shine, R.; Thompson, M.B.; Grueber, C.E. Understanding the evolution of viviparity using intraspecific variation in reproductive mode and transitional forms of pregnancy. Biol. Rev. Camb. Philos. Soc. 2022, 97, 1179–1192. [Google Scholar] [CrossRef] [PubMed]
- Foster, C.S.P.; Thompson, M.B.; Van Dyke, J.U.; Brandley, M.C.; Whittington, C.M. Emergence of an evolutionary innovation: Gene expression differences associated with the transition between oviparity and viviparity. Mol. Ecol. 2020, 29, 1315–1327. [Google Scholar] [CrossRef] [PubMed]
- Roberts, R.M.; Green, J.A.; Schulz, L.C. The evolution of the placenta. Reproduction 2016, 152, R179–R189. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.R. Developmental morphology and evolution of extraembryonic membranes of lizards and snakes (Reptilia, Squamata). J. Morphol. 2021, 282, 973–994. [Google Scholar] [CrossRef] [PubMed]
- Larue, L.; Ohsugi, M.; Hirchenhain, J.; Kemler, R. E-cadherin null mutant embryos fail to form a trophectoderm epithelium. Proc. Natl. Acad. Sci. USA 1994, 91, 8263–8267. [Google Scholar] [CrossRef] [PubMed]
- Shirayoshi, Y.; Hatta, K.; Hosoda, M.; Tsunasawa, S.; Sakiyama, F.; Takeichi, M. Cadherin cell adhesion molecules with distinct binding specificities share a common structure. Embo J. 1986, 5, 2485–2488. [Google Scholar] [CrossRef]
- Ohsugi, M.; Larue, L.; Schwarz, H.; Kemler, R. Cell-junctional and cytoskeletal organization in mouse blastocysts lacking E-cadherin. Dev. Biol. 1997, 185, 261–271. [Google Scholar] [CrossRef]
- Tsukita, S.; Yamazaki, Y.; Katsuno, T.; Tamura, A.; Tsukita, S. Tight junction-based epithelial microenvironment and cell proliferation. Oncogene 2008, 27, 6930–6938. [Google Scholar] [CrossRef]
- Mess, A.; Carter, A.M. Evolutionary transformations of fetal membrane characters in Eutheria with special reference to Afrotheria. J. Exp. Zool B Mol. Dev. Evol. 2006, 306, 140–163. [Google Scholar] [CrossRef]
- Mess, A. Evolutionary transformations of chorioallantoic placental characters in rodentia with special reference to hystricognath species. J. Exp. Zool A Comp. Exp. Biol. 2003, 299, 78–98. [Google Scholar] [CrossRef]
- Amoroso, E.C. Comparative anatomy of the placenta. Ann. N. Y. Acad. Sci. 1959, 75, 855–872. [Google Scholar] [CrossRef]
- Enders, A.C.; Carter, A.M. What can comparative studies of placental structure tell us?—A review. Placenta 2004, 25 (Suppl. A), S3–S9. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef]
- Mi, S.; Lee, X.; Li, X.; Veldman, G.M.; Finnerty, H.; Racie, L.; LaVallie, E.; Tang, X.Y.; Edouard, P.; Howes, S.; et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature 2000, 403, 785–789. [Google Scholar] [CrossRef]
- Blaise, S.; de Parseval, N.; Bénit, L.; Heidmann, T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 13013–13018. [Google Scholar] [CrossRef]
- Dupressoir, A.; Marceau, G.; Vernochet, C.; Bénit, L.; Kanellopoulos, C.; Sapin, V.; Heidmann, T. Syncytin-A and syncytin-B, two fusogenic placenta-specific murine envelope genes of retroviral origin conserved in Muridae. Proc. Natl. Acad. Sci. USA 2005, 102, 725–730. [Google Scholar] [CrossRef]
- Cornelis, G.; Heidmann, O.; Degrelle, S.A.; Vernochet, C.; Lavialle, C.; Letzelter, C.; Bernard-Stoecklin, S.; Hassanin, A.; Mulot, B.; Guillomot, M.; et al. Captured retroviral envelope syncytin gene associated with the unique placental structure of higher ruminants. Proc. Natl. Acad. Sci. USA 2013, 110, E828–E837. [Google Scholar] [CrossRef]
