Contrasting Roles of Ethylene Response Factors in Pathogen Response and Ripening in Fleshy Fruit
Abstract
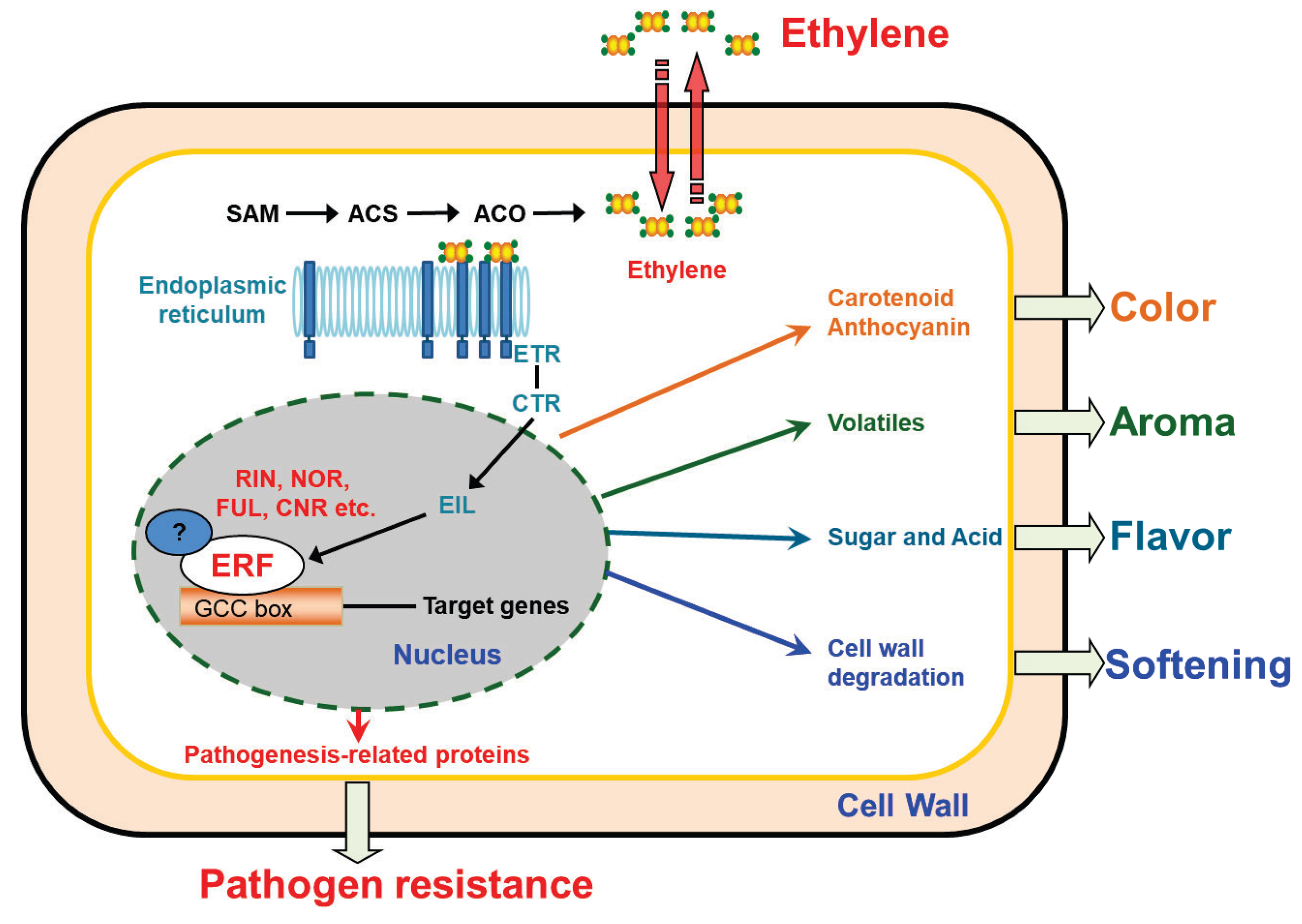
:1. Introduction
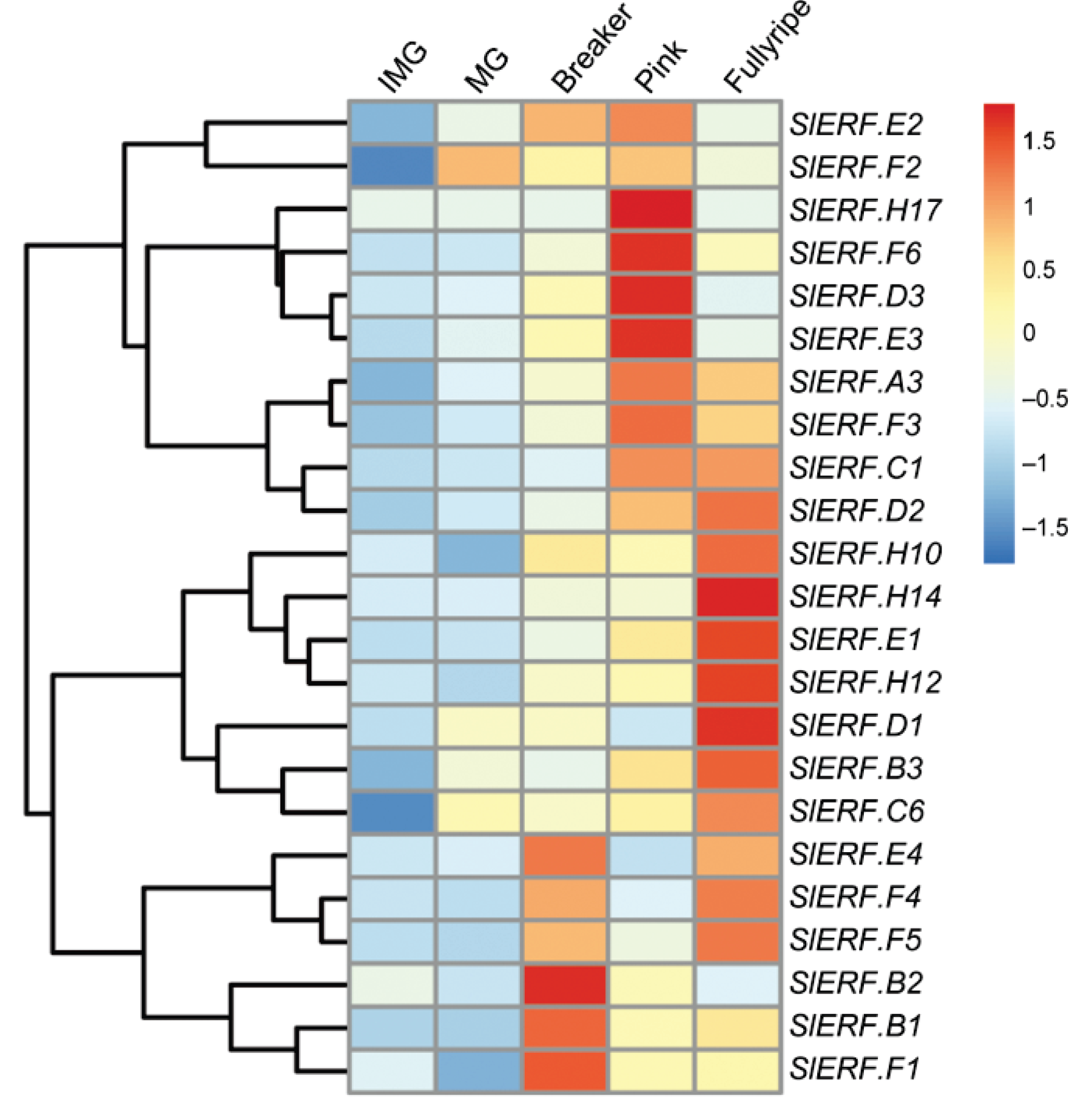
2. ERF Transcription Factors in Fruit Ripening
3. Fruit Responses to Pathogen Infection
3.1. General Responses to Infection and the Involvement of ERFs
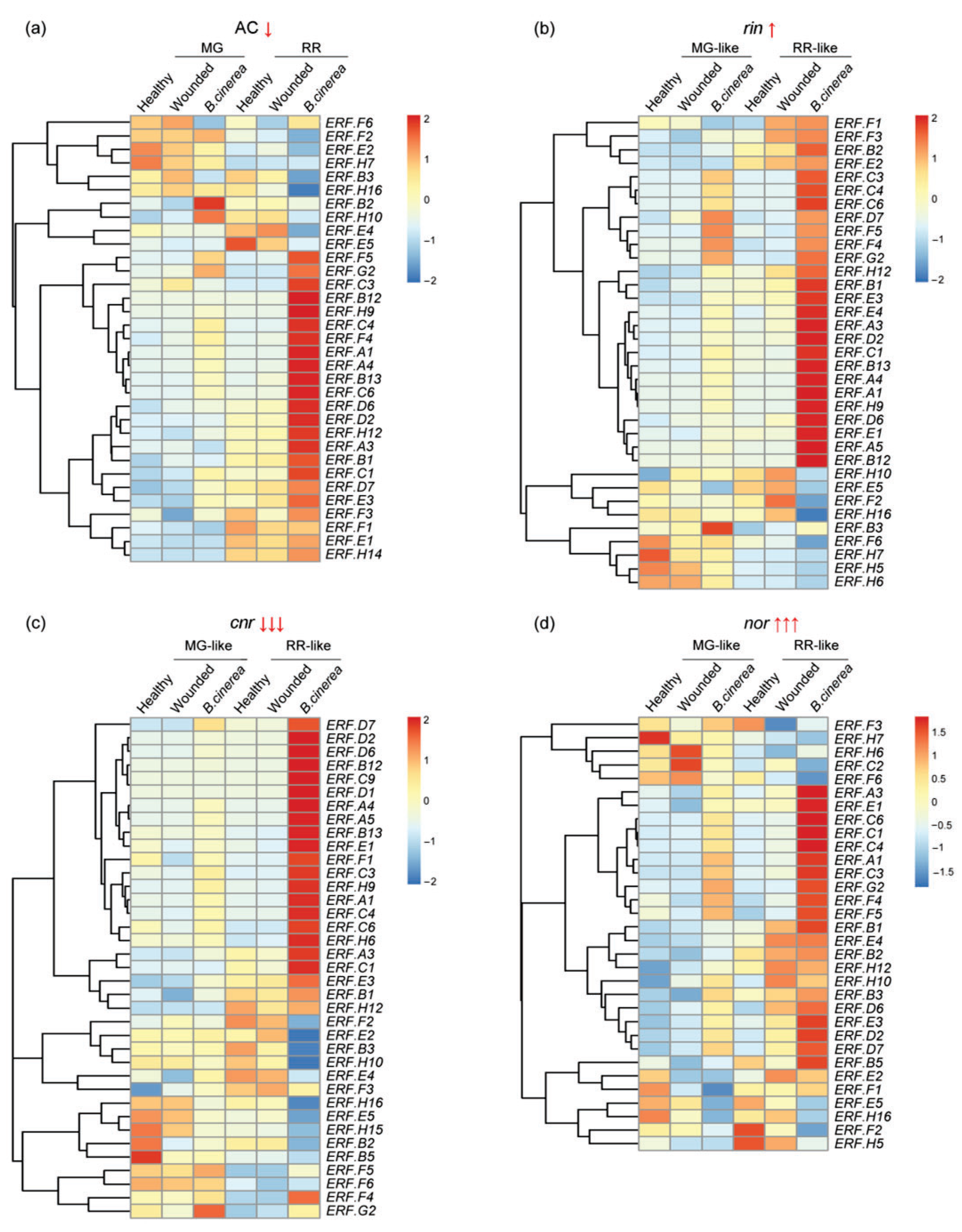
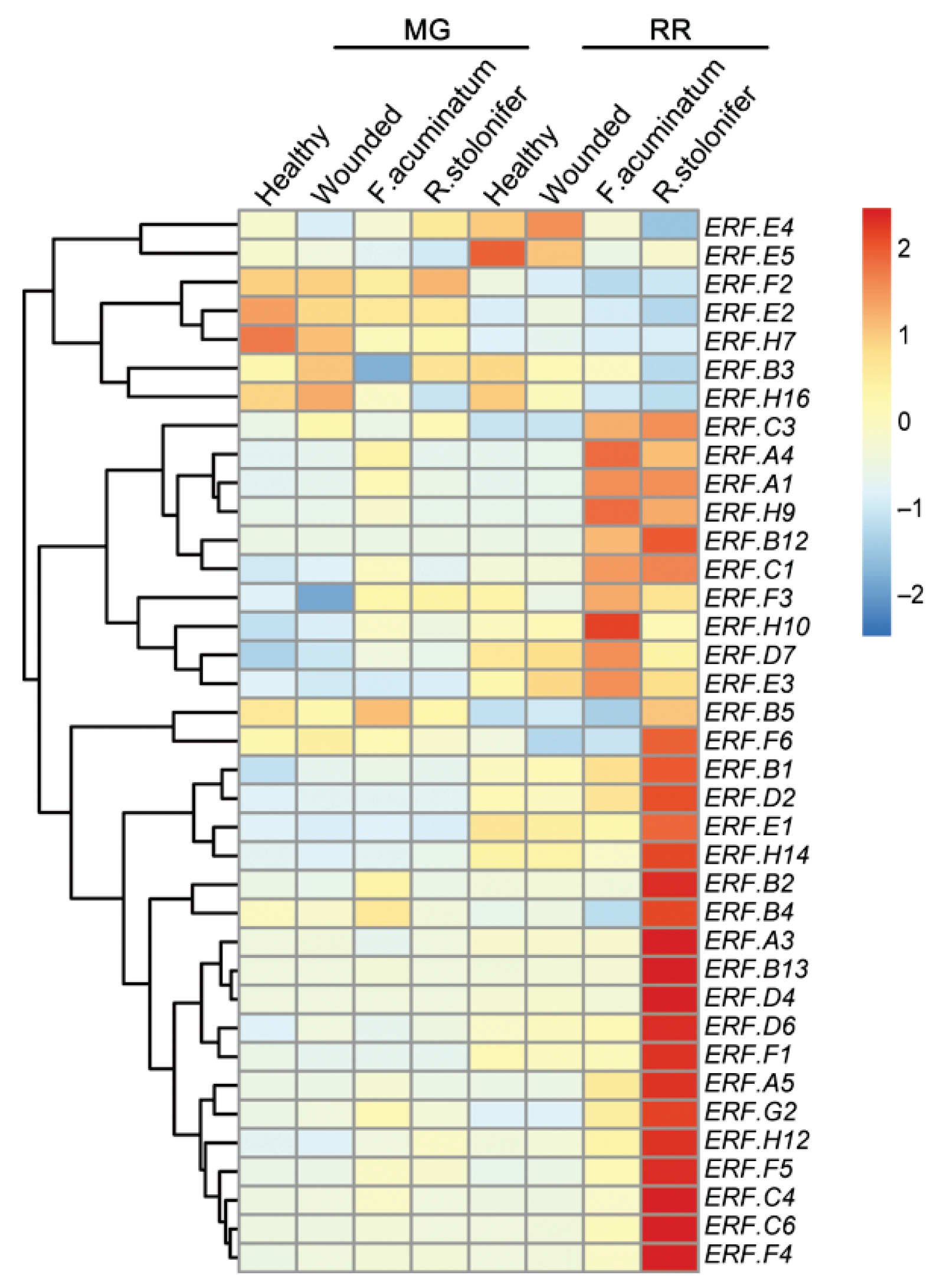
3.2. ERF Factors Induced in Fruit Responses to B. cinerea
3.3. ERFs Interact with Other Ripening Regulators and Affect Fruit Resistance to B. cinerea Infection
3.4. Tomato ERF Factors Function in Fruit Response to Other Pathogens
3.5. ERF Factors Function in Other Fleshy Fruits in Response to Pathogens
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Cantu, D.; Blanco-Ulate, B.; Yang, L.; Labavitch, J.M.; Bennett, A.B.; Powell, A.L. Ripening-regulated susceptibility of tomato fruit to Botrytis cinerea requires NOR but not RIN or ethylene. Plant Physiol. 2009, 150, 1434–1449. [Google Scholar] [CrossRef] [Green Version]
- Florlani, S.; Masiero, S.; Mizzotti, C. Fruit ripening: The role of hormones, cell wall modifications, and their relationship with pathogens. J. Exp. Bot. 2019, 70, 2993–3006. [Google Scholar] [CrossRef] [PubMed]
- Matas, A.J.; Gapper, N.E.; Chung, M.Y.; Giovannoni, J.J.; Rose, J.K. Biology and genetic engineering of fruit maturation for enhanced quality and shelf-life. Curr. Opin. Biotechnol. 2009, 20, 197–203. [Google Scholar] [CrossRef]
- Grierson, D. Ethylene and the control of fruit ripening. In The Molecular Biology and Biochemistry of Fruit Ripening; Seymour, G.B., Poole, M., Giovannoni, J.J., Tucker, G.A., Eds.; John Wiley and Sons: Hoboken, NJ, USA, 2013. [Google Scholar] [CrossRef]
- Li, S.; Chen, K.; Grierson, D. Molecular and hormonal mechanisms regulating fleshy fruit ripening. Cells 2021, 10, 1136. [Google Scholar] [CrossRef] [PubMed]
- Qin, G.; Wang, Y.; Cao, B.; Wang, W.; Tian, S. Unraveling the regulatory network of the MADS box transcription factor RIN in fruit ripening. Plant J. 2012, 70, 243–255. [Google Scholar] [CrossRef]
- Liu, Z.; Wu, X.; Liu, H.; Zhang, M.; Liao, W. DNA methylation in tomato fruit ripening. Physiol. Plant. 2022, 174, e13627. [Google Scholar] [CrossRef]
- Zhou, L.; Tian, S.; Qin, G. RNA methylomes reveal the m6A-mediated regulation of DNA demethylase gene SlDML2 in tomato fruit ripening. Genome Biol. 2019, 20, 156. [Google Scholar] [CrossRef] [Green Version]
- Giovannoni, J.; Nguyen, C.; Ampofo, B.; Zhong, S.; Fei, Z. The Epigenome and Transcriptional Dynamics of Fruit Ripening. Annu. Rev. Plant Biol. 2017, 68, 61–84. [Google Scholar] [CrossRef]
- Fillinger, S.; Elad, Y. (Eds.) Botrytis—The Fungus, the Pathogen and Its Management in Agricultural Systems; Springer International Publishing: Cham, Switzerland, 2016. [Google Scholar] [CrossRef]
- Zhang, L.; Zhu, M.; Ren, L.; Li, A.; Chen, G.; Hu, Z. The SlFSR gene controls fruit shelf-life in tomato. J. Exp. Bot. 2018, 69, 2897–2909. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Zhang, Z.; Xu, Z.; Wang, L.; Chen, C.; Ren, Z. Overexpression of SlMYB75 enhances resistance to Botrytis cinerea and prolongs fruit storage life in tomato. Plant Cell Rep. 2021, 40, 43–58. [Google Scholar] [PubMed]
- Min, D.; Li, F.; Cui, X.; Zhou, J.; Li, J.; Ai, W.; Shu, P.; Zhang, X.; Li, X.; Meng, D.; et al. SlMYC2 are required for methyl jasmonate-induced tomato fruit resistance to Botrytis cinerea. Food Chem. 2020, 310, 125901. [Google Scholar] [CrossRef] [PubMed]
- Du, M.M.; Zhao, J.H.; Tzeng, D.T.W.; Liu, Y.Y.; Deng, L.; Yang, T.X.; Zhai, Q.Z.; Wu, F.M.; Huang, Z.; Zhou, M.; et al. MYC2 orchestrates a hierarchical transcriptional cascade that regulates Jasmonate-mediated plant immunity in tomato. Plant Cell 2017, 29, 1883–1906. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buxdorf, K.; Rubinsky, G.; Barda, O.; Burdman, S.; Aharoni, A.; Levy, M. The transcription factor SlSHINE3 modulates defense responses in tomato plants. Plant Mol. Biol. 2013, 84, 37–47. [Google Scholar] [CrossRef] [PubMed]
- Powell, A.L.; Nguyen, C.V.; Hill, T.; Cheng, K.L.; Figueroa-Balderas, R.; Aktas, H.; Ashrafi, H.; Pons, C.; Fernández-Muñoz, R.; Vicente, A.; et al. Uniform ripening encodes a Golden 2-like transcription factor regulating tomato fruit chloroplast development. Science 2012, 336, 1711–1715. [Google Scholar] [CrossRef] [Green Version]
- Pan, H.; Bradley, G.; Pyke, K. Network inference analysis identifies and APRR2-Like gene linked to pigment accumulation in tomato and pepper fruits. Plant Physiol. 2013, 161, 1476–1485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadakudti, S.S.; Holdsworth, W.L.; Klein, C.L.; Barry, C.S. KNOX genes influence a gradient of fruit chloroplast development through regulation of GOLDEN2-LIKE expression in tomato. Plant J. 2014, 78, 1022–1033. [Google Scholar] [CrossRef]
