Developing a Reliable Holographic Flow Cyto-Tomography Apparatus by Optimizing the Experimental Layout and Computational Processing
Abstract
:1. Introduction
2. Materials and Methods
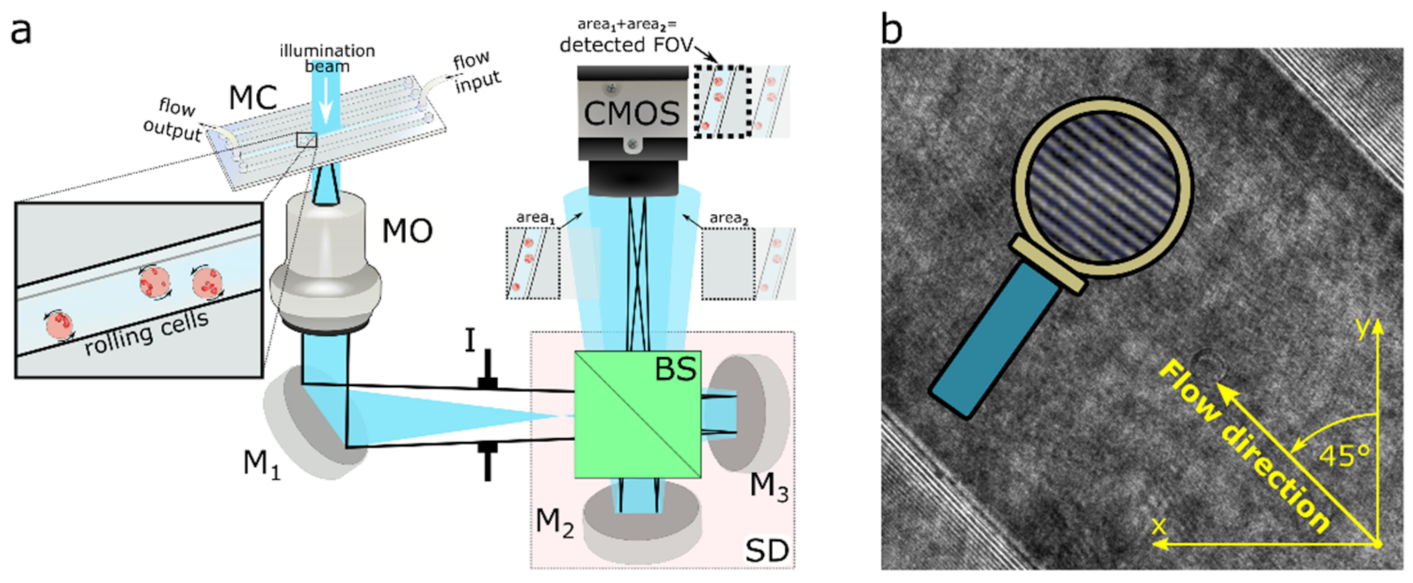
2.1. Experimental Setup
2.2. Sample Preparation
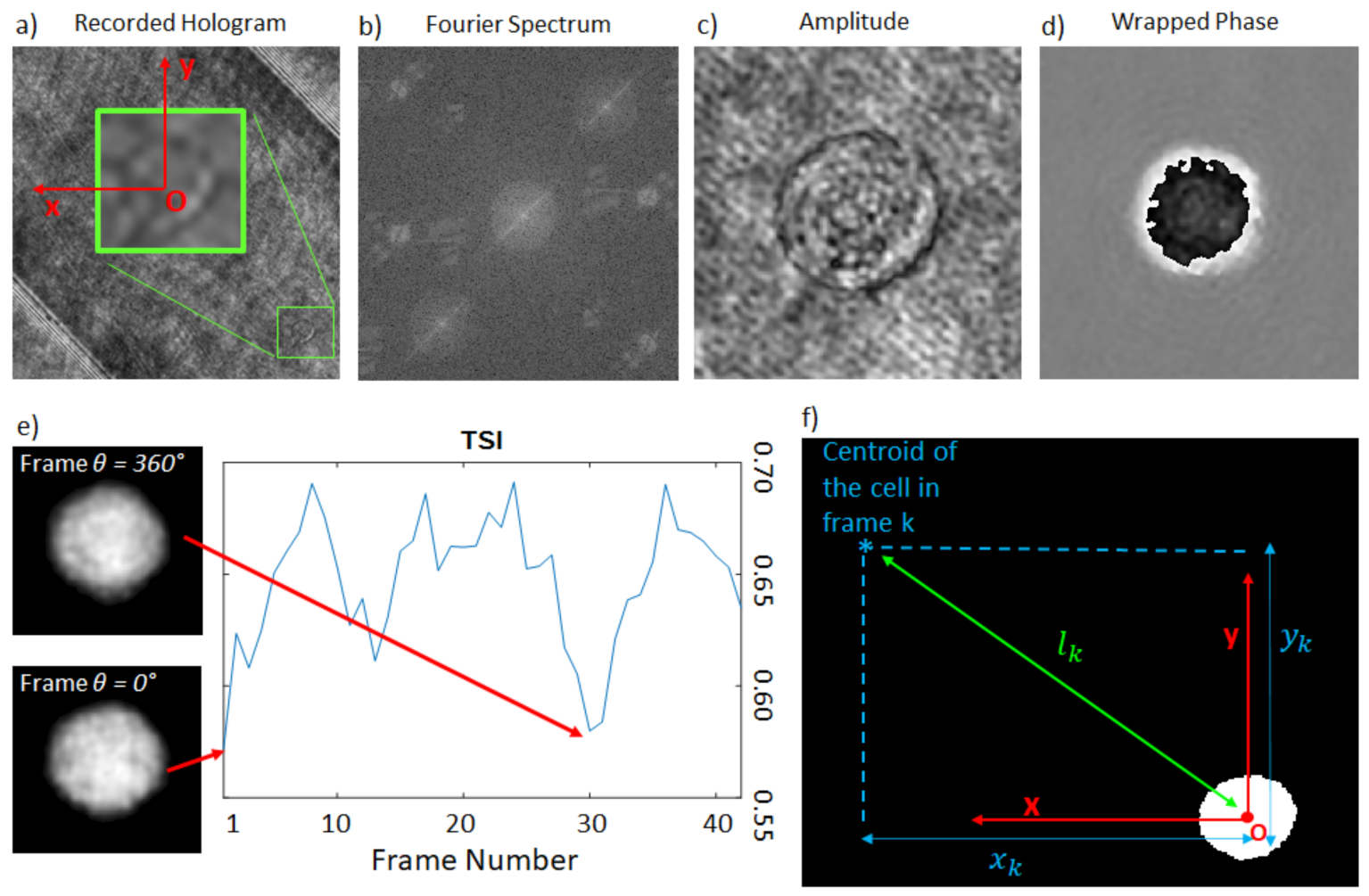
2.3. Hologram Processing
2.4. The Rolling Angle Recovery
2.5. Reconstructions by Total Variation Minimization
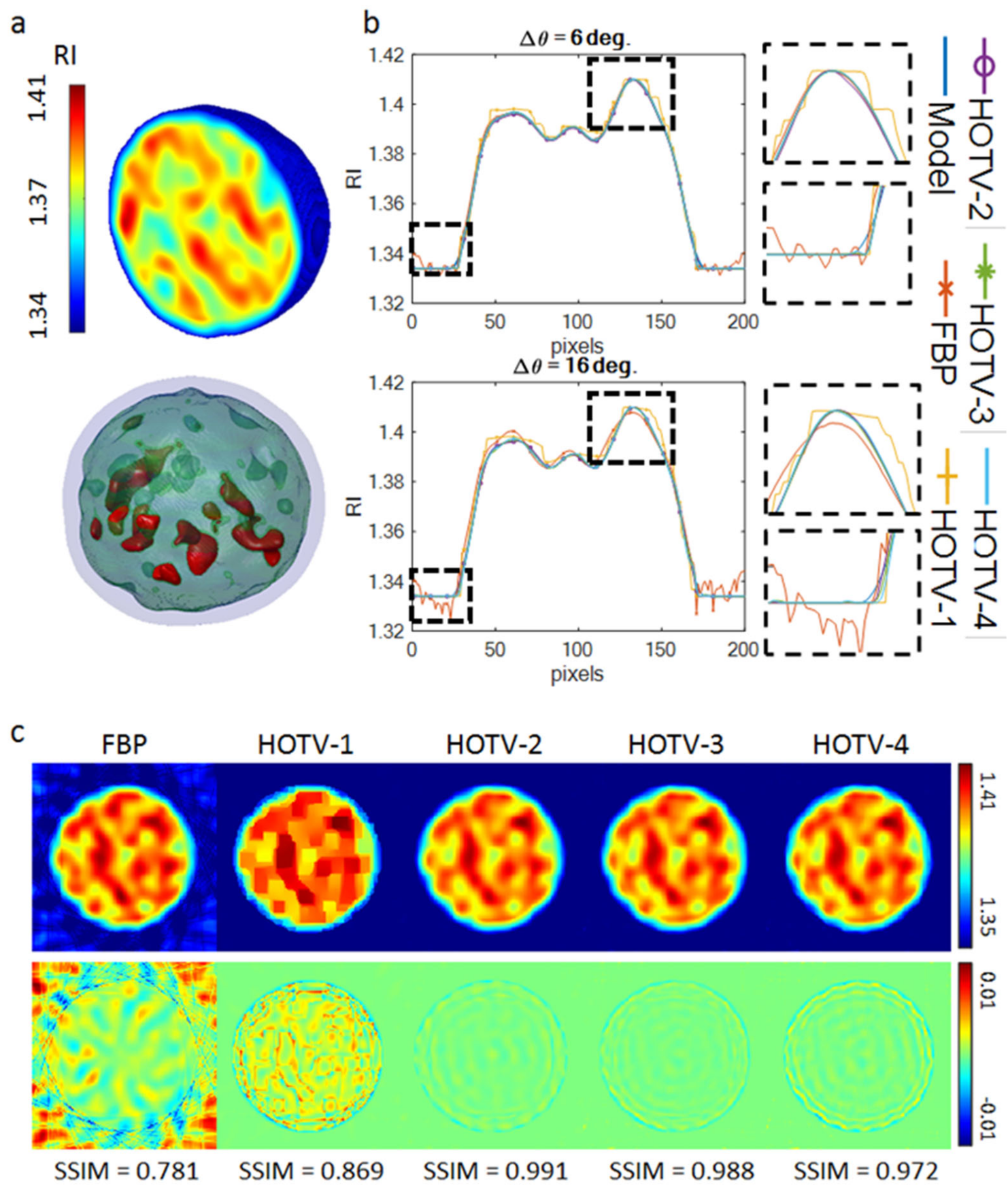
2.6. Numerical Assessment of the Performance of HOTV
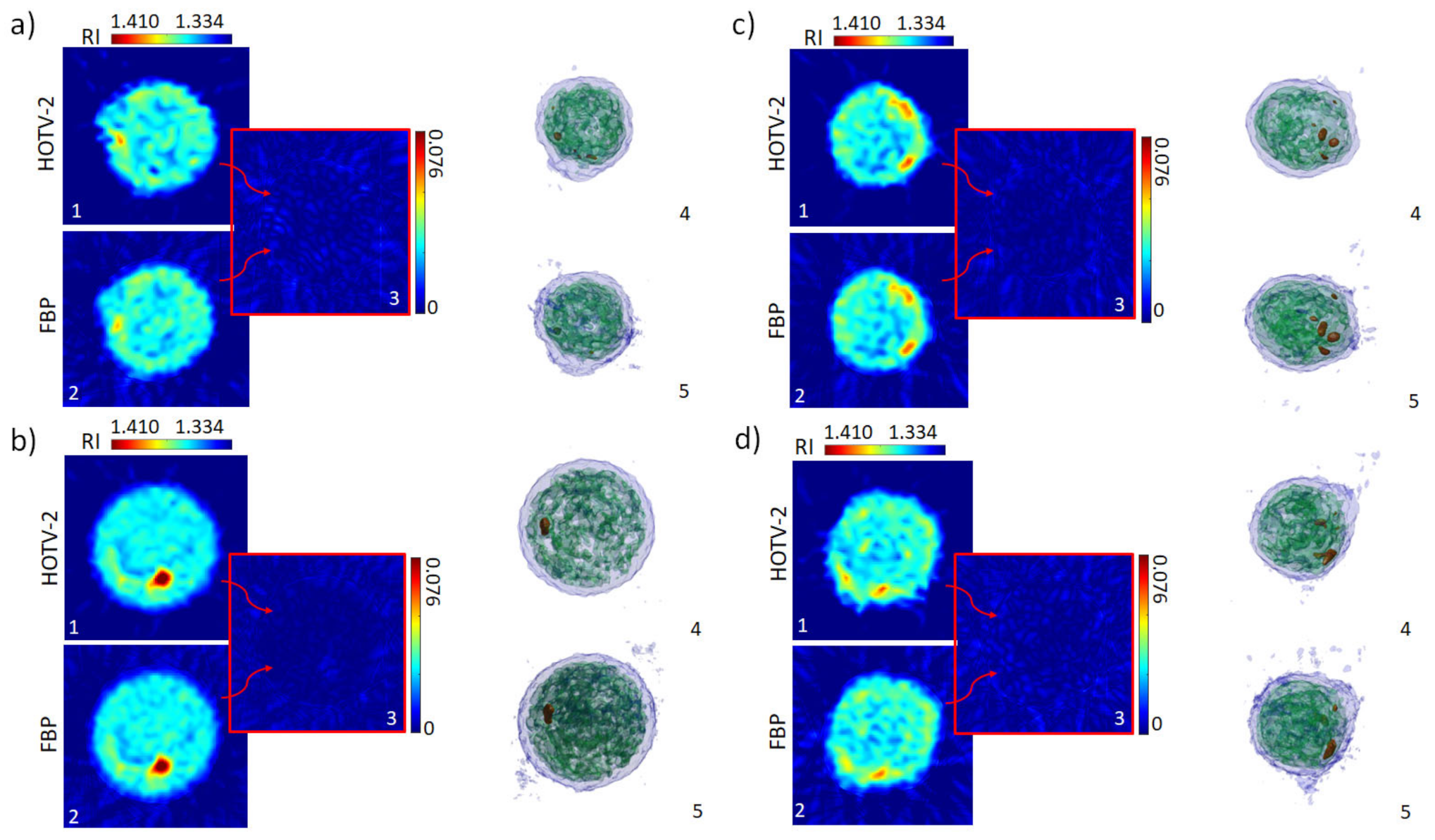
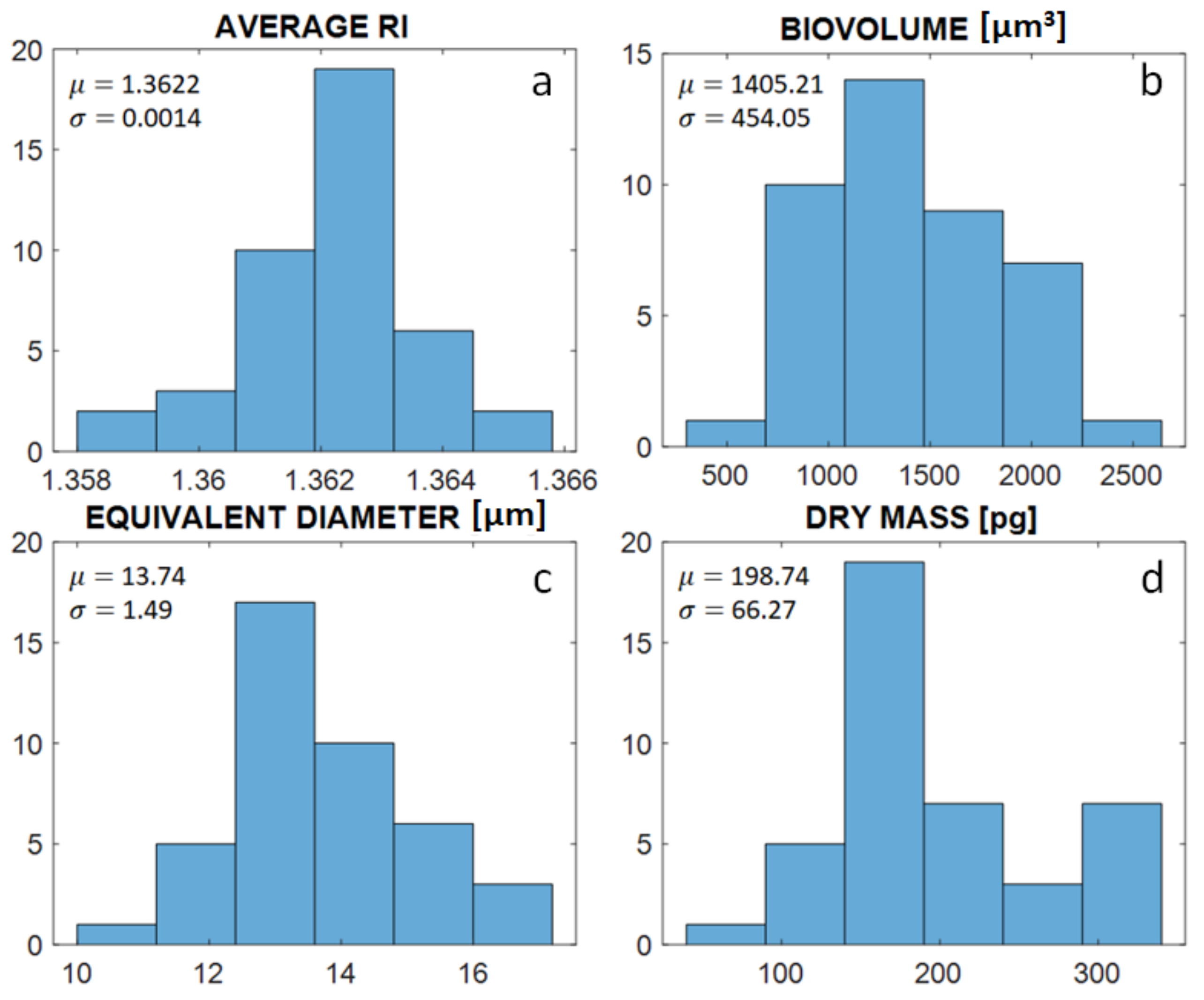
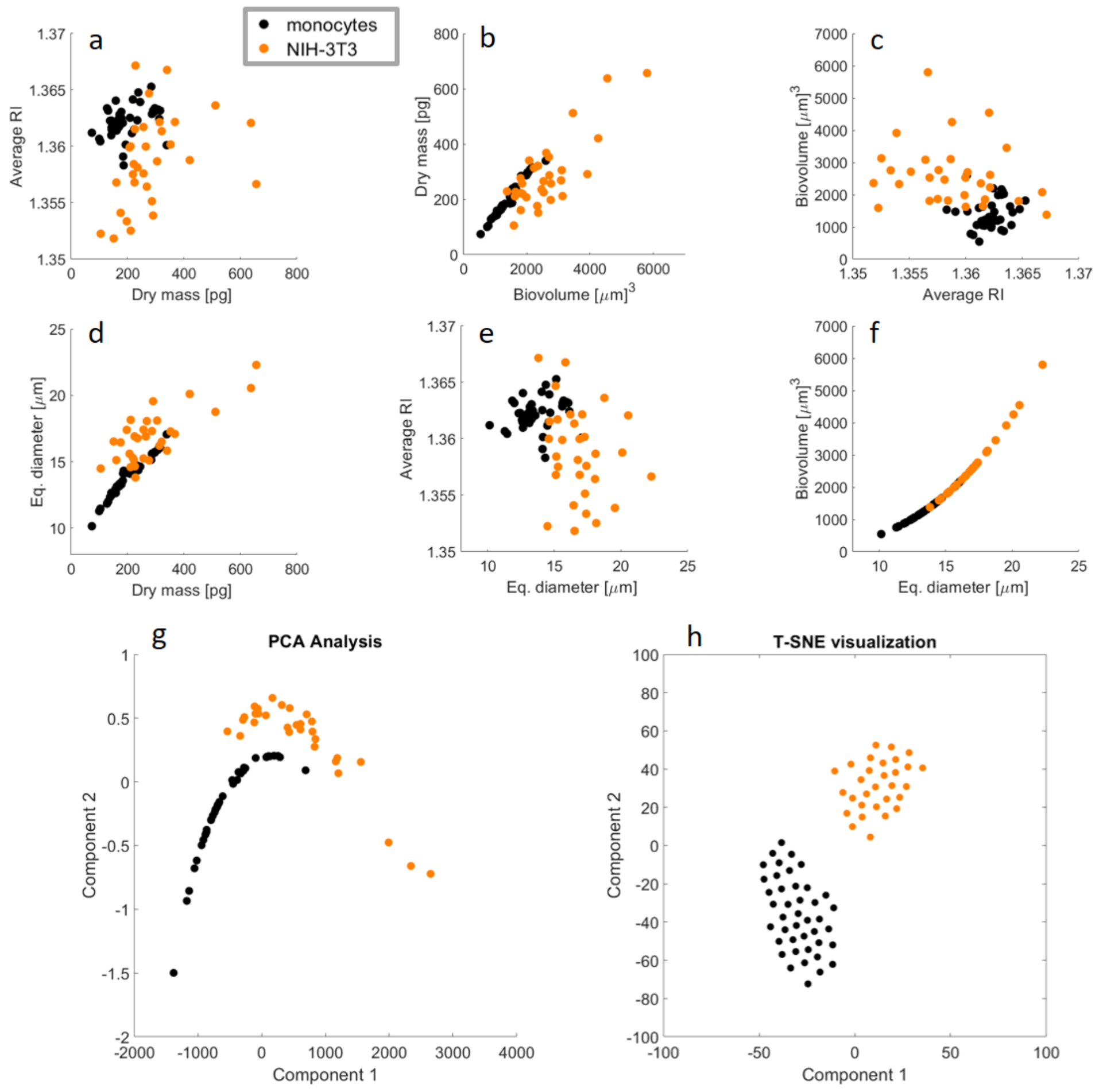
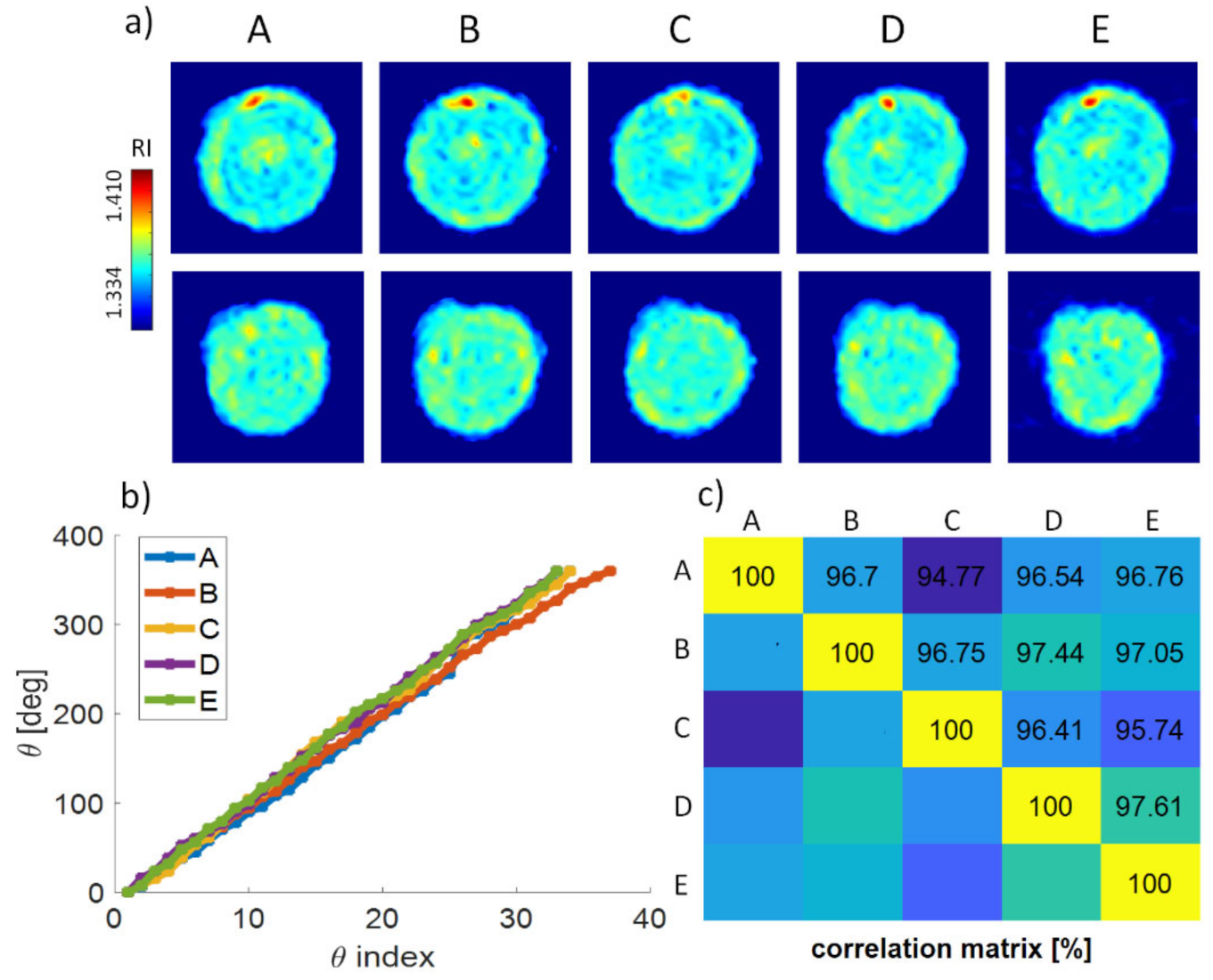
3. Results
Repeatability of the Experimental Reconstruction Process
4. Discussion and Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shields IV, C.W.; Reyes, C.D.; López, G.P. Microfluidic cell sorting: A review of the advances in the separation of cells from debulking to rare cell isolation. Lab Chip 2015, 15, 1230–1249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galanzha, E.I.; Shashkov, E.V.; Tuchin, V.V.; Zharov, V.P. In vivo multispectral, multiparameter, photoacoustic lymph flow cytometry with natural cell focusing, label-free detection and multicolor nanoparticle probes. Cytom. A 2008, 73, 884–894. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, A.C.; Gu, Y.; Han, Y.; Mei, Z.; Chiu, Y.J.; Geng, L.; Cho, S.H.; Lo, Y.H. Computational cell analysis for label-free detection of cell properties in a microfluidic laminar flow. Analyst 2016, 141, 4142–4150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, C.; Jin, T.; Yan, R.; Qi, W.; Huang, T.; Ding, H.; Tan, S.H.; Nguyen, N.T.; Xi, L. Opto-acousto-fluidic microscopy for three-dimensional label-free detection of droplets and cells in microchannels. Lab. Chip. 2018, 18, 1292–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golichenari, B.; Velonia, K.; Nosrati, R.; Nezami, A.; Farokhi-Fard, A.; Abnous, K.; Behravan, J.; Tsatsakis, A.M. Label-free nano-biosensing on the road to tuberculosis detection. Biosens. Bioelectron. 2018, 113, 124–135. [Google Scholar] [CrossRef] [PubMed]