- Nakaya, Y.; Koshi, K.; Nakagawa, S.; Hashizume, K.; Miyazawa, T. Fematrin-1 is involved in fetomaternal cell-to-cell fusion in Bovinae placenta and has contributed to diversity of ruminant placentation. J. Virol. 2013, 87, 10563–10572. [Google Scholar] [CrossRef]
- Dunlap, K.A.; Palmarini, M.; Varela, M.; Burghardt, R.C.; Hayashi, K.; Farmer, J.L.; Spencer, T.E. Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proc. Natl. Acad. Sci. USA 2006, 103, 14390–14395. [Google Scholar] [CrossRef]
- Redelsperger, F.; Cornelis, G.; Vernochet, C.; Tennant, B.C.; Catzeflis, F.; Mulot, B.; Heidmann, O.; Heidmann, T.; Dupressoir, A. Capture of syncytin-Mar1, a fusogenic endogenous retroviral envelope gene involved in placentation in the Rodentia squirrel-related clade. J. Virol. 2014, 88, 7915–7928. [Google Scholar] [CrossRef] [PubMed]
- Heidmann, O.; Vernochet, C.; Dupressoir, A.; Heidmann, T. Identification of an endogenous retroviral envelope gene with fusogenic activity and placenta-specific expression in the rabbit: A new "syncytin" in a third order of mammals. Retrovirology 2009, 6, 107. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, G.; Vernochet, C.; Carradec, Q.; Souquere, S.; Mulot, B.; Catzeflis, F.; Nilsson, M.A.; Menzies, B.R.; Renfree, M.B.; Pierron, G.; et al. Retroviral envelope gene captures and syncytin exaptation for placentation in marsupials. Proc. Natl. Acad. Sci. USA 2015, 112, E487–E496. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, G.; Heidmann, O.; Bernard-Stoecklin, S.; Reynaud, K.; Véron, G.; Mulot, B.; Dupressoir, A.; Heidmann, T. Ancestral capture of syncytin-Car1, a fusogenic endogenous retroviral envelope gene involved in placentation and conserved in Carnivora. Proc. Natl. Acad. Sci. USA 2012, 109, E432–E441. [Google Scholar] [CrossRef]
- Cornelis, G.; Funk, M.; Vernochet, C.; Leal, F.; Tarazona, O.A.; Meurice, G.; Heidmann, O.; Dupressoir, A.; Miralles, A.; Ramirez-Pinilla, M.P.; et al. An endogenous retroviral envelope syncytin and its cognate receptor identified in the viviparous placental Mabuya lizard. Proc. Natl. Acad. Sci. USA 2017, 114, E10991–E11000. [Google Scholar] [CrossRef]
- Esnault, C.; Cornelis, G.; Heidmann, O.; Heidmann, T. Differential evolutionary fate of an ancestral primate endogenous retrovirus envelope gene, the EnvV syncytin, captured for a function in placentation. PLoS Genet. 2013, 9, e1003400. [Google Scholar] [CrossRef]
- Cornelis, G.; Vernochet, C.; Malicorne, S.; Souquere, S.; Tzika, A.C.; Goodman, S.M.; Catzeflis, F.; Robinson, T.J.; Milinkovitch, M.C.; Pierron, G.; et al. Retroviral envelope syncytin capture in an ancestrally diverged mammalian clade for placentation in the primitive Afrotherian tenrecs. Proc. Natl. Acad. Sci. USA 2014, 111, E4332–E4341. [Google Scholar] [CrossRef]
- Heidmann, O.; Béguin, A.; Paternina, J.; Berthier, R.; Deloger, M.; Bawa, O.; Heidmann, T. HEMO, an ancestral endogenous retroviral envelope protein shed in the blood of pregnant women and expressed in pluripotent stem cells and tumors. Proc. Natl. Acad. Sci. USA 2017, 114, E6642–E6651. [Google Scholar] [CrossRef]
