Methyltransferase Setdb1 Promotes Osteoblast Proliferation by Epigenetically Silencing Macrod2 with the Assistance of Atf7ip
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Cell Transfection
2.3. HLU Model
2.4. Targeted Silencing of Setdb1 Model
2.5. 2D Clinorotation
2.6. Micro-CT Analysis
2.7. Three-Point Bending Test
2.8. qRT–PCR
2.9. Western Blotting
2.10. Chromatin Immunoprecipitation (ChIP) Assay
2.11. Immunoprecipitation Assay
2.12. Histological Analysis
2.13. Immunofluorescence
2.14. CCK-8
2.15. EdU Labeling
2.16. Luciferase Assay
2.17. Statistical Analysis
3. Results
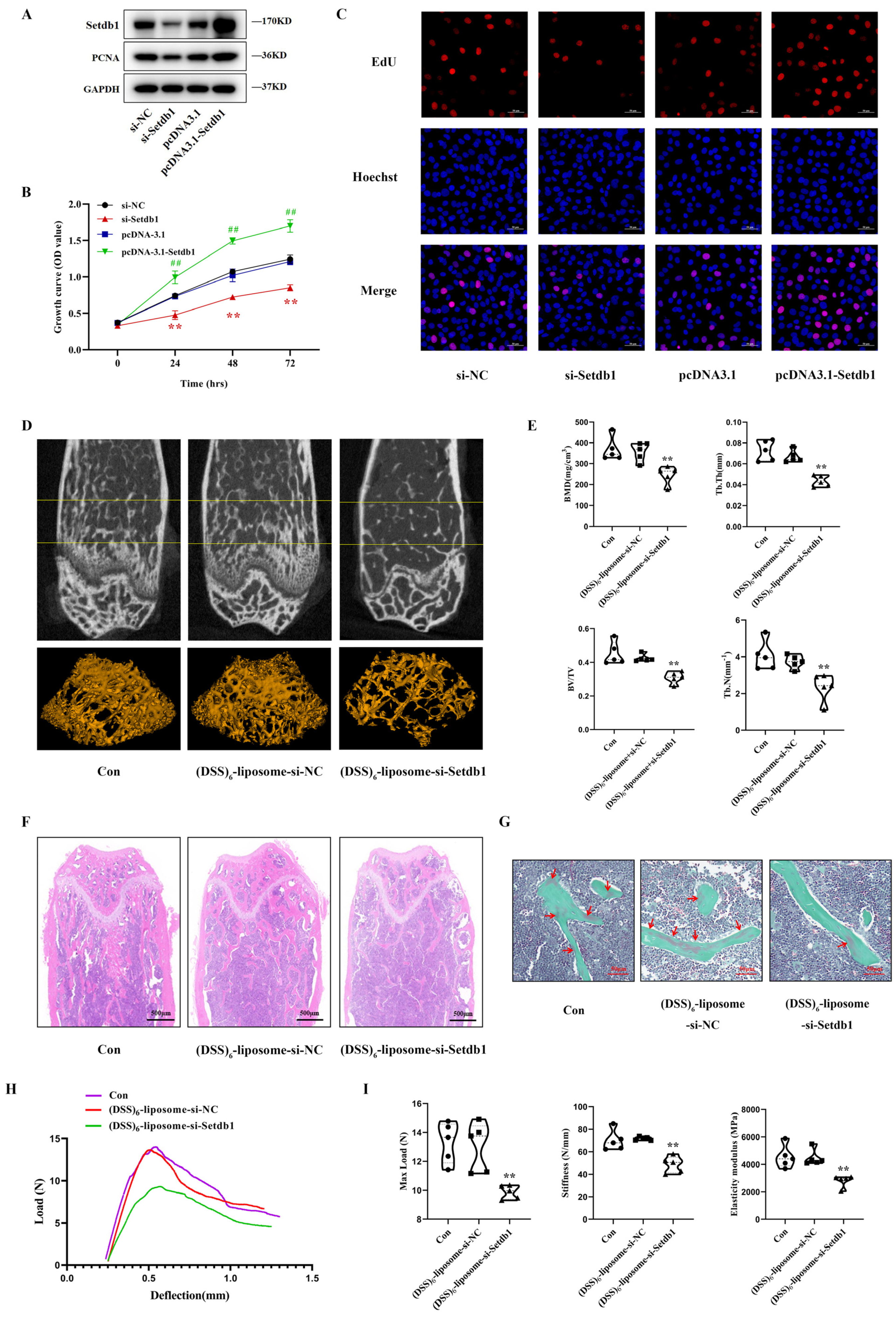
3.1. Setdb1 Is Downregulated In Vivo and In Vitro under Mechanical Unloading
3.2. Setdb1 Regulates Osteoblast Proliferation and Bone Formation
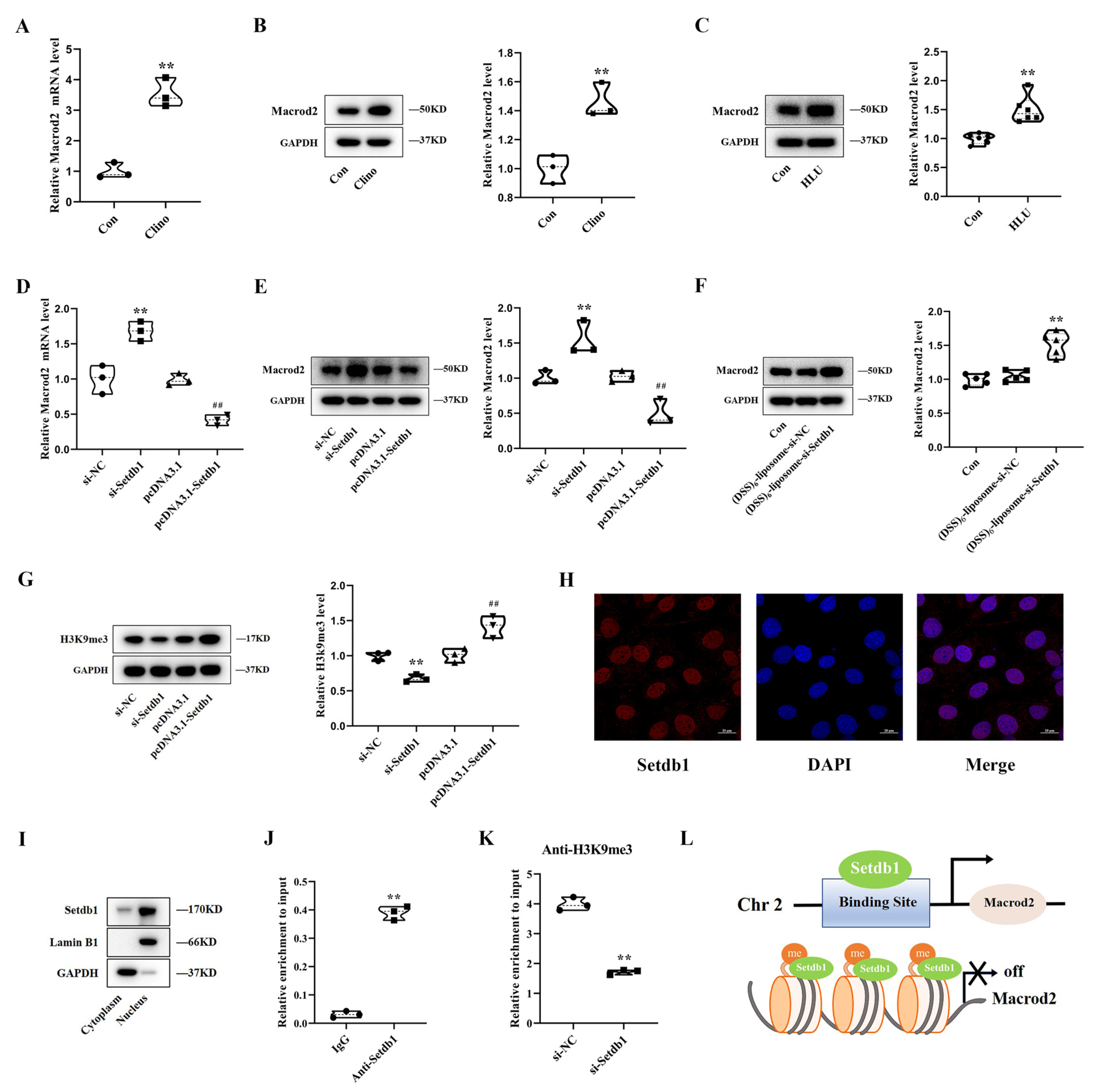
3.3. Setdb1 Silences Macrod2 Expression by Catalyzing the Formation of H3K9me3 in the Promoter Region
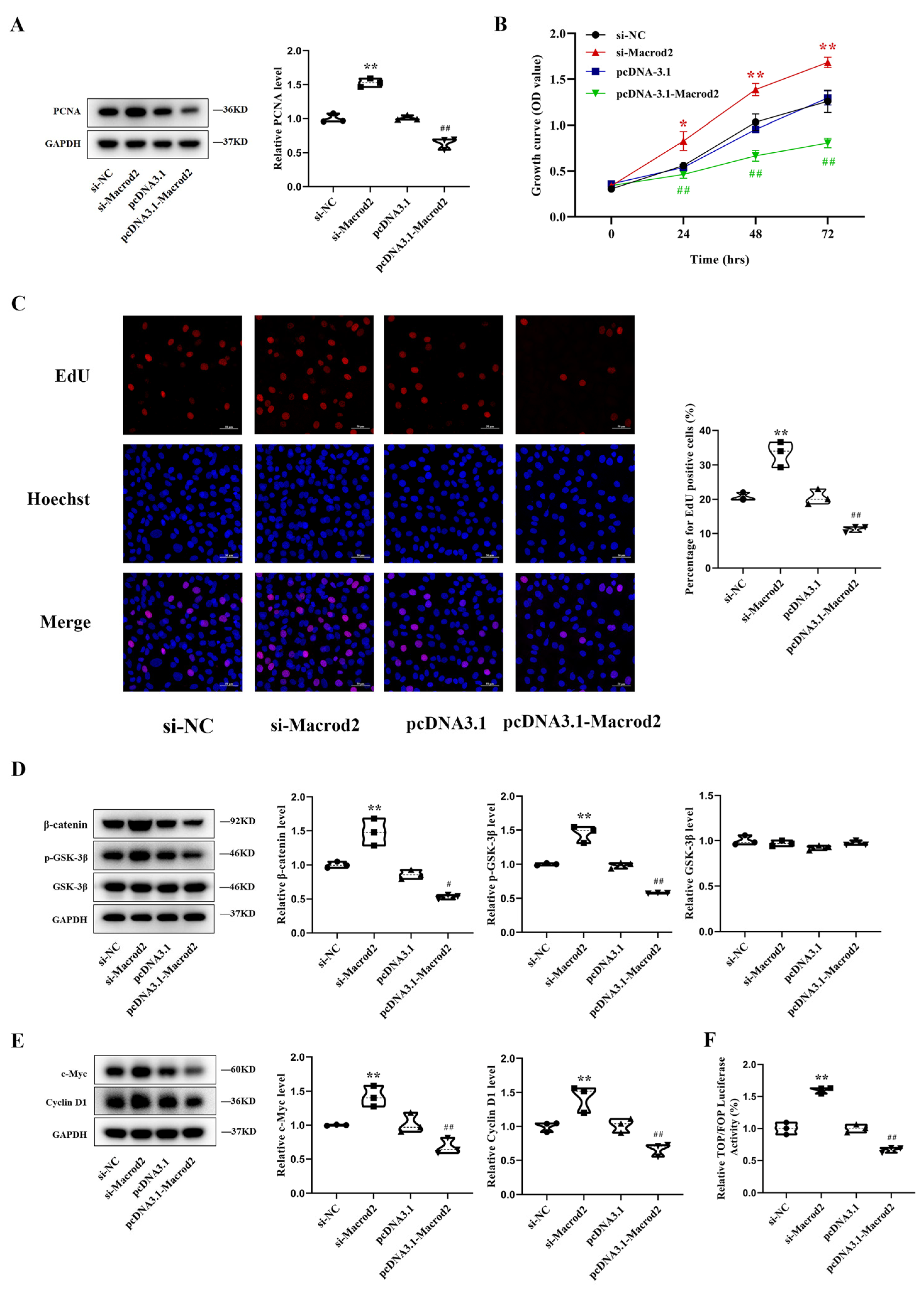
3.4. Macrod2 Inhibits Osteoblast Proliferation in MC3T3-E1 Cells
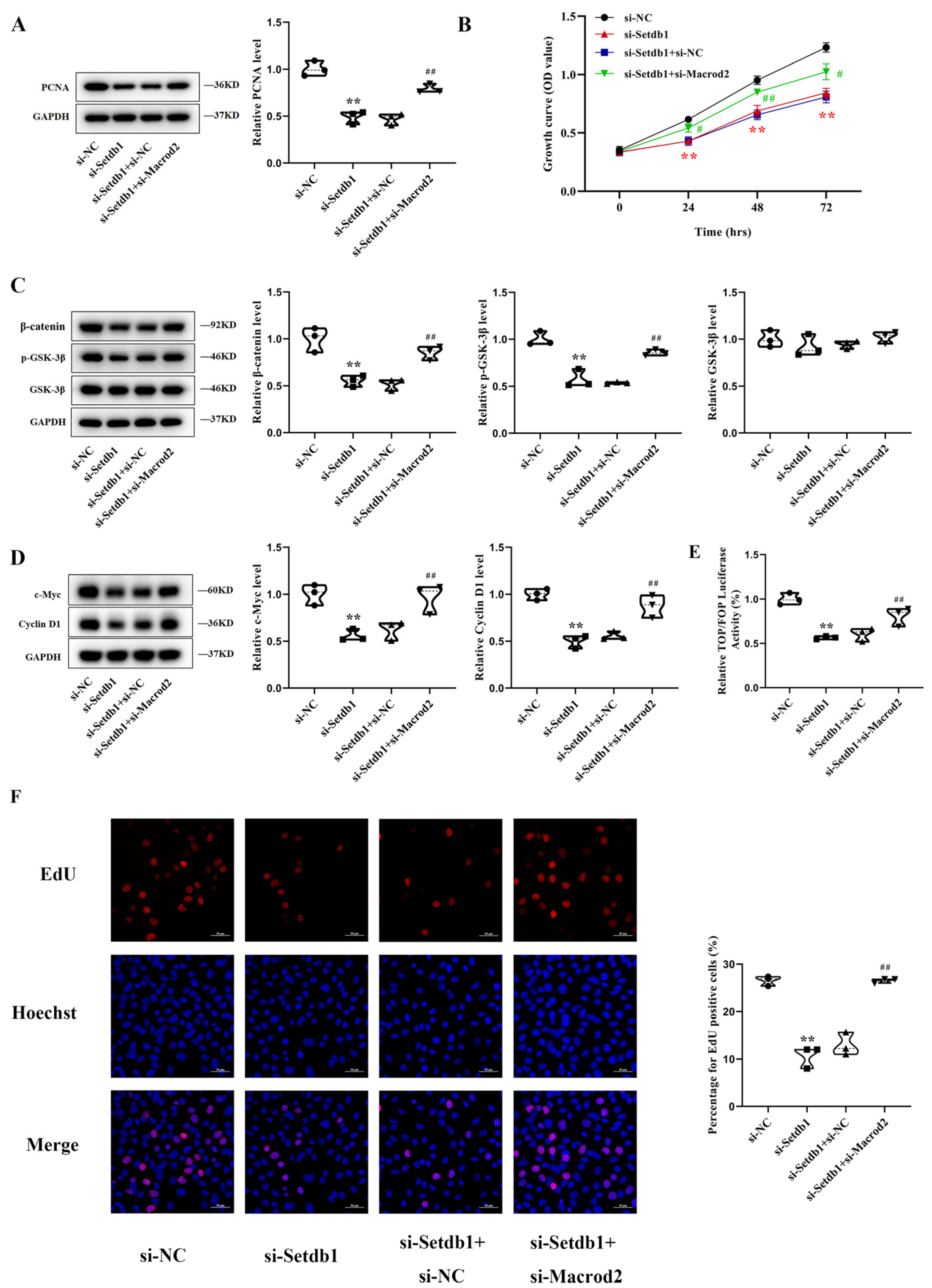
3.5. Knockdown of Macrod2 Attenuates the Inhibitory Effect of Setdb1 Knockdown on Osteoblast Proliferation in MC3T3-E1 Cells
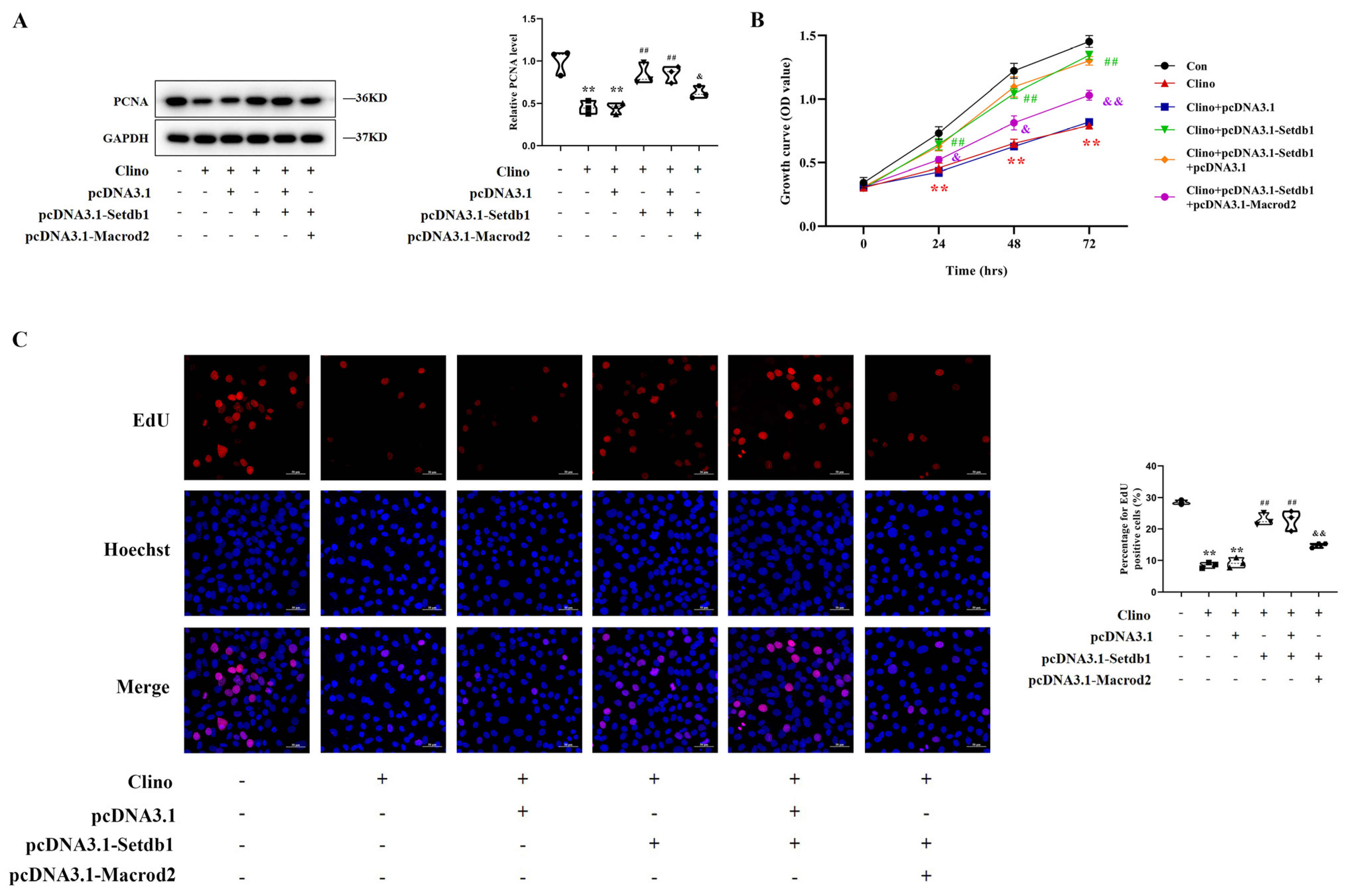
3.6. Setdb1 Blocks the Inhibition of Osteoblast Proliferation under Mechanical Unloading by Targeting Macrod2
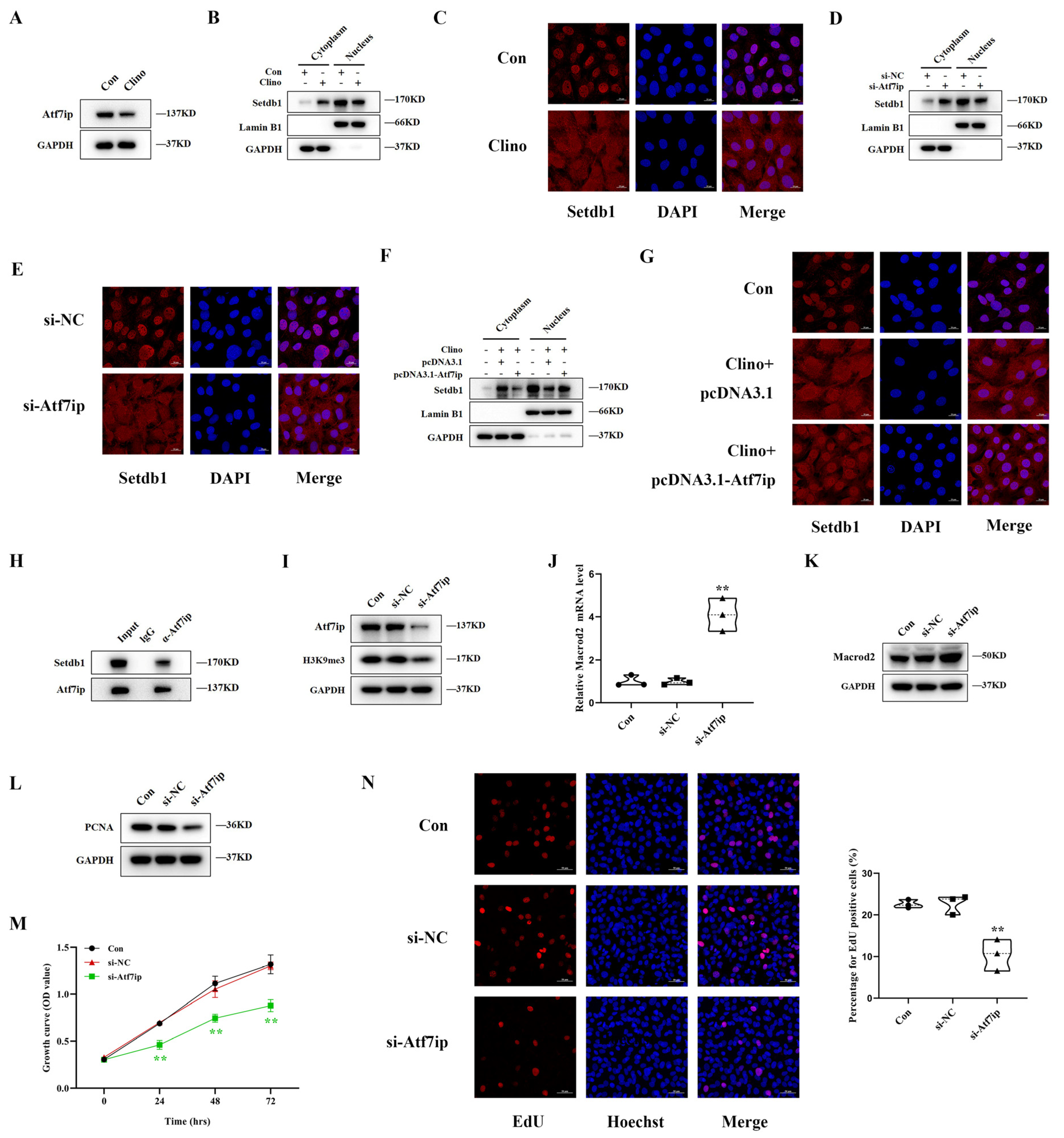
3.7. Atf7ip Regulates the Nuclear Localization of Setdb1 and Exhibits a Similar Effect on Osteoblast Proliferation under Mechanical Unloading
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rodan, G.A.; Martin, T.J. Therapeutic approaches to bone diseases. Science 2000, 289, 1508–1514. [Google Scholar] [CrossRef]
- Vico, L.; Collet, P.; Guignandon, A.; Lafage-Proust, M.; Thomas, T.; Rehailia, M.; Alexandre, C. Effects of long-term microgravity exposure on cancellous and cortical weight-bearing bones of cosmonauts. Lancet 2000, 355, 1607–1611. [Google Scholar] [CrossRef]