- Nguyen, C.V.; Vrebalov, J.T.; Gapper, N.E.; Zheng, Y.; Zhong, S.L.; Fei, Z.J.; Giovannoni, J.J. Tomato GOLDEN2-LIKE transcription factors reveal molecular gradients that function during fruit development and ripening. Plant Cell 2014, 26, 585–601. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Butelli, E.; De Stefano, R.; Schoonbeek, H.J.; Magusin, A.; Pagliarani, C.; Wellner, N.; Hill, L.; Orzaez, D.; Granell, A.; et al. Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr. Biol. 2013, 23, 1094–1100. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; De Stefano, R.; Robine, M.; Butelli, E.; Bulling, K.; Hill, L.; Rejzek, M.; Martin, C.; Schoonbeek, H.J. Different reactive oxygen species scavenging properties of flavonoids determine their abilities to extend the shelf life of tomato. Plant Physiol. 2015, 169, 1568–1583. [Google Scholar]
- Bassolino, L.; Zhang, Y.; Schoonbeek, H.; Kiferle, C.; Perata, P.; Martin, C. Accumulation of anthocyanins in tomato skin extends shelf life. New Phytol. 2013, 200, 650–655. [Google Scholar] [CrossRef] [Green Version]
- Sharma, R.R.; Singh, D.; Singh, R. Biological control of postharvest diseases of fruits and vegetables by microbial antagonists: A review. Biol. Control 2009, 50, 205–221. [Google Scholar] [CrossRef]
- Elad, Y.; Williamson, B.; Tudzynski, P.; Delen, N. Botrytis spp. and diseases they cause in agricultural systems—An introduction. In Botrytis: Biology, Pathology and Control; Elad, Y., Williamson, B., Tudzynski, P., Delen, N., Eds.; Springer: Dordrecht, The Netherlands, 2007. [Google Scholar] [CrossRef]
- Weiberg, A.; Wang, M.; Lin, F.M.; Zhao, H.; Zhang, Z.; Kaloshian, I.; Huang, H.D.; Jin, H. Fungal small RNAs suppress plant immunity by hijacking host RNA interference pathways. Science 2013, 42, 118–123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, A.E.; Swinburne, T.R. The resistance of immature banana fruits to anthracnose [Colletotrichum musae (Berk. & Curt.) Arx.]. J. Phytopathol. 1980, 99, 70–80. [Google Scholar]
- Alkan, N.; Friedlander, G.; Ment, D.; Prusky, D.; Fluhr, R. Simultaneous transcriptome analysis of Colletotrichum gloeosporioides and tomato fruit pathosystem reveals novel fungal pathogenicity and fruit defense strategies. New Phytol. 2015, 205, 801–815. [Google Scholar] [CrossRef] [PubMed]
- Hong, K.; Gong, D.; Zhang, L.; Hu, H.; Jia, Z.; Gu, H.; Song, K. Transcriptome characterization and expression profiles of the related defense genes in postharvest mango fruit against Colletotrichum gloeosporioides. Gene 2016, 576, 275–283. [Google Scholar] [CrossRef] [PubMed]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Deng, H.; Chen, Y.; Liu, Z.Y.; Liu, Z.Q.; Shu, P.; Wang, R.C.; Hao, Y.W.; Su, D.; Pirrello, J.; Liu, Y.S.; et al. SlERF.F12 modulates the transition to ripening in tomato fruit by recruiting the co-repressor TOPLESS and histone deacetylases to repress key ripening genes. Plant Cell 2022, 34, 1250–1272. [Google Scholar] [CrossRef]
- Ju, C.; Chang, C. Mechanistic insights in ethylene perception and signal transduction. Plant Physiol. 2015, 169, 85–95. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.C.; Pirrello, J.; Chervin, C.; Roustan, J.P.; Bouzayen, M. Ethylene control of fruit ripening: Revisiting the complex network of transcriptional regulation. Plant Physiol. 2015, 169, 2380–2390. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.C.; Gomes, B.L.; Mila, I.; Purgatto, E.; Peres, L.; Frasse, P.; Maza, E.; Zouine, M.; Roustan, J.P.; Bouzayen, M.; et al. Comprehensive profiling of ethylene response factor expression identifies ripening-associated ERF genes and their link to key regulators of fruit ripening in tomato. Plant Physiol. 2016, 170, 1732–1744. [Google Scholar] [CrossRef] [Green Version]
- Müller, M.; Munné-Bosch, S. Ethylene Response Factors: A Key Regulatory Hub in Hormone and Stress Signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, W.; Zhao, R.; Sheng, J.; Shen, L. SlERF2 is associated with methyl jasmonate-mediated defense response against Botrytis cinerea in tomato fruit. J. Agric. Food Chem. 2018, 66, 9923–9932. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, Z.; Liu, S.; Huang, L.; Hong, Y.B.; Li, X.H.; Huang, L.; Zhang, Y.F.; Zhang, H.J.; Li, D.Y.; Song, F.M. Tomato SlERF.A1, SlERF.B4, SlERF.C3 and SlERF.A3, members of B3 group of ERF family, are required for resistance to Botrytis cinerea. Front. Plant Sci. 2016, 7, 1964. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Zhu, B.; Pirrello, J.; Xu, C.; Zhang, B.; Bouzayen, M.; Chen, K.; Grierson, D. Roles of RIN and ethylene in tomato fruit ripening and ripening-associated traits. New Phytol. 2020, 226, 460–475. [Google Scholar] [CrossRef]
- Li, B.J.; Grierson, D.; Shi, Y.N.; Chen, K.S. Roles of abscisic acid in regulating ripening and quality of strawberry, a model non-climacteric fruit. Hortic. Res. 2022, 9, uhac089. [Google Scholar] [CrossRef] [PubMed]
- McMurchie, E.J.; McGlasson, W.B.; Eaks, I.L. Treatment of fruit with propylene gives information about the biogenesis of ethylene. Nature 1972, 237, 235–236. [Google Scholar] [CrossRef]
- Xie, X.L.; Yin, X.R.; Chen, K.S. Roles of APETALA2/Ethylene responsive factors in regulation of fruit quality. Crit. Rev. Plant Sci. 2016, 35, 120–130. [Google Scholar] [CrossRef]