- Matsuda, T.; Sasaki, M.; Kato, H.; Yamada, H.; Cohen, M.; Barrett, J.C.; Oshimura, M.; Wake, N. Human chromosome 7 carries a putative tumor suppressor gene(s) involved in choriocarcinoma. Oncogene 1997, 15, 2773–2781. [Google Scholar] [CrossRef]
- Sugimoto, J.; Sugimoto, M.; Bernstein, H.; Jinno, Y.; Schust, D. A novel human endogenous retroviral protein inhibits cell-cell fusion. Sci. Rep. 2013, 3, 1462. [Google Scholar] [CrossRef]
- Boso, G.; Fleck, K.; Carley, S.; Liu, Q.; Buckler-White, A.; Kozak, C.A. The Oldest Co-opted gag Gene of a Human Endogenous Retrovirus Shows Placenta-Specific Expression and Is Upregulated in Diffuse Large B-Cell Lymphomas. Mol. Biol. Evol. 2021, 38, 5453–5471. [Google Scholar] [CrossRef]
- Ono, R.; Nakamura, K.; Inoue, K.; Naruse, M.; Usami, T.; Wakisaka-Saito, N.; Hino, T.; Suzuki-Migishima, R.; Ogonuki, N.; Miki, H.; et al. Deletion of Peg10, an imprinted gene acquired from a retrotransposon, causes early embryonic lethality. Nat. Genet. 2006, 38, 101–106. [Google Scholar] [CrossRef]
- Sekita, Y.; Wagatsuma, H.; Nakamura, K.; Ono, R.; Kagami, M.; Wakisaka, N.; Hino, T.; Suzuki-Migishima, R.; Kohda, T.; Ogura, A.; et al. Role of retrotransposon-derived imprinted gene, Rtl1, in the feto-maternal interface of mouse placenta. Nat. Genet. 2008, 40, 243–248. [Google Scholar] [CrossRef]
- Naruse, M.; Ono, R.; Irie, M.; Nakamura, K.; Furuse, T.; Hino, T.; Oda, K.; Kashimura, M.; Yamada, I.; Wakana, S.; et al. Sirh7/Ldoc1 knockout mice exhibit placental P4 overproduction and delayed parturition. Development 2014, 141, 4763–4771. [Google Scholar] [CrossRef]
- Vernochet, C.; Heidmann, O.; Dupressoir, A.; Cornelis, G.; Dessen, P.; Catzeflis, F.; Heidmann, T. A syncytin-like endogenous retrovirus envelope gene of the guinea pig specifically expressed in the placenta junctional zone and conserved in Caviomorpha. Placenta 2011, 32, 885–892. [Google Scholar] [CrossRef]
- Nakagawa, S.; Bai, H.; Sakurai, T.; Nakaya, Y.; Konno, T.; Miyazawa, T.; Gojobori, T.; Imakawa, K. Dynamic evolution of endogenous retrovirus-derived genes expressed in bovine conceptuses during the period of placentation. Genome Biol. Evol. 2013, 5, 296–306. [Google Scholar] [CrossRef]
- Baba, K.; Nakaya, Y.; Shojima, T.; Muroi, Y.; Kizaki, K.; Hashizume, K.; Imakawa, K.; Miyazawa, T. Identification of novel endogenous betaretroviruses which are transcribed in the bovine placenta. J. Virol. 2011, 85, 1237–1245. [Google Scholar] [CrossRef]
- Sakurai, T.; Nakagawa, S.; Bai, H.; Bai, R.; Kusama, K.; Ideta, A.; Aoyagi, Y.; Kaneko, K.; Iga, K.; Yasuda, J.; et al. Novel endogenous retrovirus-derived transcript expressed in the bovine placenta is regulated by WNT signaling. Biochem. J. 2017, 474, 3499–3512. [Google Scholar] [CrossRef]
- Funk, M.; Cornelis, G.; Vernochet, C.; Heidmann, O.; Dupressoir, A.; Conley, A.; Glickman, S.; Heidmann, T. Capture of a Hyena-Specific Retroviral Envelope Gene with Placental Expression Associated in Evolution with the Unique Emergence among Carnivorans of Hemochorial Placentation in Hyaenidae. J. Virol. 2019, 93, e01811-18. [Google Scholar] [CrossRef]
- Imakawa, K.; Bai, R.; Fujiwara, H.; Ideta, A.; Aoyagi, Y.; Kusama, K. Continuous model of conceptus implantation to the maternal endometrium. J. Endocrinol. 2017, 233, R53–R65. [Google Scholar] [CrossRef]
- Biggers, J.D.; Bell, J.E.; Benos, D.J. Mammalian blastocyst: Transport functions in a developing epithelium. Am. J. Physiol. 1988, 255, C419–C432. [Google Scholar] [CrossRef]