- Cabahug-Zuckerman, P.; Frikha-Benayed, D.; Majeska, R.J.; Tuthill, A.; Yakar, S.; Judex, S.; Schaffler, M.B. Osteocyte apoptosis caused by hindlimb unloading is required to trigger osteocyte rankl production and subsequent resorption of cortical and trabecular bone in mice femurs. J. Bone Min. Res. 2016, 31, 1356–1365. [Google Scholar] [CrossRef]
- Sun, Z.Y.; Cao, X.S.; Hu, Z.B.; Zhang, L.C.; Wang, H.; Zhou, H.; Li, D.T.; Zhang, S.; Xie, M. MiR-103 inhibits osteoblast proliferation mainly through suppressing Cav1.2 expression in simulated microgravity. Bone 2015, 76, 121–128. [Google Scholar] [CrossRef]
- Kim, K.T.; Lee, Y.S.; Han, I. The role of epigenomics in osteoporosis and osteoporotic vertebral fracture. Int. J. Mol. Sci. 2020, 21, 9455. [Google Scholar] [CrossRef] [PubMed]
- Park-Min, K.H. Epigenetic regulation of bone cells. Connect. Tissue Res. 2017, 58, 76–89. [Google Scholar] [CrossRef]
- Ideno, H.; Nakashima, K.; Komatsu, K.; Araki, R.; Abe, M.; Arai, Y.; Kimura, H.; Shinkai, Y.; Tachibana, M.; Nifuji, A. G9a is involved in the regulation of cranial bone formation through activation of Runx2 function during development. Bone 2020, 137, 115332. [Google Scholar] [CrossRef] [PubMed]
- Yi, S.J.; Jang, Y.J.; Kim, H.J.; Lee, K.; Lee, H.; Kim, Y.; Kim, J.; Hwang, S.Y.; Song, J.S.; Okada, H.; et al. The KDM4B-CCAR1-MED1 axis is a critical regulator of osteoclast differentiation and bone homeostasis. Bone Res. 2021, 9, 27. [Google Scholar] [CrossRef]
- Markouli, M.; Strepkos, D.; Chlamydas, S.; Piperi, C. Histone lysine methyltransferase SETDB1 as a novel target for central nervous system diseases. Prog. Neurobiol. 2021, 200, 101968. [Google Scholar] [CrossRef]
- Ogawa, S.; Fukuda, A.; Matsumoto, Y.; Hanyu, Y.; Sono, M.; Fukunaga, Y.; Masuda, T.; Araki, O.; Nagao, M.; Yoshikawa, T.; et al. SETDB1 Inhibits p53-Mediated apoptosis and is required for formation of pancreatic ductal adenocarcinomas in mice. Gastroenterology 2020, 159, 682–696. [Google Scholar] [CrossRef]