- Reid, M.S.; Staby, G.L. A brief history of 1-methylcyclopropene. HortScience 2008, 43, 83–85. [Google Scholar] [CrossRef] [Green Version]
- Sisler, E.C.; Serek, M. Inhibitors of ethylene responses in plants at the receptor level: Recent development. Physiol. Plant. 1997, 100, 577–582. [Google Scholar] [CrossRef]
- Sisler, E.C. The discovery and development of compounds counteracting ethylene at the receptor level. Biotechnol. Adv. 2006, 24, 357–367. [Google Scholar] [CrossRef]
- Pirrello, J.; Prasad, B.C.; Zhang, W.; Chen, K.; Mila, I.; Zouine, M.; Latché, A.; Pech, J.C.; Ohme-Takagi, M.; Regad, F.; et al. Functional analysis and binding affinity of tomato ethylene response factors provide insight on the molecular bases of plant differential responses to ethylene. BMC Plant Biol. 2012, 12, 190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kagale, S.; Rozwadowski, K. EAR motif-mediated transcriptional repression in plants: An underlying mechanism for epigenetic regulation of gene expression. Epigenetics 2011, 6, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Zouine, M.; Maza, E.; Djari, A.; Lauvernier, M.; Frasse, P.; Smouni, A.; Pirrello, J.; Bouzayen, M. TomExpress, a unified tomato RNA-Seq platform for visualization of expression data, clustering and correlation networks. Plant J. 2017, 92, 727–735. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.M.; Joung, J.G.; McQuinn, R.; Chung, M.Y.; Fei, Z.J.; Tieman, D.; Klee, H.; Giovannoni, J. Combined transcriptome, genetic diversity and metabolite profiling in tomato fruit reveals that the ethylene response factor SlERF6 plays an important role in ripening and carotenoid accumulation. Plant J. 2012, 70, 191–204. [Google Scholar] [CrossRef]
- Li, Y.; Zhu, B.; Xu, W.; Zhu, H.; Chen, A.; Xie, Y.; Shao, Y.; Luo, Y. LeERF1 positively modulated ethylene triple response on etiolated seedling, plant development and fruit ripening and softening in tomato. Plant Cell Rep. 2007, 26, 1999–2008. [Google Scholar] [CrossRef]
- Liu, M.; Diretto, G.; Pirrello, J.; Roustan, J.P.; Li, Z.; Giuliano, G.; Regad, F.; Bouzayen, M. The chimeric repressor version of an Ethylene Response Factor (ERF) family member, Sl-ERF.B3, shows contrasting effects on tomato fruit ripening. New Phytol. 2014, 203, 206–218. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Hu, Z.; Grierson, D. Differential regulation of tomato ethylene responsive factor LeERF3b, a putative repressor, and the activator Pti4 in ripening mutants and in response to environmental stresses. J. Plant Physiol. 2008, 165, 662–670. [Google Scholar] [CrossRef]
- Ye, J.; Hu, T.; Yang, C.; Li, H.; Yang, M.; Ijaz, R.; Ye, Z.; Zhang, Y. Transcriptome profiling of tomato fruit development reveals transcription factors associated with ascorbic acid, carotenoid and flavonoid biosynthesis. PLoS ONE 2015, 10, e0130885. [Google Scholar] [CrossRef]
- Sun, Y.; Liang, B.; Wang, J.; Kai, W.; Chen, P.; Jiang, L.; Du, Y.; Leng, P. SlPti4 affects regulation of fruit ripening, seed germination and stress responses by modulating ABA signaling in tomato. Plant Cell Physiol. 2018, 59, 1956–1965. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zeng, W.; Ding, Y.; Wang, Y.; Niu, L.; Yao, J.L.; Pan, L.; Lu, Z.; Cui, G.; Li, G.; et al. PpERF3 positively regulates ABA biosynthesis by activating PpNCED2/3 transcription during fruit ripening in peach. Hortic. Res. 2019, 6, 19. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Zeng, W.; Ding, Y.; Wang, Y.; Niu, L.; Yao, J.L.; Pan, L.; Lu, Z.; Cui, G.; Li, G.; et al. Peach ethylene response factor PpeERF2 represses the expression of ABA biosynthesis and cell wall degradation genes during fruit ripening. Plant Sci. 2019, 283, 116–126. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Liu, J.; Wang, X.; Wang, Y.; Yuan, Y.; Yang, S. PpERF/ABR1 functions as an activator to regulate PpPG expression resulting in fruit softening during storage in peach (Prunus persica). Postharvest Biol. Technol. 2022, 189, 111919. [Google Scholar] [CrossRef]
- Khaksar, G.; Sirikantaramas, S. Transcriptome-wide identification and expression profiling of the ERF gene family suggest roles as transcriptional activators and repressors of fruit ripening in durian. PLoS ONE 2021, 16, e0252367. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.Y.; Chen, J.Y.; Kuang, J.F.; Shan, W.; Xie, H.; Jiang, Y.M.; Lu, W.J. Banana ethylene response factors are involved in fruit ripening through their interactions with ethylene biosynthesis genes. J. Exp. Bot. 2013, 64, 2499–2510. [Google Scholar] [CrossRef] [Green Version]
- Wang, A.; Tan, D.; Takahashi, A.; Li, T.Z.; Harada, T. MdERFs, two ethylene-response factors involved in apple fruit ripening. J. Exp. Bot. 2007, 58, 3743–3748. [Google Scholar] [CrossRef]
- Li, T.; Jiang, Z.; Zhang, L.; Tan, D.; Wei, Y.; Yuan, H.; Li, T.; Wang, A. Apple (Malus domestica) MdERF2 negatively affects ethylene biosynthesis during fruit ripening by suppressing MdACS1 transcription. Plant J. 2016, 88, 735–748. [Google Scholar] [CrossRef]