- Lv, N.; Zhang, L.; Jiang, L.; Muhammad, A.; Wang, H.; Yuan, L. A Design of Microfluidic Chip with Quasi-Bessel Beam Waveguide for Scattering Detection of Label-Free Cancer Cells. Cytometry 2020, 97, 78–90. [Google Scholar] [CrossRef]
- Popescu, G.; Ikeda, T.; Dasari, R.R.; Feld, M.S. Diffraction phase microscopy for quantifying cell structure and dynamics. Opt. Lett. 2006, 31, 775–777. [Google Scholar] [CrossRef]
- Shaked, N.T. Quantitative phase microscopy of biological samples using a portable interferometer. Opt. Lett. 2012, 37, 2016–2018. [Google Scholar] [CrossRef]
- Zangle, T.A.; Teitell, M.A. Live-cell mass profiling: An emerging approach in quantitative biophysics. Nat. Methods 2014, 11, 1221–1228. [Google Scholar] [CrossRef] [Green Version]
- Kasprowicz, R.; Suman, R.; O’Toole, P. Characterising live cell behaviour: Traditional label-free and quantitative phase imaging approaches. Int. J. Biochem. Cell Biol. 2017, 84, 89–95. [Google Scholar] [CrossRef]
- Mahjoubfar, A.; Chen, C.; Niazi, K.R.; Rabizadeh, S.; Jalali, B. Label-free high-throughput cell screening in flow. Biomed. Opt. Express 2013, 4, 1618–1625. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lau, A.K.S.; Shum, H.C.; Wong, K.K.Y.; Tsia, K.K. Optofluidic time-stretch imaging—An emerging tool for high-throughput imaging flow cytometry. Lab Chip 2016, 16, 1743–1756. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.K.; Ahrens, C.C.; Li, W.; Vanapalli, S.A. Label-free, high-throughput holographic screening and enumeration of tumor cells in blood. Lab Chip 2017, 17, 2920. [Google Scholar] [CrossRef] [PubMed]
- Yamada, H.; Hirotsu, A.; Yamashita, D.; Yasuhiko, O.; Yamauchi, T.; Kayou, T.; Suzuki, H.; Okazaki, S.; Kikuchi, H.; Takeuchi, H.; et al. Label-free imaging flow cytometer for analyzing large cell populations by line-field quantitative phase microscopy with digital refocusing. Biomed. Opt. Express 2020, 11, 2213–2223. [Google Scholar] [CrossRef] [PubMed]
- Vinoth, B.; Lai, X.J.; Lin, Y.C.; Tu, H.Y.; Cheng, C.J. Integrated dual-tomography for refractive index analysis of free-floating single living cell with isotropic superresolution. Sci. Rep. 2018, 8, 5943. [Google Scholar] [CrossRef] [Green Version]