- Sutherland, A. Mechanisms of implantation in the mouse: Differentiation and functional importance of trophoblast giant cell behavior. Dev. Biol. 2003, 258, 241–251. [Google Scholar] [CrossRef][Green Version]
- Kokkinos, M.I.; Murthi, P.; Wafai, R.; Thompson, E.W.; Newgreen, D.F. Cadherins in the human placenta--epithelial-mesenchymal transition (EMT) and placental development. Placenta 2010, 31, 747–755. [Google Scholar] [CrossRef]
- Jordan, N.V.; Johnson, G.L.; Abell, A.N. Tracking the intermediate stages of epithelial-mesenchymal transition in epithelial stem cells and cancer. Cell Cycle 2011, 10, 2865–2873. [Google Scholar] [CrossRef]
- Floridon, C.; Nielsen, O.; Holund, B.; Sunde, L.; Westergaard, J.G.; Thomsen, S.G.; Teisner, B. Localization of E-cadherin in villous, extravillous and vascular trophoblasts during intrauterine, ectopic and molar pregnancy. Mol. Hum. Reprod. 2000, 6, 943–950. [Google Scholar] [CrossRef]
- Lavialle, C.; Cornelis, G.; Dupressoir, A.; Esnault, C.; Heidmann, O.; Vernochet, C.; Heidmann, T. Paleovirology of ’syncytins’, retroviral env genes exapted for a role in placentation. Philos Trans. R Soc. Lond. B Biol. Sci. 2013, 368, 20120507. [Google Scholar] [CrossRef]
- Kemp, B.; Kertschanska, S.; Kadyrov, M.; Rath, W.; Kaufmann, P.; Huppertz, B. Invasive depth of extravillous trophoblast correlates with cellular phenotype: A comparison of intra- and extrauterine implantation sites. Histochem. Cell Biol. 2002, 117, 401–414. [Google Scholar] [CrossRef]
- Yamakoshi, S.; Bai, R.; Chaen, T.; Ideta, A.; Aoyagi, Y.; Sakurai, T.; Konno, T.; Imakawa, K. Expression of mesenchymal-related genes by the bovine trophectoderm following conceptus attachment to the endometrial epithelium. Reproduction 2012, 143, 377–387. [Google Scholar] [CrossRef]
- Talbot, N.C.; Caperna, T.J.; Edwards, J.L.; Garrett, W.; Wells, K.D.; Ealy, A.D. Bovine blastocyst-derived trophectoderm and endoderm cell cultures: Interferon tau and transferrin expression as respective in vitro markers. Biol. Reprod. 2000, 62, 235–247. [Google Scholar] [CrossRef]
- Skarzynski, D.J.; Miyamoto, Y.; Okuda, K. Production of prostaglandin f(2alpha) by cultured bovine endometrial cells in response to tumor necrosis factor alpha: Cell type specificity and intracellular mechanisms. Biol. Reprod. 2000, 62, 1116–1120. [Google Scholar] [CrossRef]
- Bai, R.; Kusama, K.; Nakamura, K.; Sakurai, T.; Kimura, K.; Ideta, A.; Aoyagi, Y.; Imakawa, K. Down-regulation of transcription factor OVOL2 contributes to epithelial-mesenchymal transition in a noninvasive type of trophoblast implantation to the maternal endometrium. FASEB J. 2018, 32, 3371–3384. [Google Scholar] [CrossRef] [PubMed]
- Kusama, K.; Bai, R.; Ideta, A.; Aoyagi, Y.; Okuda, K.; Imakawa, K. Regulation of epithelial to mesenchymal transition in bovine conceptuses through the interaction between follistatin and activin A. Mol. Cell Endocrinol. 2016, 434, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Moustakas, A.; Heldin, P. TGFβ and matrix-regulated epithelial to mesenchymal transition. Biochim. Biophys. Acta 2014, 1840, 2621–2634. [Google Scholar] [CrossRef] [PubMed]
- Wooding, F.B.; Beckers, J.F. Trinucleate cells and the ultrastructural localisation of bovine placental lactogen. Cell Tissue Res. 1987, 247, 667–673. [Google Scholar] [CrossRef]