- Han, Y.C.; Kuang, J.F.; Chen, J.Y.; Liu, X.C.; Xiao, Y.Y.; Fu, C.C.; Wang, J.N.; Wu, K.Q.; Lu, W.J. Banana transcription factor MaERF11 recruits histone deacetylase MaHDA1 and represses the expression of MaACO1 and expansins during fruit ripening. Plant Physiol. 2016, 171, 1070–1084. [Google Scholar] [CrossRef] [Green Version]
- Ampomah-Dwamena, C.; Driedonks, N.; Lewis, D.; Shumskaya, M.; Chen, X.; Wurtzel, E.T.; Espley, R.V.; Allan, A.C. The Phytoene synthase gene family of apple (Malus x domestica) and its role in controlling fruit carotenoid content. BMC Plant Biol. 2015, 28, 185. [Google Scholar] [CrossRef] [Green Version]
- Dang, Q.; Sha, H.; Nie, J.; Wang, Y.; Yuan, Y.; Jia, D. An apple (Malus domestica) AP2/ERF transcription factor modulates carotenoid accumulation. Hortic. Res. 2021, 8, 223. [Google Scholar] [CrossRef]
- Hu, X.; Li, S.; Lin, X.; Fang, H.; Shi, Y.; Grierson, D.; Chen, K. Transcription Factor CitERF16 Is Involved in Citrus Fruit Sucrose Accumulation by Activating CitSWEET11d. Front. Plant Sci. 2021, 12, 809619. [Google Scholar] [CrossRef]
- Shen, S.L.; Yin, X.R.; Zhang, B.; Xie, X.L.; Jiang, Q.; Grierson, D.; Chen, K.S. CitAP2.10 activation of the terpene synthase CsTPS1 is associated with the synthesis of (+)-valencene in ‘Newhall’ orange. J. Exp. Bot. 2016, 67, 4105–4115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Xu, Y.; Shen, S.; Yin, X.; Klee, H.; Zhang, B.; Chen, K.; Hancock, R. Transcription factor CitERF71 activates the terpene synthase gene CitTPS16 involved in the synthesis of E-geraniol in sweet orange fruit. J. Exp. Bot. 2017, 68, 4929–4938. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Yin, X.; Xiao, Y.; Zhang, Z.; Li, S.; Liu, X.; Zhang, B.; Yang, X.; Grierson, D.; Jiang, G.; et al. An ETHYLENE RESPONSE FACTOR-MYB transcription complex regulates furaneol biosynthesis by activating QUINONE OXIDOREDUCTASE expression in strawberry. Plant Physiol. 2018, 178, 189–201. [Google Scholar] [CrossRef] [Green Version]
- Zhou, M.; Guo, S.; Tian, S.; Zhang, J.; Ren, Y.; Gong, G.; Li, C.; Zhang, H.; Xu, Y. Overexpression of the watermelon ethylene response factor ClERF069 in transgenic tomato resulted in delayed fruit ripening. Hortic. Plant J. 2020, 6, 247–256. [Google Scholar] [CrossRef]
- Li, H.; Chen, Y.; Zhang, Z.Q.; Li, B.Q.; Qin, G.Z.; Tian, S.P. Pathogenic mechanisms and control strategies of Botrytis cinerea causing post-harvest decay in fruits and vegetables. Food Qual. Saf. 2018, 2, 111–119. [Google Scholar]
- Tian, S.; Qin, G.; Li, B. Reactive oxygen species involved in regulating fruit senescence and fungal pathogenicity. Plant Mol. Biol. 2013, 82, 593–602. [Google Scholar] [CrossRef]
- Cantu, D.; Vicente, A.R.; Greve, L.C.; Dewey, F.M.; Bennett, A.B.; Labavith, J.M.; Powell, A.L.T. The intersection between cell wall disassembly, ripening, and fruit susceptibility to Botrytis cinerea. Proc. Natl. Acad. Sci. USA 2008, 105, 859–864. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Li, B.; Su, G.; Zhang, M.; Grierson, D.; Chen, K. Transcriptional regulation of fleshy fruit texture. J. Integr. Plant Biol. 2022. accepted. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Huang, W.; Xiong, F.; Xian, Z.; Su, D.; Ren, M.; Li, Z. Silencing of SlPL, which encodes a pectate lyase in tomato, confers enhanced fruit firmness, prolonged shelf-life and reduced susceptibility to grey mould. Plant Biotechnol. J. 2017, 15, 1544–1555. [Google Scholar] [CrossRef] [Green Version]
- Silva, C.J.; van den Abeele, C.; Ortega-Salazar, I.; Papin, V.; Adaskaveg, J.A.; Wang, D.; Casteel, C.L.; Seymour, G.B.; Blanco-Ulate, B. Host susceptibility factors render ripe tomato fruit vulnerable to fungal disease despite active immune responses. J. Exp. Bot. 2021, 72, 2696–2709. [Google Scholar] [CrossRef] [PubMed]
- Campos, M.D.; Félix, M.D.R.; Patanita, M.; Materatski, P.; Albuquerque, A.; Ribeiro, J.A.; Varanda, C. Defense Strategies: The Role of transcription factors in tomato-pathogen interaction. Biology 2022, 11, 235. [Google Scholar] [CrossRef]
- Van Loon, L.C.; Rep, M.; Pieterse, C.M.J. Significance of inducible defense- related proteins in infected plants. Annu. Rev. Phytopathol. 2006, 44, 135–162. [Google Scholar] [CrossRef] [Green Version]
- Bari, R.; Jones, J.D. Role of plant hormones in plant defence responses. Plant Mol. Biol. 2009, 69, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Heller, J.; Tudzynski, P. Reactive oxygen species in phytopathogenic fungi: Signaling development, and disease. Annu. Rev. Phytopathol. 2011, 49, 369–390. [Google Scholar] [CrossRef]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D.G. Hormone crosstalk in plant disease and defense: More than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef]
- Li, J.; Brader, G.; Kariola, T.; Palva, E.T. WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 2006, 46, 477–491. [Google Scholar] [CrossRef]
- Mao, P.; Duan, M.R.; Wei, C.H.; Li, Y. WRKY62 transcription factor acts downstream of cytosolic NPR1 and negatively regulates jasmonate-responsive gene expression. Plant Cell Physiol. 2007, 48, 833–842. [Google Scholar] [CrossRef] [Green Version]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef]