- Lauer, V. New approach to optical diffraction tomography yielding a vector equation of diffraction tomography and a novel tomographic microscope. J. Microsc. 2002, 205, 165–176. [Google Scholar] [CrossRef]
- Choi, W.; Fang-Yen, C.; Badizadegan, K.; Oh, S.; Lue, N.; Dasari, R.R.; Feld, M.S. Tomographic phase microscopy. Nat. Methods 2007, 4, 717–719. [Google Scholar] [CrossRef] [PubMed]
- Debailleul, M.; Georges, V.; Simon, B.; Morin, R.; Haeberlé, O. High-resolution three-dimensional tomographic diffractive microscopy of transparent inorganic and biological samples. Opt. Lett. 2009, 34, 79–81. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.; Yoon, H.; Diez-Silva, M.; Dao, M.; Dasari, R.R.; Park, Y. High-resolution three-dimensional imaging of red blood cells parasitized by Plasmodium falciparum and in situ hemozoin crystals using optical diffraction tomography. J. Biomed. Opt. 2014, 19, 011005. [Google Scholar] [CrossRef] [Green Version]
- Gorski, W.; Osten, W. Tomographic imaging of photonic crystal fibers. Opt. Lett. 2007, 32, 1977–1979. [Google Scholar] [CrossRef]
- Charrière, F.; Pavillon, N.; Colomb, T.; Depeursinge, C.; Heger, T.J.; Mitchell, E.A.; Marquet, P.; Rappaz, B. Living specimen tomography by digital holographic microscopy: Morphometry of testate amoeba. Opt. Express 2006, 14, 7005–7013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Merola, F.; Memmolo, P.; Miccio, L.; Savoia, R.; Mugnano, M.; Fontana, A.; D’Ippolito, G.; Sardo, A.; Iolascon, A.; Gambale, A.; et al. Tomographic flow cytometry by digital holography. Light Sci. Appl. 2017, 6, e16241. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villone, M.M.; Memmolo, P.; Merola, F.; Mugnano, M.; Miccio, L.; Maffettone, P.L.; Ferraro, P. Full-angle tomographic phase microscopy of flowing quasi-spherical cells. Lab Chip 2018, 18, 126–131. [Google Scholar] [CrossRef] [PubMed]
- Kleiber, A.; Kraus, D.; Henkel, T.; Fritzsche, W. Review: Tomographic imaging flow cytometry. Lab Chip 2021, 21, 3655–3666. [Google Scholar] [CrossRef]
- Pirone, D.; Mugnano, M.; Memmolo, P.; Merola, F.; Lama, G.C.; Castaldo, R.; Miccio, L.; Bianco, V.; Grilli, S.; Ferraro, P. Three-Dimensional Quantitative Intracellular Visualization of Graphene Oxide Nanoparticles by Tomographic Flow Cytometry. Nano Lett. 2021, 21, 5958–5966. [Google Scholar] [CrossRef]