- Seo, H.; Bazer, F.W.; Burghardt, R.C.; Johnson, G.A. Immunohistochemical Examination of Trophoblast Syncytialization during Early Placentation in Sheep. Int. J. Mol. Sci. 2019, 20, 4530. [Google Scholar] [CrossRef]
- Yamada, A.; Ohtsuki, K.; Shiga, N.; Green, J.A.; Matsuno, Y.; Imakawa, K. Epithelial-mesenchymal transition and bi- and multi-nucleated trophoblast cell formation in ovine conceptuses during the peri-implantation period. J. Reprod. Dev. 2022, 68, 110–117. [Google Scholar] [CrossRef]
- Bischof, P.; Irminger-Finger, I. The human cytotrophoblastic cell, a mononuclear chameleon. Int. J. Biochem. Cell Biol. 2005, 37, 1–16. [Google Scholar] [CrossRef]
- Moffett, A.; Loke, C. Immunology of placentation in eutherian mammals. Nat. Rev. Immunol. 2006, 6, 584–594. [Google Scholar] [CrossRef]
- Bhiwgade, D.A.; Singh, A.B.; Manekar, A.P.; Menon, S.N. Ultrastructural development of chorioallantoic placenta in the Indian Miniopterus bat, Miniopterus schreibersii fuliginosus (Hodgson). Acta Anat. 1992, 145, 248–264. [Google Scholar] [CrossRef]
- DeSesso, J.M.; Williams, A.L.; Ahuja, A.; Bowman, C.J.; Hurtt, M.E. The placenta, transfer of immunoglobulins, and safety assessment of biopharmaceuticals in pregnancy. Crit. Rev. Toxicol. 2012, 42, 185–210. [Google Scholar] [CrossRef]
- King, B.F.; Mossman, H.W. The fetal membranes and unusual giant cell placenta of the jerboa (Jaculus) and jumping mouse (Zapus). Am. J. Anat. 1974, 140, 405–431. [Google Scholar] [CrossRef]
- Oduor-Okelo, D.; Katema, R.M.; Carter, A.M. Placenta and fetal membranes of the four-toed elephant shrew, Petrodromus tetradactylus. Placenta 2004, 25, 803–809. [Google Scholar] [CrossRef]
- Oduor-Okelo, D.; Musewe, V.O.; Gombe, S. Electron microscopic study of the chorioallantoic placenta of the rock hyrax (Heterohyrax brucei). J. Reprod. Fertil. 1983, 68, 311–316. [Google Scholar] [CrossRef]
- Wooding, F.B. Role of binucleate cells in fetomaternal cell fusion at implantation in the sheep. Am. J. Anat. 1984, 170, 233–250. [Google Scholar] [CrossRef]
- Haig, D. Retroviruses and the placenta. Curr. Biol. 2012, 22, R609–R613. [Google Scholar] [CrossRef]
- Bininda-Emonds, O.R.; Cardillo, M.; Jones, K.E.; MacPhee, R.D.; Beck, R.M.; Grenyer, R.; Price, S.A.; Vos, R.A.; Gittleman, J.L.; Purvis, A. The delayed rise of present-day mammals. Nature 2007, 446, 507–512. [Google Scholar] [CrossRef]
- Ono, R.; Kobayashi, S.; Wagatsuma, H.; Aisaka, K.; Kohda, T.; Kaneko-Ishino, T.; Ishino, F. A retrotransposon-derived gene, PEG10, is a novel imprinted gene located on human chromosome 7q21. Genomics 2001, 73, 232–237. [Google Scholar] [CrossRef]
- Suzuki, S.; Ono, R.; Narita, T.; Pask, A.J.; Shaw, G.; Wang, C.; Kohda, T.; Alsop, A.E.; Marshall Graves, J.A.; Kohara, Y.; et al. Retrotransposon silencing by DNA methylation can drive mammalian genomic imprinting. PLoS Genet. 2007, 3, e55. [Google Scholar] [CrossRef]
- Charlier, C.; Segers, K.; Wagenaar, D.; Karim, L.; Berghmans, S.; Jaillon, O.; Shay, T.; Weissenbach, J.; Cockett, N.; Gyapay, G.; et al. Human-ovine comparative sequencing of a 250-kb imprinted domain encompassing the callipyge (clpg) locus and identification of six imprinted transcripts: DLK1, DAT, GTL2, PEG11, antiPEG11, and MEG8. Genome. Res. 2001, 11, 850–862. [Google Scholar] [CrossRef]
