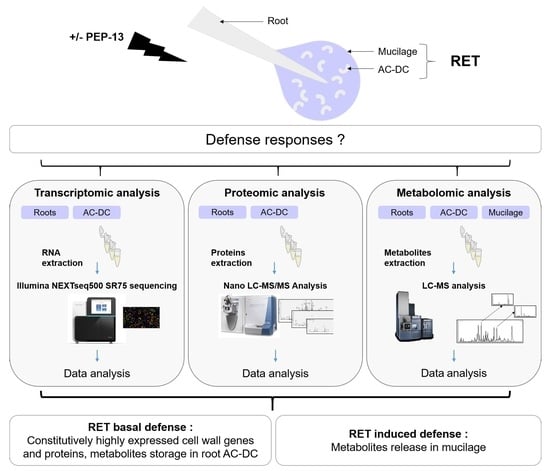
Elicitation of Roots and AC-DC with PEP-13 Peptide Shows Differential Defense Responses in Multi-Omics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material, PEP-13 Elicitation and RET Collection
2.2. Transcriptomic Analysis
2.2.1. Sample Preparation and Sequencing
2.2.2. Data Analysis
2.3. Proteomic Analysis
2.3.1. Sample Preparation and Analysis
2.3.2. Data Analysis
2.4. Metabolomic Analysis
2.4.1. Sample Preparation and Analysis