- Pirone, D.; Sirico, D.; Miccio, L.; Bianco, V.; Mugnano, M.; Ferraro, P.; Memmolo, P. Speeding up reconstruction of 3D tomograms in holographic flow cytometry via deep learning. Lab Chip 2022, 22, 793–804. [Google Scholar] [CrossRef]
- Pirone, D.; Memmolo, P.; Merola, F.; Miccio, L.; Mugano, M.; Capozzoli, A.; Curcio, C.; Liseno, A.; Ferraro, P. Rolling angle recovery of flowing cells in holographic tomography exploiting the phase similarity. Appl. Opt. 2021, 60, A277–A284. [Google Scholar] [CrossRef]
- Hsu, W.C.; Su, J.W.; Tseng, T.Y.; Sung, K.B. Tomographic diffractive microscopy of living cells based on a common-path configuration. Opt. Lett. 2014, 39, 2210–2213. [Google Scholar] [CrossRef]
- Kim, K.; Yaqoob, Z.; Lee, K.; Kang, J.W.; Choi, Y.; Hosseini, P.; So, P.T.C.; Park, Y. Diffraction optical tomography using a quantitative phase imaging unit. Opt. Lett. 2014, 39, 6935–6938. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Shim, H.; Kim, K.; Park, H.; Heo, J.H.; Yoon, J.; Choi, C.; Jang, S.; Park, Y. Common-path diffraction optical tomography for investigation of three-dimensional structures and dynamics of biological cells. Opt. Express 2014, 22, 10398–10407. [Google Scholar] [CrossRef]
- Zhang, J.; Dai, S.; Ma, C.; Xi, T.; Di, J.; Zhao, J. A review of common-path off-axis digital holography: Towards high stable optical instrument manufacturing. Light Adv. Manuf. 2021, 2, 333–349. [Google Scholar] [CrossRef]
- Patel, N.; Rawat, S.; Joglekar, M.; Chhaniwal, V.K.; Dubey, S.K.; O’Connor, T.; Javidi, B.; Anand, A. Compact and low-cost instrument for digital holographic microscopy of immobilized micro-particles. Opt. Lasers Eng. 2021, 137, 106397. [Google Scholar] [CrossRef]
- O’Connor, T.; Anand, A.; Andemariam, B.; Javidi, B. Deep learning-based cell identification and disease diagnosis using spatio-temporal cellular dynamics in compact digital holographic microscopy. Biomed. Opt. Express 2020, 11, 4491–4508. [Google Scholar] [CrossRef] [PubMed]
- Ebrahimi, S.; Dashtdar, M.; Anand, A.; Javidi, B. Common-path lensless digital holographic microscope employing a Fresnel biprism. Opt. Lasers Eng. 2020, 128, 106014. [Google Scholar] [CrossRef]