- Kim, A.; Terzian, C.; Santamaria, P.; Pélisson, A.; Purd’homme, N.; Bucheton, A. Retroviruses in invertebrates: The gypsy retrotransposon is apparently an infectious retrovirus of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1994, 91, 1285–1289. [Google Scholar] [CrossRef]
- Song, S.U.; Gerasimova, T.; Kurkulos, M.; Boeke, J.D.; Corces, V.G. An env-like protein encoded by a Drosophila retroelement: Evidence that gypsy is an infectious retrovirus. Genes Dev. 1994, 8, 2046–2057. [Google Scholar] [CrossRef] [PubMed]
- Kaneko-Ishino, T.; Ishino, F. The role of genes domesticated from LTR retrotransposons and retroviruses in mammals. Front. Microbiol. 2012, 3, 262. [Google Scholar] [CrossRef] [PubMed]
- Kaneko-Ishino, T.; Ishino, F. The Evolutionary Advantage in Mammals of the Complementary Monoallelic Expression Mechanism of Genomic Imprinting and Its Emergence From a Defense Against the Insertion Into the Host Genome. Front. Genet. 2022, 13, 832983. [Google Scholar] [CrossRef] [PubMed]
- Kitazawa, M.; Tamura, M.; Kaneko-Ishino, T.; Ishino, F. Severe damage to the placental fetal capillary network causes mid- to late fetal lethality and reduction in placental size in Peg11/Rtl1 KO mice. Genes Cells 2017, 22, 174–188. [Google Scholar] [CrossRef]
- Shiura, H.; Ono, R.; Tachibana, S.; Kohda, T.; Kaneko-Ishino, T.; Ishino, F. PEG10 viral aspartic protease domain is essential for the maintenance of fetal capillary structure in the mouse placenta. Development 2021, 148, dev199564. [Google Scholar] [CrossRef] [PubMed]
- Malassiné, A.; Frendo, J.L.; Evain-Brion, D. A comparison of placental development and endocrine functions between the human and mouse model. Hum. Reprod. Update 2003, 9, 531–539. [Google Scholar] [CrossRef] [PubMed]
- Murr, S.M.; Stabenfeldt, G.H.; Bradford, G.E.; Geschwind, I.I. Plasma progesterone during pregnancy in the mouse. Endocrinology 1974, 94, 1209–1211. [Google Scholar] [CrossRef]
- Virgo, B.B.; Bellward, G.D. Serum progesterone levels in the pregnant and postpartum laboratory mouse. Endocrinology 1974, 95, 1486–1490. [Google Scholar] [CrossRef]
- Renaud, S.J.; Karim Rumi, M.A.; Soares, M.J. Review: Genetic manipulation of the rodent placenta. Placenta 2011, 32 (Suppl. 2), S130–S135. [Google Scholar] [CrossRef][Green Version]
- De Parseval, N.; Lazar, V.; Casella, J.F.; Benit, L.; Heidmann, T. Survey of human genes of retroviral origin: Identification and transcriptome of the genes with coding capacity for complete envelope proteins. J. Virol. 2003, 77, 10414–10422. [Google Scholar] [CrossRef]
- Chang, C.; Chen, P.T.; Chang, G.D.; Huang, C.J.; Chen, H. Functional characterization of the placental fusogenic membrane protein syncytin. Biol. Reprod. 2004, 71, 1956–1962. [Google Scholar] [CrossRef]
- Chen, C.P.; Chen, L.F.; Yang, S.R.; Chen, C.Y.; Ko, C.C.; Chang, G.D.; Chen, H. Functional characterization of the human placental fusogenic membrane protein syncytin 2. Biol. Reprod. 2008, 79, 815–823. [Google Scholar] [CrossRef]
- Okahara, G.; Matsubara, S.; Oda, T.; Sugimoto, J.; Jinno, Y.; Kanaya, F. Expression analyses of human endogenous retroviruses (HERVs): Tissue-specific and developmental stage-dependent expression of HERVs. Genomics 2004, 84, 982–990. [Google Scholar] [CrossRef]