- Sun, Q.Y.; Ding, L.W.; Xiao, J.F.; Chien, W.; Lim, S.L.; Hattori, N.; Goodglick, L.; Chia, D.; Mah, V.; Alavi, M.; et al. SETDB1 accelerates tumourigenesis by regulating the WNT signalling pathway. J. Pathol. 2015, 235, 559–570. [Google Scholar] [CrossRef] [PubMed]
- Karanth, A.V.; Maniswami, R.R.; Prashanth, S.; Govindaraj, H.; Padmavathy, R.; Jegatheesan, S.K.; Mullangi, R.; Rajagopal, S. Emerging role of SETDB1 as a therapeutic target. Expert Opin. Ther. Targets 2017, 21, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Minkovsky, A.; Sahakyan, A.; Rankin-Gee, E.; Bonora, G.; Patel, S.; Plath, K. The Mbd1-Atf7ip-Setdb1 pathway contributes to the maintenance of X chromosome inactivation. Epigenetics Chromatin 2014, 7, 12. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.L.; Nishi, M.; Ohtsuka, T.; Matsui, T.; Takemoto, K.; Kamio-Miura, A.; Aburatani, H.; Shinkai, Y.; Kageyama, R. Essential roles of the histone methyltransferase ESET in the epigenetic control of neural progenitor cells during development. Development 2012, 139, 3806–3816. [Google Scholar] [CrossRef]
- Liu, R.; Liu, Z.; Guo, M.; Zeng, W.; Zheng, Y. SETDB1 regulates porcine spermatogonial adhesion and proliferation through modulating mmp3/10 transcription. Cells 2022, 11, 370. [Google Scholar] [CrossRef]
- Takada, I.; Kouzmenko, A.P.; Kato, S. Wnt and PPARgamma signaling in osteoblastogenesis and adipogenesis. Nat. Rev. Rheumatol. 2009, 5, 442–447. [Google Scholar] [CrossRef]
- Takada, I.; Suzawa, M.; Matsumoto, K.; Kato, S. Suppression of PPAR transactivation switches cell fate of bone marrow stem cells from adipocytes into osteoblasts. Ann. N. Y. Acad. Sci. 2007, 1116, 182–195. [Google Scholar] [CrossRef]
- Zhang, G.; Guo, B.; Wu, H.; Tang, T.; Zhang, B.T.; Zheng, L.; He, Y.; Yang, Z.; Pan, X.; Chow, H.; et al. A delivery system targeting bone formation surfaces to facilitate RNAi-based anabolic therapy. Nat. Med. 2012, 18, 307–314. [Google Scholar] [CrossRef]