- Amorim, L.L.B.; da Fonseca Dos Santos, R.; Neto, J.P.B.; Guida-Santos, M.; Crovella, S.; Benko-Iseppon, A.M. Transcription Factors Involved in Plant Resistance to Pathogens. Curr. Protein Pept. Sci. 2017, 18, 335–351. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, B.L.; Sun, S.; Xing, G.M.; Wang, F.; Li, M.Y.; Tian, Y.S.; Xiong, A.S. AP2/ERF transcription factors involved in response to tomato yellow leaf curly virus in tomato. Plant Genome 2016, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Camehl, I.; Oelmüller, R. Do ethylene response factors9 and -14 repress PR gene expression in the interaction between Piriformospora indica and Arabidopsis? Plant Signal Behav. 2010, 5, 932–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soltis, N.E.; Atwell, S.; Shi, G.; Fordyce, R.; Gwinner, R.; Gao, D.; Shafi, A.; Kliebenstein, D.J. Interactions of tomato and Botrytis cinerea genetic diversity: Parsing the contributions of host differentiation, domestication, and pathogen variation. Plant Cell 2009, 31, 502–519. [Google Scholar] [CrossRef] [PubMed]
- Blanco-Ulate, B.; Vincenti, E.; Powell, A.L.; Cantu, D. Tomato transcriptome and mutant analyses suggest a role for plant stress hormones in the interaction between fruit and Botrytis cinerea. Front. Plant Sci. 2013, 14, 142. [Google Scholar] [CrossRef] [Green Version]
- Gu, Y.Q.; Wildermuth, M.C.; Chakravarthy, S.; Loh, Y.T.; Yang, C.; He, X.; Han, Y.; Martin, G.B. Tomato transcription factors pti4, pti5, and pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 2002, 14, 817–831. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, K.; Tian, L.; Hollingworth, J.; Brown, D.C.; Miki, B. Functional analysis of tomato Pti4 in Arabidopsis. Plant Physiol. 2002, 128, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Li, C.W.; Su, R.C.; Cheng, C.P.; Sanjaya; You, S.J.; Hsieh, T.H.; Chao, T.C.; Chan, M.T. Tomato RAV transcription factor is a pivotal modulator involved in the AP2/EREBP-mediated defense pathway. Plant Physiol. 2011, 156, 213–227. [Google Scholar] [CrossRef] [Green Version]
- Shi, X.L.; Gupta, S.; Rashotte, A.M. Characterization of two tomato AP2/ERF genes, SlCRF1 and SlCRF2 in hormone and stress responses. Plant Cell Rep. 2014, 33, 35–45. [Google Scholar] [CrossRef] [PubMed]
- Chakravarthy, S.; Tuori, R.P.; D’Ascenzo, M.D.; Fobert, P.R.; Despres, C.; Martin, G.B. The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell 2003, 15, 3033–3050. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhang, D.; Chen, J.; Yang, Y.; Huang, Z.; Huang, D.; Wang, X.C.; Huang, R. Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum. Plant Mol. Biol. 2004, 55, 825–834. [Google Scholar] [CrossRef]
- Liu, A.C.; Cheng, C.P. Pathogen-induced ERF68 regulates hypersensitive cell death in tomato. Mol. Plant Pathol. 2017, 18, 1062–1074. [Google Scholar] [CrossRef]
- Álvarez-Gómez, T.B.; Ramírez-Trujillo, J.A.; Ramírez-Yáñez, M.; Suárez-Rodríguez, R. Overexpression of SlERF3b and SlERF5 in transgenic tomato alters fruit size, number of seeds and promotes early flowering, tolerance to abiotic stress and resistance to Botrytis cinerea infection. Ann. Appl. Biol. 2021, 179, 382–394. [Google Scholar] [CrossRef]
- Czekus, Z.; Poor, P.; Tari, I.; Ordog, A. Effects of light and daytime on the regulation of chitosan-induced stomatal responses and defence in tomato plants. Plants 2020, 9, 59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Shen, F.; Wang, H.; Zhao, T.; Zhang, H.; Jiang, J.; Xu, X.; Li, J. Functional analysis of the SlERF01 gene in disease resistance to S. lycopersici. BMC Plant Biol. 2020, 20, 376. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, W.; Chen, J.; Yang, Y.; Zhang, Z.; Zhang, H.; Wang, X.C.; Huang, R. Transcriptional activator TSRF1 reversely regulates pathogen resistance and osmotic stress tolerance in tobacco. Plant Mol. Biol. 2007, 63, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Sun, Y.; Wang, H.; Zhao, T.; Xu, X.; Jiang, J.; Li, J. Genome-wide identification and functional analysis of the ERF2 gene family in response to disease resistance against Stemphylium lycopersici in tomato. BMC Plant Biol. 2021, 21, 72. [Google Scholar] [CrossRef]
- Li, Z.; Tian, Y.; Xu, J.; Fu, X.Y.; Gao, J.J.; Wang, B.; Han, H.J.; Wang, L.J.; Peng, R.H.; Yao, Q.H. A tomato ERF transcription factor, SlERF84, confers enhanced tolerance to drought and salt stress but negatively regulates immunity against Pseudomonas syringae pv. tomato DC3000. Plant Physiol. Biochem. 2018, 132, 683–695. [Google Scholar] [CrossRef]
- Pan, X.Q.; Fu, D.Q.; Zhu, B.Z.; Lu, C.W.; Luo, Y.B. Overexpression of the ethylene response factor SlERF1 gene enhances resistance of tomato fruit to Rhizopus nigricans. Postharvest Biol. Technol. 2013, 75, 28–36. [Google Scholar] [CrossRef]
- Giovannoni, J.J. Genetic regulation of fruit development and ripening. Plant Cell 2004, 16, S170–S180. [Google Scholar] [CrossRef] [Green Version]