- Hayward, M.D.; Pötgens, A.J.; Drewlo, S.; Kaufmann, P.; Rasko, J.E. Distribution of human endogenous retrovirus type W receptor in normal human villous placenta. Pathology 2007, 39, 406–412. [Google Scholar] [CrossRef]
- Esnault, C.; Priet, S.; Ribet, D.; Vernochet, C.; Bruls, T.; Lavialle, C.; Weissenbach, J.; Heidmann, T. A placenta-specific receptor for the fusogenic, endogenous retrovirus-derived, human syncytin-2. Proc. Natl. Acad. Sci. USA 2008, 105, 17532–17537. [Google Scholar] [CrossRef]
- Blaise, S.; de Parseval, N.; Heidmann, T. Functional characterization of two newly identified Human Endogenous Retrovirus coding envelope genes. Retrovirology 2005, 2, 19. [Google Scholar] [CrossRef]
- Cáceres, M.; Thomas, J.W. The gene of retroviral origin Syncytin 1 is specific to hominoids and is inactive in Old World monkeys. J. Hered. 2006, 97, 100–106. [Google Scholar] [CrossRef]
- Pötgens, A.J.; Schmitz, U.; Bose, P.; Versmold, A.; Kaufmann, P.; Frank, H.G. Mechanisms of syncytial fusion: A review. Placenta 2002, 23 (Suppl. A), S107–S113. [Google Scholar] [CrossRef]
- Yu, C.; Shen, K.; Lin, M.; Chen, P.; Lin, C.; Chang, G.D.; Chen, H. GCMa regulates the syncytin-mediated trophoblastic fusion. J. Biol. Chem. 2002, 277, 50062–50068. [Google Scholar] [CrossRef]
- Le Naour, F.; Rubinstein, E.; Jasmin, C.; Prenant, M.; Boucheix, C. Severely reduced female fertility in CD9-deficient mice. Science 2000, 287, 319–321. [Google Scholar] [CrossRef]
- Tachibana, I.; Hemler, M.E. Role of transmembrane 4 superfamily (TM4SF) proteins CD9 and CD81 in muscle cell fusion and myotube maintenance. J. Cell Biol. 1999, 146, 893–904. [Google Scholar] [CrossRef] [PubMed]
- Muroi, Y.; Sakurai, T.; Hanashi, A.; Kubota, K.; Nagaoka, K.; Imakawa, K. CD9 regulates transcription factor GCM1 and ERVWE1 expression through the cAMP/protein kinase A signaling pathway. Reproduction 2009, 138, 945–951. [Google Scholar] [CrossRef] [PubMed]
- Dunn-Fletcher, C.E.; Muglia, L.M.; Pavlicev, M.; Wolf, G.; Sun, M.A.; Hu, Y.C.; Huffman, E.; Tumukuntala, S.; Thiele, K.; Mukherjee, A.; et al. Anthropoid primate-specific retroviral element THE1B controls expression of CRH in placenta and alters gestation length. PLoS Biol. 2018, 16, e2006337. [Google Scholar] [CrossRef] [PubMed]
- Kitao, K.; Tanikaga, T.; Miyazawa, T. Identification of a post-transcriptional regulatory element in the human endogenous retroviral syncytin-1. J. Gen. Virol. 2019, 100, 662–668. [Google Scholar] [CrossRef]
- Kitao, K.; Nakagawa, S.; Miyazawa, T. An ancient retroviral RNA element hidden in mammalian genomes and its involvement in co-opted retroviral gene regulation. Retrovirology 2021, 18, 36. [Google Scholar] [CrossRef]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science 2016, 351, 1083–1087. [Google Scholar] [CrossRef]
- Sun, M.A.; Wolf, G.; Wang, Y.; Senft, A.D.; Ralls, S.; Jin, J.; Dunn-Fletcher, C.E.; Muglia, L.J.; Macfarlan, T.S. Endogenous Retroviruses Drive Lineage-Specific Regulatory Evolution across Primate and Rodent Placentae. Mol. Biol. Evol. 2021, 38, 4992–5004. [Google Scholar] [CrossRef]
- Imakawa, K.; Nakagawa, S.; Miyazawa, T. Baton pass hypothesis: Successive incorporation of unconserved endogenous retroviral genes for placentation during mammalian evolution. Genes Cells 2015, 20, 771–788. [Google Scholar] [CrossRef]