- Wang, X.; Guo, B.; Li, Q.; Peng, J.; Yang, Z.; Wang, A.; Li, D.; Hou, Z.; Lv, K.; Kan, G.; et al. miR-214 targets ATF4 to inhibit bone formation. Nat. Med. 2013, 19, 93–100. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, K.; Hu, Z.; Zhou, H.; Zhang, L.; Wang, H.; Li, G.; Zhang, S.; Cao, X.; Shi, F. MicroRNA-139-3p regulates osteoblast differentiation and apoptosis by targeting ELK1 and interacting with long noncoding RNA ODSM. Cell Death Dis. 2018, 9, 1107. [Google Scholar] [CrossRef]
- Hashimoto, H.; Vertino, P.M.; Cheng, X. Molecular coupling of DNA methylation and histone methylation. Epigenomics 2010, 2, 657–669. [Google Scholar] [CrossRef]
- Karimi, M.M.; Goyal, P.; Maksakova, I.A.; Bilenky, M.; Leung, D.; XinTang, J.; Shinkai, Y.; Mager, D.L.; Jones, S.; Hirst, M.; et al. DNA methylation and SETDB1/H3K9me3 regulate predominantly distinct sets of genes, retroelements, and chimeric transcripts in mESCs. Cell Stem Cell 2011, 8, 676–687. [Google Scholar] [CrossRef]
- Everson, T.M.; Marsit, C.J.; O’Shea, T.M.; Burt, A.; Hermetz, K.; Carter, B.S.; Helderman, J.; Hofheimer, J.A.; McGowan, E.C.; Neal, C.R.; et al. Epigenome-wide Analysis Identifies Genes and Pathways Linked to Neurobehavioral Variation in Preterm Infants. Sci. Rep. 2019, 9, 6322. [Google Scholar] [CrossRef] [PubMed]
- Kebir, O.; Chaumette, B.; Krebs, M.O. Epigenetic variability in conversion to psychosis: Novel findings from an innovative longitudinal methylomic analysis. Transl. Psychiatry 2018, 8, 93. [Google Scholar] [CrossRef] [PubMed]
- Pei, Y.F.; Hu, W.Z.; Yan, M.W.; Lie, C.W.; Liu, L.; Yang, X.L.; Hai, R.; Wang, X.Y.; Shen, H.; Tian, Q.; et al. Joint study of two genome-wide association meta-analyses identified 20p12.1 and 20q13.33 for bone mineral density. Bone 2018, 110, 378–385. [Google Scholar] [CrossRef]