- Vrebalov, J.; Ruezinsky, D.; Padmanabhan, V.; White, R.; Medrano, D.; Drake, R.; Schuch, W.; Giovannoni, J. A MADS-box gene necessary for fruit ripening at the tomato ripening-inhibitor (rin) locus. Science 2002, 296, 343–346. [Google Scholar] [CrossRef]
- Klee, H.J.; Giovannoni, J.J. Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 2011, 45, 41–59. [Google Scholar] [CrossRef]
- Manning, K.; Tor, M.; Poole, M.; Hong, Y.; Thompson, A.J.; King, G.J. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006, 38, 948–952. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, J.Q.; Lanahan, M.B.; Yen, H.S.; Giovannoni, J.J.; Klee, H.J. An ethylene-inducible component of signal transduction encoded by Never-ripe. Science 1995, 270, 1807–1809. [Google Scholar] [CrossRef] [PubMed]
- Zhong, S.L.; Fei, Z.J.; Chen, Y.R.; Zheng, Y.; Huang, M.Y.; Vrebalov, J.; McQuinn, R.; Gapper, N.; Liu, B.; Xiang, J.; et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 2013, 31, 154–159. [Google Scholar] [CrossRef]
- Ito, Y.; Nishizawa-Yokoi, A.; Endo, M.; Mikami, M.; Shima, Y.; Nakamura, N.; Kotake-Nara, E.; Kawasaki, S.; Toki, S. Re-evaluation of the rin mutation and the role of RIN in the induction of tomato ripening. Nat. Plants 2017, 3, 866–874. [Google Scholar] [CrossRef]
- Li, S.; Xu, H.; Ju, Z.; Cao, D.; Zhu, H.; Fu, D.; Grierson, D.; Qin, G.; Luo, Y.; Zhu, B. The RIN-MC fusion of MADS-Box transcription factors has transcriptional activity and modulates expression of many ripening genes. Plant Physiol. 2018, 176, 891–909. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Zhu, N.; Zhu, X.; Wu, M.; Jiang, C.; Grierson, D.; Luo, Y.; Shen, W.; Zhong, S.; Fu, D.; et al. Diversity and redundancy of the ripening regulatory networks revealed by the fruitENCODE and the new CRISPR/Cas9 CNR and NOR mutants. Hortic. Res. 2019, 6, 39. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Wei, W.; Fan, Z.; Zhao, X.; Zhang, Y.; Jing, Y.; Zhu, B.; Zhu, H.; Shan, W.; Chen, J.; et al. Re-evaluation of the nor mutation and the role of the NAC-NOR transcription factor in tomato fruit ripening. J. Exp. Bot. 2020, 71, 3560–3574. [Google Scholar] [CrossRef]
- Wang, R.; Tavano, E.C.D.R.; Lammers, M.; Martinelli, A.P.; Angenent, G.C.; de Maagd, R.A. Re-evaluation of transcription factor function in tomato fruit development and ripening with CRISPR/Cas9-mutagenesis. Sci. Rep. 2019, 9, 1696. [Google Scholar] [CrossRef]
- Zheng, H.; Jin, R.; Liu, Z.M.; Sun, C.; Shi, Y.N.; Grierson, D.; Zhu, C.Q.; Li, S.; Ferguson, I.; Chen, K.S. Role of the tomato fruit ripening regulator MADS-RIN in resistance to Botrytis cinerea infection. Food Qual. Saf. 2021, 5, fyab028. [Google Scholar] [CrossRef]
- Petrasch, S.; Silva, C.J.; Mesquida-Pesci, S.D.; Gallegos, K.; van den Abeele, C.; Papinm, V.; Fernandez-Acerom, F.J.; Knapp, S.J.; BlancoUlate, B. Infection strategies deployed by Botrytis cinerea, Fusarium acuminatum, and Rhizopus stolonifer as a function of tomato fruit ripening stage. Front. Plant Sci. 2019, 10, 223. [Google Scholar] [CrossRef] [Green Version]
- Bautista-Baños, S. (Ed.) Postharvest Decay: Control Strategies; Academic Press: London, UK; Waltham, MA, USA; San Diego, CA, USA, 2014; pp. 1–37. [Google Scholar]
- Coppola, M.; Diretto, G.; Digilio, M.C.; Woo, S.L.; Giuliano, G.; Molisso, D.; Pennacchio, F.; Lorito, M.; Rao, R. Transcriptome and metabolome reprogramming in tomato plants by Trichoderma harzianum strain T22 primes and enhances defense responses against Aphids. Front. Physiol. 2019, 21, 745. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Xu, Y.P.; Zhang, Z.X.; Cao, W.Y.; Li, F.; Zhou, X.P.; Chen, G.Y.; Cai, X.Z. Identification of genes required for nonhost resistance to Xanthomonas oryzae pv. oryzae reveals novel signaling components. PLoS ONE 2012, 7, e42796. [Google Scholar]
- Luan, Y.; Cui, J.; Li, J.; Jiang, N.; Liu, P.; Meng, J. Effective enhancement of resistance to Phytophthora infestans by overexpression of miR172a and b in Solanum lycopersicum. Planta 2018, 247, 127–138. [Google Scholar] [CrossRef]
- Huang, P.Y.; Catinot, J.; Zimmerli, L. Ethylene response factors in Arabidopsis immunity. J. Exp. Bot. 2016, 67, 1231–1241. [Google Scholar] [CrossRef] [Green Version]
- Haile, Z.M.; Nagpala-De Guzman, E.G.; Moretto, M.; Sonego, P.; Engelen, K.; Zoli, L.; Moser, C.; Baraldi, E. Transcriptome profiles of strawberry (Fragaria vesca) fruit interacting with Botrytis cinerea at different ripening stages. Front. Plant Sci. 2019, 18, 1131. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Lee, J.G.; Min, K.; Choi, J.H.; Lim, S.; Lee, E.J. Transcriptome analysis of the fruit of Two strawberry cultivars “Sunnyberry” and “Kingsberry” that show different susceptibility to Botrytis cinerea after harvest. Int. J. Mol. Sci. 2021, 22, 1518. [Google Scholar] [CrossRef]