- Mangeney, M.; Renard, M.; Schlecht-Louf, G.; Bouallaga, I.; Heidmann, O.; Letzelter, C.; Richaud, A.; Ducos, B.; Heidmann, T. Placental syncytins: Genetic disjunction between the fusogenic and immunosuppressive activity of retroviral envelope proteins. Proc. Natl. Acad. Sci. USA 2007, 104, 20534–20539. [Google Scholar] [CrossRef]
- Dewannieux, M.; Heidmann, T. Endogenous retroviruses: Acquisition, amplification and taming of genome invaders. Curr Opin Virol 2013, 3, 646–656. [Google Scholar] [CrossRef]
- Chuong, E.B. The placenta goes viral: Retroviruses control gene expression in pregnancy. PLoS Biol. 2018, 16, e3000028. [Google Scholar] [CrossRef]
- Chuong, E.B.; Rumi, M.A.; Soares, M.J.; Baker, J.C. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat. Genet. 2013, 45, 325–329. [Google Scholar] [CrossRef]
- Kaneko-Ishino, T.; Ishino, F. Mammalian-specific genomic functions: Newly acquired traits generated by genomic imprinting and LTR retrotransposon-derived genes in mammals. Proc. Jpn Acad. Ser. B Phys. Biol. Sci. 2015, 91, 511–538. [Google Scholar] [CrossRef]


| Gene Name | LTR/ERV | Taxonomy | Fusion Activity | Reference |
|---|---|---|---|---|
| syncytin-1 | HERV-W | Hominidae (Catarrhini) | yes | [15] |
| syncytin-2 | HERV-FRD | Simiiformes | yes | [16] |
| syncytin-A | - | Muridae | yes | [17] |
| syncytin-B | - | Muridae | yes | [17] |
| syncytin-Rum1 | - | Ruminantia | yes | [18] |
| Fematrin-1 | BERV-K1 | Bovinae | yes | [19] |
| enJSRV env | endogenous JSRV | Caprinae | yes | [20] |
| syncytin-Mar1 | - | Squirrel | yes | [21] |
| syncyitn-Ory1 | - | Leporidae | yes | [22] |
| syncytin-Opo1 | - | Monodelphis | yes | [23] |
| syncytin-Car1 | - | Carnivora | yes | [24] |
| syncytin-Mab1 | - | Mabuya | yes | [25] |
| mac-syncytin-3 | ERV-V2 | Simiiformes | yes | [26] |
| syncytin-Ten1 | - | Tenrecidae | yes | [27] |
| HEMO | MER34 | Simiiformes (Boreoeutheria) | No | [28] |
| ERV3-1 | HERV-R | Simiiformes | No | [29] |
| suppressyn | HERV-Fb1 | Catarrhini | No | [30] |
| gagV1 | HERV-V1 | Simiiformes | No | [31] |
| PEG10 | sushi-ichi | Theria | No | [32] |
| PEG11/RTL1 | sushi-ichi | Placentalia | No | [33] |
| SIRH7/LDOC1 | sushi-ichi | Placentalia | No | [34] |
| env-Cav1 | - | Caviomorpha | No | [35] |
| BERV-P | BERV-P | Bovinae | No | [36] |
| BERV-K2 | BERV-K2 | Bovinae | No | [37] |
| BERV-K3 | BERV-K3 | Bovinae | No | [38] |
| hyena-Env2 | - | Hyaenidae | No | [39] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imakawa, K.; Kusama, K.; Kaneko-Ishino, T.; Nakagawa, S.; Kitao, K.; Miyazawa, T.; Ishino, F. Endogenous Retroviruses and Placental Evolution, Development, and Diversity. Cells 2022, 11, 2458. https://doi.org/10.3390/cells11152458
Imakawa K, Kusama K, Kaneko-Ishino T, Nakagawa S, Kitao K, Miyazawa T, Ishino F. Endogenous Retroviruses and Placental Evolution, Development, and Diversity. Cells. 2022; 11(15):2458. https://doi.org/10.3390/cells11152458
Chicago/Turabian StyleImakawa, Kazuhiko, Kazuya Kusama, Tomoko Kaneko-Ishino, So Nakagawa, Koichi Kitao, Takayuki Miyazawa, and Fumitoshi Ishino. 2022. "Endogenous Retroviruses and Placental Evolution, Development, and Diversity" Cells 11, no. 15: 2458. https://doi.org/10.3390/cells11152458
APA StyleImakawa, K., Kusama, K., Kaneko-Ishino, T., Nakagawa, S., Kitao, K., Miyazawa, T., & Ishino, F. (2022). Endogenous Retroviruses and Placental Evolution, Development, and Diversity. Cells, 11(15), 2458. https://doi.org/10.3390/cells11152458