- Yaghoubi, S.H.S.; Ebrahimi, S.; Dashtdar, M.; Doblas, A.; Javidi, B. Quantitative phase imaging based on Fresnel diffraction from a phase plate. Appl. Phys. Lett. 2019, 114, 183701. [Google Scholar] [CrossRef]
- Kemper, B.; Vollmer, A.; Bally, G.; Rommel, C.E.; Schnekenburger, J. Simplified approach for quantitative digital holographic phase contrast imaging of living cells. J. Biomed. Opt. 2011, 16, 026014. [Google Scholar] [CrossRef] [Green Version]
- Schubert, R.; Vollmer, A.; Ketelhut, S.I.; Kemper, B. Quantitative phase microscopy for evaluation of intestinal inflammation and wound healing utilizing label-free biophysical markers. Biomed. Opt. Express 2014, 5, 4213–4222. [Google Scholar] [CrossRef] [Green Version]
- Ma, C.; Li, Y.; Zhang, J.; Li, P.; Xi, T.; Di, J.; Zhao, J. Lateral shearing common-path digital holographic microscopy based on a slightly trapezoid Sagnac interferometer. Opt. Express 2017, 25, 13659–13667. [Google Scholar] [CrossRef]
- Singh, A.S.G.; Anand, A.; Leitgeb, R.A.; Javidi, B. Lateral shearing digital holographic imaging of small biological specimens. Opt. Express 2012, 20, 23617–23622. [Google Scholar] [CrossRef]
- Di, J.; Li, Y.; Xie, M.; Zhang, J.; Ma, C.; Xi, T.; Li, E.; Zhao, J. Dual-wavelength common-path digital holographic microscopy for quantitative phase imaging based on lateral shearing interferometry. Appl. Opt. 2016, 55, 7287–7293. [Google Scholar] [CrossRef]
- Kim, B.M.; Park, S.J.; Kim, E.S. Single-shot digital holographic microscopy with a modified lateral-shearing interferometer based on computational telecentricity. Opt. Express 2017, 25, 6151–6168. [Google Scholar] [CrossRef] [PubMed]
- Qu, W.; Yu, Y.; Choo, C.O.; Anand, A. Digital holographic microscopy with physical phase compensation. Opt. Lett. 2009, 34, 1276–1278. [Google Scholar] [CrossRef]
- Lee, K.; Park, Y. Quantitative phase imaging unit. Opt. Lett. 2014, 39, 3630–3633. [Google Scholar] [CrossRef] [PubMed]
- Di, J.; Li, Y.; Wang, K.; Zhao, J. Quantitative and Dynamic Phase Imaging of Biological Cells by the Use of the Digital Holographic Microscopy Based on a Beam Displacer Unit. IEEE Photon. J. 2018, 10, 1–10. [Google Scholar] [CrossRef]