- Zhou, Z.J.; Luo, C.B.; Xin, H.Y.; Hu, Z.Q.; Zhu, G.Q.; Li, J.; Zhou, S.L. MACROD2 deficiency promotes hepatocellular carcinoma growth and metastasis by activating GSK-3β/β-catenin signaling. NPJ Genom. Med. 2020, 5, 15. [Google Scholar] [CrossRef]
- Rosenthal, F.; Feijs, K.L.; Frugier, E.; Bonalli, M.; Forst, A.H.; Imhof, R.; Winkler, H.C.; Fischer, D.; Caflisch, A.; Hassa, P.O.; et al. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat. Struct. Mol. Biol. 2013, 20, 502–507. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Sun, Z.; Wang, Y.; Hu, Z.; Zhou, H.; Zhang, L.; Hong, B.; Zhang, S.; Cao, X. miR-33-5p, a novel mechano-sensitive microRNA promotes osteoblast differentiation by targeting Hmga2. Sci. Rep. 2016, 6, 23170. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, K.; Zhang, L.; Tan, Y.; Hu, Z.; Dang, L.; Zhou, H.; Li, G.; Wang, H.; Zhang, S.; et al. Targeted overexpression of the long noncoding RNA ODSM can regulate osteoblast function in vitro and in vivo. Cell Death Dis. 2020, 11, 133. [Google Scholar] [CrossRef]
- Sun, Z.; Li, Y.; Wang, H.; Cai, M.; Gao, S.; Liu, J.; Tong, L.; Hu, Z.; Wang, Y.; Wang, K.; et al. miR-181c-5p mediates simulated microgravity-induced impaired osteoblast proliferation by promoting cell cycle arrested in the G2 phase. J. Cell Mol. Med. 2019, 23, 3302–3316. [Google Scholar] [CrossRef]
- Wang, K.; Wang, Y.; Hu, Z.; Zhang, L.; Li, G.; Dang, L.; Tan, Y.; Cao, X.; Shi, F.; Zhang, S.; et al. Bone-targeted lncRNA OGRU alleviates unloading-induced bone loss via miR-320-3p/Hoxa10 axis. Cell Death Dis. 2020, 11, 382. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Lawson, K.A.; Teteak, C.J.; Zou, J.; Hacquebord, J.; Patterson, D.; Ghatan, A.C.; Mei, Q.; Zielinska-Kwiatkowska, A.; Bain, S.D.; et al. ESET histone methyltransferase is essential to hypertrophic differentiation of growth plate chondrocytes and formation of epiphyseal plates. Dev. Biol. 2013, 380, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Lawson, K.A.; Teteak, C.J.; Gao, J.; Li, N.; Hacquebord, J.; Ghatan, A.; Zielinska-Kwiatkowska, A.; Song, G.; Chansky, H.A.; Yang, L. ESET histone methyltransferase regulates osteoblastic differentiation of mesenchymal stem cells during postnatal bone development. FEBS Lett. 2013, 587, 3961–3967. [Google Scholar] [CrossRef]
- Yahiro, K.; Higashihori, N.; Moriyama, K. Histone methyltransferase Setdb1 is indispensable for Meckel’s cartilage development. Biochem. Biophys. Res. Commun. 2017, 482, 883–888. [Google Scholar] [CrossRef] [PubMed]
- Munnur, D.; Ahel, I. Reversible mono-ADP-ribosylation of DNA breaks. FEBS J. 2017, 284, 4002–4016. [Google Scholar] [CrossRef]
- Sakthianandeswaren, A.; Parsons, M.J.; Mouradov, D.; MacKinnon, R.N.; Catimel, B.; Liu, S.; Palmieri, M.; Love, C.; Jorissen, R.N.; Li, S.; et al. MACROD2 haploinsufficiency impairs catalytic activity of PARP1 and promotes chromosome instability and growth of intestinal tumors. Cancer Discov. 2018, 8, 988–1005. [Google Scholar] [CrossRef]
- Lombardo, B.; Esposito, D.; Iossa, S.; Vitale, A.; Verdesca, F.; Perrotta, C.; Di Leo, L.; Costa, V.; Pastore, L.; Franzé, A. Intragenic deletion in MACROD2: A family with complex phenotypes including microcephaly, intellectual disability, polydactyly, renal and pancreatic malformations. Cytogenet. Genome Res. 2019, 158, 25–31. [Google Scholar] [CrossRef]
- Lahm, H.; Jia, M.; Dreßen, M.; Wirth, F.; Puluca, N.; Gilsbach, R.; Keavney, B.D.; Cleuziou, J.; Beck, N.; Bondareva, O.; et al. Congenital heart disease risk loci identified by genome-wide association study in European patients. J. Clin. Investig. 2021, 131, e141837. [Google Scholar] [CrossRef]
- Kim, H.R.; Jin, H.S.; Eom, Y.B. Association of MACROD2 gene variants with obesity and physical activity in a Korean population. Mol. Genet. Genom. Med. 2021, 9, e1635. [Google Scholar] [CrossRef]