- Akagi, A.; Dandekar, A.M.; Stotz, H.U. Resistance of Malus domestica fruit to Botrytis cinerea depends on endogenous ethylene biosynthesis. Phytopathology 2011, 101, 1311–1321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.H.; Gu, K.D.; Han, P.L.; Yu, J.Q.; Wang, C.K.; Zhang, Q.Y.; You, C.X.; Hu, D.G.; Hao, Y.J. Apple ethylene response factor MdERF11 confers resistance to fungal pathogen Botryosphaeria dothidea. Plant Sci. 2020, 291, 110351. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, L.; Ma, H.; Zhang, Y.; Zhang, X.; Ji, M.; van Nocker, S.; Ahmad, B.; Zhao, Z.; Wang, X.; et al. Overexpression of the apple (Malus× domestica) MdERF100 in Arabidopsis increases resistance to Powdery Mildew. Int. J. Mol. Sci. 2021, 22, 5713. [Google Scholar] [CrossRef]
- Dong, T.; Zheng, T.; Fu, W.; Guan, L.; Jia, H.; Fang, J. The Effect of Ethylene on the Color Change and Resistance to Botrytis cinerea Infection in ‘Kyoho’ Grape Fruits. Foods 2020, 9, 892. [Google Scholar] [CrossRef]
- Wang, L.; Liu, W.; Wang, Y. Heterologous expression of Chinese wild grapevine VqERFs in Arabidopsis thaliana enhance resistance to Pseudomonas syringae pv. tomato DC3000 and to Botrytis cinerea. Plant Sci. 2020, 293, 110421. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Li, Y.; Zhang, S.; Zhang, X.; Yao, J.; Luo, Q.; Sun, F.; Wang, X. Genome-wide identification and expression analysis reveal the potential function of ethylene responsive factor gene family in response to Botrytis cinerea infection and ovule development in grapes (Vitis vinifera L.). Plant Biol. 2019, 21, 571–584. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Shi, J.; Xu, W.; Li, H.; He, M.; Xu, Y.; Xu, T.; Yang, Y.; Cao, J.; Wang, Y. Three ERF transcription factors from Chinese wild grapevine Vitis pseudoreticulata participate in different biotic and abiotic stress-responsive pathways. J. Plant Physiol. 2013, 170, 923–933. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Wang, A.; Zhu, S.; Zhang, L. Expression of ACO1, ERS1 and ERF1 genes in harvested bananas in relation to heat-induced defense against Colletotrichum musae. J. Plant Physiol. 2011, 168, 1634–1640. [Google Scholar] [CrossRef]
- Li, X.; Zhu, X.; Mao, J.; Zou, Y.; Fu, D.; Chen, W.; Lu, W. Isolation and characterization of ethylene response factor family genes during development, ethylene regulation and stress treatments in papaya fruit. Plant Physiol. Biochem. 2013, 70, 81–92. [Google Scholar] [CrossRef]
- He, Y.; Jia, R.; Qi, J.; Chen, S.; Lei, T.; Xu, L.; Peng, A.; Yao, L.; Long, Q.; Li, Z.; et al. Functional analysis of citrus AP2 transcription factors identified CsAP2-09 involved in citrus canker disease response and tolerance. Gene 2019, 707, 178–188. [Google Scholar] [CrossRef]
- Shinde, B.A.; Dholakia, B.B.; Hussain, K.; Aharoni, A.; Giri, A.P.; Kamble, A.C. WRKY1 acts as a key component improving resistance against Alternaria solani in wild tomato, Solanum arcanum Peralta. Plant Biotechnol. J. 2018, 16, 1502–1513. [Google Scholar] [CrossRef] [Green Version]


| ERF | Other Name | Pathogen | Reported Function | Reference |
|---|---|---|---|---|
| ERF.A1 | ERF68 | Pseudomonas syringae pv. Tomato (Pst) DC3000; Xanthomonas euvesicatoria (Xeu); B. cinerea | Activation of hypersensitive cell death and disease defense involving modulation of ethylene, SA, jasmonic acid (JA) and hypersensitive response (HR) pathways ERF.A1 silencing resulted in increased susceptibility to B. cinerea, attenuated the B. cinerea-induced expression of JA/ethylene-mediated signaling responsive defense genes and promoted the B. cinerea-induced H2O2 accumulation | [36,93] |
| ERF.A2 | ERF1 | B. cinerea | Expression of ERF1 was upregulated in fruit after B. cinerea infection at MG and RR stage | [86] |
| ERF.A3 | Pti4 | Pst DC3000; B. cinerea; | ERF.A3 silencing decreased the resistance against Pst DC3000, but increased susceptibility to B. cinerea; Similar to ERF.A1, ERF.A3 silencing affected expression of genes involved in JA/ethylene-mediated signaling responsive defense genes and B. cinerea-induced H2O2 accumulation | [36] |
| ERF.B1 | Tomato yellow leaf curly virus (TYLCV) | Transcript of genes were affected in response to TYLCY infection in different cultivated tomato either resistant or susceptible to the virus | [83] | |
| ERF.B2 | SlERF5 | TYLCV; B. cinerea | Similar to ERF.B1, the transcript of ERF.B2 was affected in response to TYLCY infection; SlERF5 overexpression transgenic tomato plants enhances the resistance to B. cinerea | [83,94] |
| ERF.B3 | LeERF4 | TYLCV | Similar to ERF.B1 and B2, the transcript was affected in response to TYLCY infection | [83] |
| ERF.B4 | B. cinerea | ERF.B4 silencing increased the susceptibility to B. cinerea, which affected expression of genes involved in JA/ethylene-mediated signaling responsive defense genes and B. cinerea-induced H2O2 accumulation | [36] | |
| ERF.C1 | TERF1; JERF2; SlERF1 | ERF1 was involved in chitosan (CHT)-induced systemic acquired resistance (SAR) response | [95] | |
| ERF.C3 | B. cinerea | ERF.C3 silencing increased the susceptibility to B. cinerea, which affected expression of genes involved in JA/ethylene-mediated signaling responsive defense genes and B. cinerea-induced H2O2 accumulation | [36] | |