- Ebrahimi, S.; Dashtdar, M.; Sánchez-Ortiga, E.; Martínez-Corral, M.; Javidi, B. Stable and simple quantitative phase-contrast imaging by Fresnel biprism. Appl. Phys. Lett. 2018, 112, 11370. [Google Scholar] [CrossRef] [Green Version]
- Guo, R.; Mirsky, S.K.; Barnea, I.; Dudaie, M.; Shaked, N.T. Coded aperture correlation holographic microscope for single-shot quantitative phase and amplitude imaging with extended field of view. Opt. Express 2020, 28, 5617–5628. [Google Scholar] [CrossRef]
- Běhal, J. Quantitative phase imaging in common-path cross-referenced holographic microscopy using double-exposure method. Sci. Rep. 2019, 9, 9801. [Google Scholar] [CrossRef]
- Schnars, U.; Jüptner, W. Digital recording and numerical reconstruction of holograms. Meas. Sci. Technol. 2002, 13, R85–R101. [Google Scholar] [CrossRef]
- Memmolo, P.; Miccio, L.; Paturzo, M.; Caprio, G.D.; Coppola, G.; Nei, P.A.; Ferraro, P. Recent advances in holographic 3D particle tracking. Adv. Opt. Photonics 2015, 7, 713–755. [Google Scholar] [CrossRef]
- Bioucas-Dias, J.M.; Valadao, G. Phase unwrapping via graph cuts. IEEE Trans. Image Processing 2007, 16, 698–709. [Google Scholar] [CrossRef]
- Kemao, Q. Windowed Fourier transform for fringe pattern analysis. Appl. Opt. 2004, 43, 2695–2702. [Google Scholar] [CrossRef] [PubMed]
- Montrésor, S.; Memmolo, P.; Bianco, V.; Ferraro, P.; Picart, P. Comparative study of multi-look processing for phase map denoising in digital Fresnel holographic interferometry. J. Opt. Soc. Am. A 2019, 36, A59–A66. [Google Scholar] [CrossRef] [PubMed]
- Rudin, L.I.; Osher, S.; Fatemi, E. Nonlinear total variation based noise removal algorithms. Phys. D Nonlinear Phenom. 1992, 60, 259–268. [Google Scholar] [CrossRef]
- Jin, D.; Renjie, Z.; Zahid, Y.; So, P.T.C. Tomographic phase microscopy: Principles and applications in bioimaging [Invited]. J. Opt. Soc. Am. B 2017, 34, B64–B77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cha, T.; Marquina, A.; Mulet, P. High-Order Total Variation-Based Image Restoration. SIAM J. Sci. Comput. 2000, 22, 503–516. [Google Scholar] [CrossRef]