- Chang, Y.C.; Hee, S.W.; Lee, W.J.; Li, H.Y.; Chang, T.J.; Lin, M.W.; Hung, Y.J.; Lee, I.-T.; Hung, K.Y.; Assimes, T.; et al. Genome-wide scan for circulating vascular adhesion protein-1 levels: MACROD2 as a potential transcriptional regulator of adipogenesis. J. Diabetes Investig. 2018, 9, 1067–1074. [Google Scholar] [CrossRef]
- Jones, R.M.; Cadby, G.; Blangero, J.; Abraham, L.J.; Whitehouse, A.; Moses, E.K. MACROD2 gene associated with autistic-like traits in a general population sample. Psychiatr. Genet. 2014, 24, 241–248. [Google Scholar] [CrossRef]
- Lionel, A.C.; Crosbie, J.; Barbosa, N.; Goodale, T.; Thiruvahindrapuram, B.; Rickaby, J.; Gazzellone, M.; Carson, A.R.; Howe, J.L.; Scherer, S.W.; et al. Rare copy number variation discovery and cross-disorder comparisons identify risk genes for ADHD. Sci. Transl. Med. 2011, 3, 95ra75. [Google Scholar] [CrossRef]
- Xu, B.; Woodroffe, A.; Rodriguez-Murillo, L. Elucidating the genetic architecture of familial schizophrenia using rare copy number variant and linkage scans. Proc. Natl. Acad. Sci. USA 2009, 106, 16746–16751. [Google Scholar] [CrossRef]
- Uchimura, Y.; Ichimura, T.; Uwada, J.; Sugahara, S.; Nakao, M.; Saitoh, H.; Saitoh, H. Involvement of SUMO modification in MBD1- and MCAF1-mediated heterochromatin formation. J. Biol. Chem. 2006, 281, 23180–23190. [Google Scholar] [CrossRef]
- Waterfield, M.; Khan, I.S.; Cortez, J.T.; Fan, U.; Metzger, T.; Greer, A.; Fasano, K.; Martinez-Llordella, M.; Pollack, J.L.; Erle, D.J.; et al. The transcriptional regulator Aire coopts the repressive ATF7ip-MBD1 complex for the induction of immunotolerance. Nat. Immunol. 2014, 15, 258–265. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Khodadadi-Jamayran, A.; Dolgalev, I.; Cho, H.; Badri, S.; Chiriboga, L.A.; Zeck, B.; De Rodas Gregorio, M.L.; Dowling, C.M.; Kristen, L.; et al. Targeting the Atf7ip-Setdb1 complex augments antitumor immunity by boosting tumor immunogenicity. Cancer Immunol. Res. 2021, 9, 1298–1315. [Google Scholar] [CrossRef] [PubMed]