| ERF.C4 | SlERF01; TSRF1 | Stemphylium lycopersici; Ralstonia solanacearum; Pst DC3000; B. cinerea; TYLCV | SlERF01 activates expression of PR1 and plays a key role in SA, JA and ROS signaling pathways, promoting resistance to S. lycopersici invasion; A transcriptional activator TSRF1, which was previously demonstrated to regulate plant resistance to R. solanacearum, reversely regulates pathogen resistance including Pst DC3000 and B. cinerea; Similar to ERF.B1- B3, the transcript was affected in response to TYLCY infection | [83,92,96,97] |
| ERF.C6 | ERF2 or Pti5 | Stemphylium lycopersici; TYLCV | ERF2 either directly or indirectly regulates Pto, PR1b1 and PR-P2 expression and enhances tomato resistance to S. lycopersici, which has a key role in multiple SA, JA and ROS signaling pathways that contribute to resistance Similar to ERF.B1–B3 and C4, the transcript was affected in response to TYLCY infection | [83,98] |
| ERF.D6 | ERF84 | Pst DC3000 | Overexpression of SlERF84 resulted in decreased plant resistance against Pst DC3000, which might due to downregulated expression of PR genes. | [99] |
| ERF.E1 | SlERF2 | B. cinerea | Tomato ERF2 (ERF.E1) participates in MeJA-mediated disease by promoting genes that encode defense enzymes including pathogenesis-related proteins | [35] |
| ERF.F5 | ERF3, SlERF3b | B. cinerea | Expression of ERF3 was upregulated in fruit after B. cinerea infection at mature green and redripe stage; SlERF3b overexpression transgenic tomato plants enhances the resistance to B. cinerea | [86,94] |
| ERF.H1 | SlERF1 | Rhizopus nigricans | Overexpression of ERF1 in tomato fruit enhanced resistance against R. nigricans, including accumulation of transcripts of PR1a, PR5, Chi1 and PAL genes | [100] |
| Species | ERF | Pathogen | Reported Function | Reference |
|---|---|---|---|---|
| Strawberry | WRI1, ERF061, ERF053 | B. cinerea | Upregulated gene expression in B. cinerea inoculated fruit | [119] |
| Strawberry | ERF2-like, ERF5 | B. cinerea | Upregulated gene expression after B. cinerea infection in fruit at Mature-red stage | [120] |
| Apple | MdERF3, -4, -5, -6 | B. cinerea | Expression of all four MdERF mRNAs is ethylene dependent and also induced by wounding or by B. cinerea infection | [121] |
| Apple | MdERF11 | Botryosphaeria dothidea (B. dothidea) | MdERF11 overexpression increases the resistance to B. dothidea infection, which act through SA synthesis pathway. | [122] |
| Apple | MdERF100 | Powdery Mildew | MdERF100 physically interacts with MdbHLH92 which mediates the powdery mildew resistance by regulating the JA and SA signaling pathways | [123] |
| Grape | VvERF1 | B. cinerea | Overexpression of VvERF1 in strawberry fruits reduced the susceptibility to B. cinerea infection | [124] |
| Grape | VqERF112, VqERF114, VqERF072 | Pst DC3000, B. cinerea | VqERF112, VqERF114 and VqERF072 in Chinese wild Vitis quinquangularis positively regulate resistance to Pst DC3000 and B. cinerea | [125] |
| Grape | VpERF1, VpERF2, VpERF3 | Ralstonia solanacearum; Phytophtora parasitica | VpERF1-3 from a highly powdery mildew (PM)-resistant Chinese wild Vitis pseudoreticulata, were positively related to resistance to both bacterial pathogen Ralstonia solanacearum and fungal pathogen Phytophtora parasitica var. nicotianae Tucker. | [126] |
| Citrus | CsAP2-09 | Xanthomonas citri subsp. (Xcc) | CsAP2-09 overexpression enhanced the resistance to Xcc, while its silence decreased the resistance | [127] |
| Banana | MaERF1 | Colletotrichum musae | Heat-induced disease resistance in harvested bananas involves up-regulation of MaERF1 expression | [128] |
| Papaya | CpERF2, CpERF4 | Colletotrichum gloeosporioides | Pathogen stress induces strong accumulation of CpERF2 and CpERF4 transcripts. Expression of CpERF2, CpERF4 increases more gradually, reaching maximal levels 14 days after inoculation, with ~5- and ~20-fold increases, respectively | [129] |
| Mango | Comp11955, comp12486, comp12577 etc. | Colletotrichum gloeosporioides | Expression levels of 13 ERF unigenes (1.5- to 85-fold) were up-regulated in infected fruits. | [28] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, S.; Wu, P.; Yu, X.; Cao, J.; Chen, X.; Gao, L.; Chen, K.; Grierson, D. Contrasting Roles of Ethylene Response Factors in Pathogen Response and Ripening in Fleshy Fruit. Cells 2022, 11, 2484. https://doi.org/10.3390/cells11162484
Li S, Wu P, Yu X, Cao J, Chen X, Gao L, Chen K, Grierson D. Contrasting Roles of Ethylene Response Factors in Pathogen Response and Ripening in Fleshy Fruit. Cells. 2022; 11(16):2484. https://doi.org/10.3390/cells11162484
Chicago/Turabian StyleLi, Shan, Pan Wu, Xiaofen Yu, Jinping Cao, Xia Chen, Lei Gao, Kunsong Chen, and Donald Grierson. 2022. "Contrasting Roles of Ethylene Response Factors in Pathogen Response and Ripening in Fleshy Fruit" Cells 11, no. 16: 2484. https://doi.org/10.3390/cells11162484
APA StyleLi, S., Wu, P., Yu, X., Cao, J., Chen, X., Gao, L., Chen, K., & Grierson, D. (2022). Contrasting Roles of Ethylene Response Factors in Pathogen Response and Ripening in Fleshy Fruit. Cells, 11(16), 2484. https://doi.org/10.3390/cells11162484