- Krauze, W.; Makowski, P.; Kujawińska, M. Total variation iterative constraint algorithm for limited-angle tomographic reconstruction of non-piecewise-constant structures. In Modeling Aspects in Optical Metrology V, Proceedings of the SPIE Optical Metrology, Munich, Germany, 21–25 June 2015; SPIE: Bellingham, WA, USA, 2015; Volume 9526, p. 95260Y. [Google Scholar] [CrossRef]
- Kus, A.; Krauze, W.; Makowski, P.; Kujawinska, M. Holographic tomography: Hardware and software solutions for 3D quantitative biomedical imaging (Invited paper). ETRI J. 2019, 41, 61–72. [Google Scholar] [CrossRef] [Green Version]
- Krauze, W.; Makowski, P.; Kujawińska, M.; Kuś, A. Generalized total variation iterative constraint strategy in limited angle optical diffraction tomography. Opt. Express 2016, 24, 4924–4936. [Google Scholar] [CrossRef]
- Kus, A.; Krauze, W.; Kujawinska, M.; Filipiak, M. Limited-angle hybrid diffraction tomography for biological samples. In Optical Micro- and Nanometrology V, Proceedings of the SPIE Photonics Europe, Brussels, Belgium, 13–17 April 2014; SPIE: Bellingham, WA, USA, 2014; Volume 9132, p. 913201. [Google Scholar] [CrossRef]
- Saders, T.; Gelb, A.; Platte, R.B.; Arslan, I.; Landskron, K. Recovering fine details from under-resolved electron tomography data using higher order total variation ℓ 1 regularization. Ultramicroscopy 2017, 174, 97–105. [Google Scholar] [CrossRef]
- Archibald, R.; Gekb, A.; Platte, R.B. Image reconstruction from undersampled Fourier data using the polynomial annihilation transform. J. Sci. Comput. 2016, 67, 432–452. [Google Scholar] [CrossRef]
- Archibald, R.; Gelb, A.; Yoon, J. Polynomial fitting for edge detection in irregularly sampled signals and images. SIAM J. Numer. Anal. 2005, 43, 259–279. [Google Scholar] [CrossRef] [Green Version]
- Sanders, T.; Matlab Imaging Algorithms: Image Reconstruction, Restoration, and Alignment, with a Focus in Tomography. Version 2. Available online: https://www.toby-sanders.com/software (accessed on 3 March 2022).
- Li, C.; Yin, W.; Jiang, H.; Zhang, Y. An efficient augmented Lagrangian method with applications to total variation minimization. Comput. Optim. Appl. 2013, 56, 507–530. [Google Scholar] [CrossRef] [Green Version]