- Tsusaka, T.; Shimura, C.; Shinkai, Y. ATF7IP regulates SETDB1 nuclear localization and increases its ubiquitination. EMBO Rep. 2019, 20, e48297. [Google Scholar] [CrossRef]
- Timms, R.T.; Tchasovnikarova, I.A.; Antrobus, R.; Dougan, G.; Lehner, P.J. ATF7IP-mediated stabilization of the histone methyltransferase SETDB1 is essential for heterochromatin formation by the HUSH complex. Cell Rep. 2016, 17, 653–659. [Google Scholar] [CrossRef]
- Tsusaka, T.; Fukuda, K.; Shimura, C.; Kato, M.; Shinkai, Y. The fibronectin type-III (FNIII) domain of ATF7IP contributes to efficient transcriptional silencing mediated by the SETDB1 complex. Epigenetics Chromatin 2020, 13, 52. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, L.; Xu, L.; Zhang, X.; Wang, K.; Tan, Y.; Li, G.; Wang, Y.; Xue, T.; Sun, Q.; Cao, X.; et al. Methyltransferase Setdb1 Promotes Osteoblast Proliferation by Epigenetically Silencing Macrod2 with the Assistance of Atf7ip. Cells 2022, 11, 2580. https://doi.org/10.3390/cells11162580
Zhang L, Xu L, Zhang X, Wang K, Tan Y, Li G, Wang Y, Xue T, Sun Q, Cao X, et al. Methyltransferase Setdb1 Promotes Osteoblast Proliferation by Epigenetically Silencing Macrod2 with the Assistance of Atf7ip. Cells. 2022; 11(16):2580. https://doi.org/10.3390/cells11162580
Chicago/Turabian StyleZhang, Lijun, Liqun Xu, Xiaoyan Zhang, Ke Wang, Yingjun Tan, Gaozhi Li, Yixuan Wang, Tong Xue, Quan Sun, Xinsheng Cao, and et al. 2022. "Methyltransferase Setdb1 Promotes Osteoblast Proliferation by Epigenetically Silencing Macrod2 with the Assistance of Atf7ip" Cells 11, no. 16: 2580. https://doi.org/10.3390/cells11162580
APA StyleZhang, L., Xu, L., Zhang, X., Wang, K., Tan, Y., Li, G., Wang, Y., Xue, T., Sun, Q., Cao, X., Zhang, G., Hu, Z., Zhang, S., & Shi, F. (2022). Methyltransferase Setdb1 Promotes Osteoblast Proliferation by Epigenetically Silencing Macrod2 with the Assistance of Atf7ip. Cells, 11(16), 2580. https://doi.org/10.3390/cells11162580