- Sanders, T. Parameter selection for HOTV regularization. Appl. Numer. Math. 2018, 125, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Liu, P.Y.; Chin, L.K.; Ser, W.; Chen, H.F.; Hsieh, C.-M.; Lee, C.-H.; Sung, K.-B.; Ayi, T.C.; Yap, P.H.; Liedberg, B.; et al. Cell refractive index for cell biology and disease diagnosis: Past, present and future. Lab Chip 2016, 16, 634–644. [Google Scholar] [CrossRef] [PubMed]
- He, Y.R.; He, S.; Kandel, M.E.; Jae Lee, Y.; Hu, C.; Sobh, N.; Anastasio, M.A.; Popescu, G. Cell Cycle Stage Classification Using Phase Imaging with Computational Specificity. ACS Photonics 2022, 9, 1264–1273. [Google Scholar] [CrossRef]
- Yoon, J.; Kim, K.; Park, H.; Choi, C.; Jang, S.; Park, Y.K. Label-free characterization of white blood cells by measuring 3D refractive index maps Biomed. Opt. Express 2015, 6, 3865–3875. [Google Scholar] [CrossRef] [Green Version]
- Dannhauser, D.; Rosi, D.; Memmolo, P.; Finizio, A.; Ferraro, P.; Netti, P.A.; Causa, F. Biophysical investigation of living monocytes in flow by collaborative coherent imaging techniques. Biomed. Opt. Express 2018, 9, 5194–5204. [Google Scholar] [CrossRef]
- Villat, A.; Abkarian, M. (Eds.) Dynamics of Blood Cell Suspensions in Microflows; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Běhal, J.; Borrelli, F.; Mugnano, M.; Bianco, V.; Capozzoli, A.; Curcio, C.; Liseno, A.; Miccio, L.; Memmolo, P.; Ferraro, P. Developing a Reliable Holographic Flow Cyto-Tomography Apparatus by Optimizing the Experimental Layout and Computational Processing. Cells 2022, 11, 2591. https://doi.org/10.3390/cells11162591
Běhal J, Borrelli F, Mugnano M, Bianco V, Capozzoli A, Curcio C, Liseno A, Miccio L, Memmolo P, Ferraro P. Developing a Reliable Holographic Flow Cyto-Tomography Apparatus by Optimizing the Experimental Layout and Computational Processing. Cells. 2022; 11(16):2591. https://doi.org/10.3390/cells11162591
Chicago/Turabian StyleBěhal, Jaromír, Francesca Borrelli, Martina Mugnano, Vittorio Bianco, Amedeo Capozzoli, Claudio Curcio, Angelo Liseno, Lisa Miccio, Pasquale Memmolo, and Pietro Ferraro. 2022. "Developing a Reliable Holographic Flow Cyto-Tomography Apparatus by Optimizing the Experimental Layout and Computational Processing" Cells 11, no. 16: 2591. https://doi.org/10.3390/cells11162591