2.4.2. Data Analysis
3. Results and Discussion
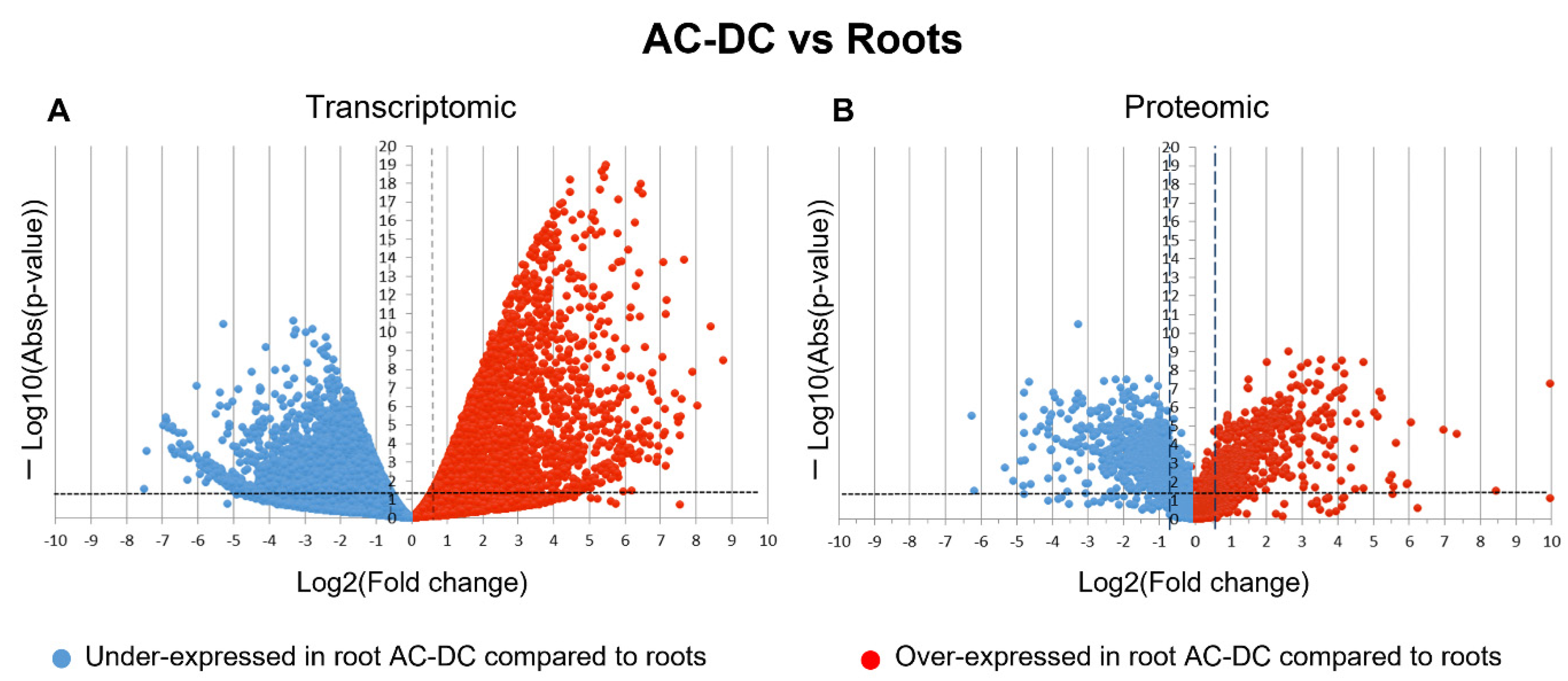
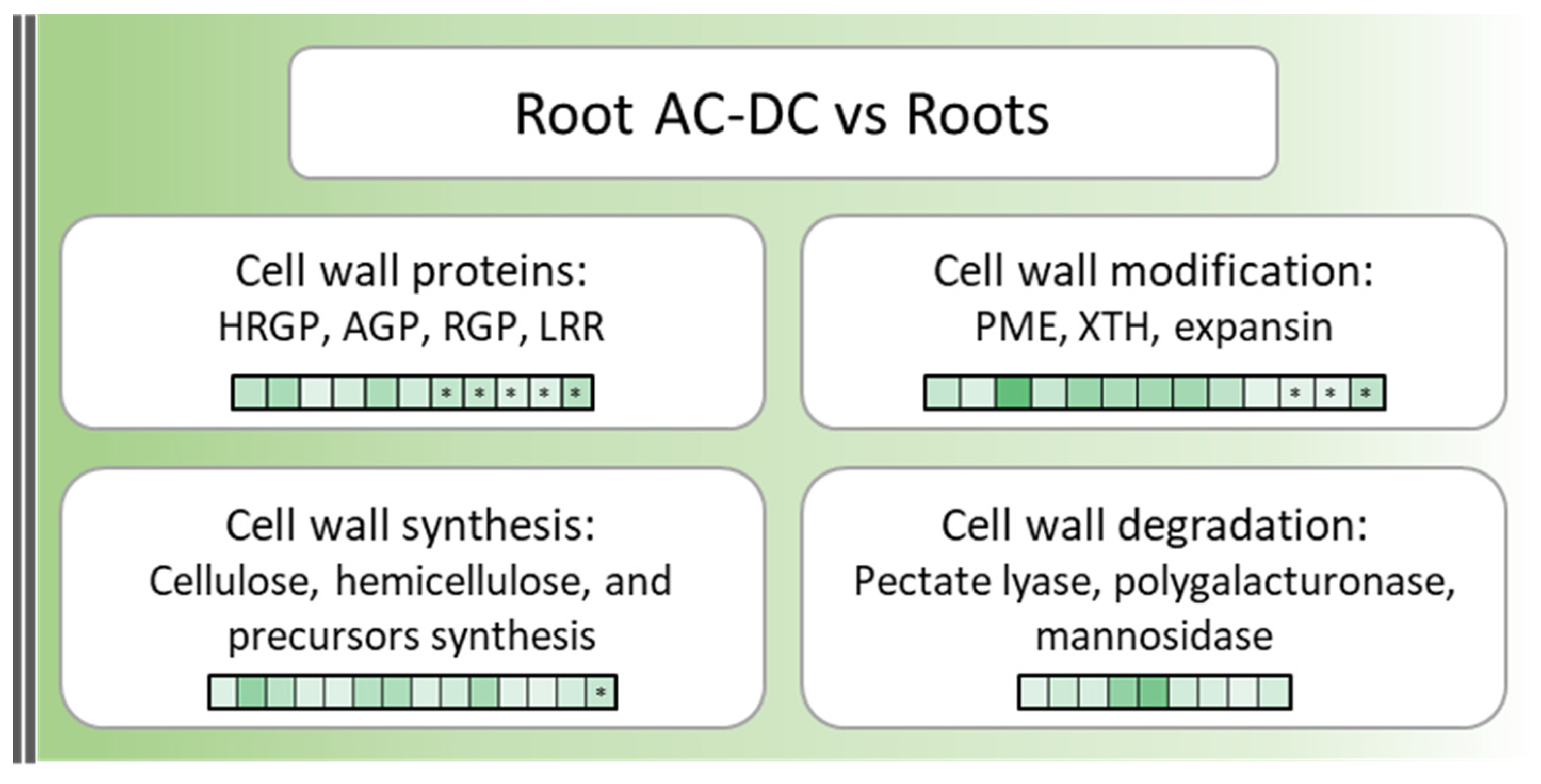
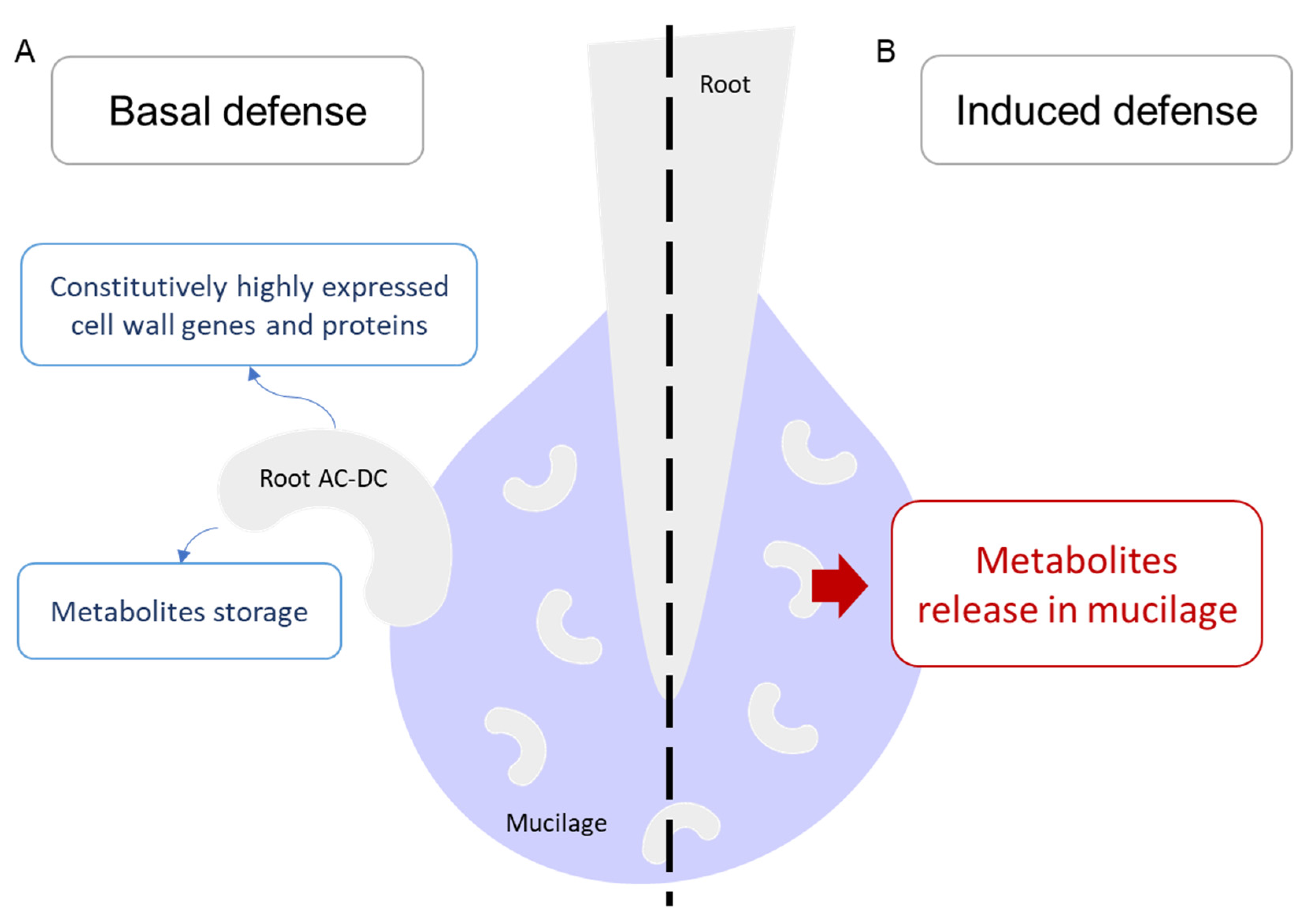
3.1. A Picture of Basal State in Root Cells and Root AC-DC
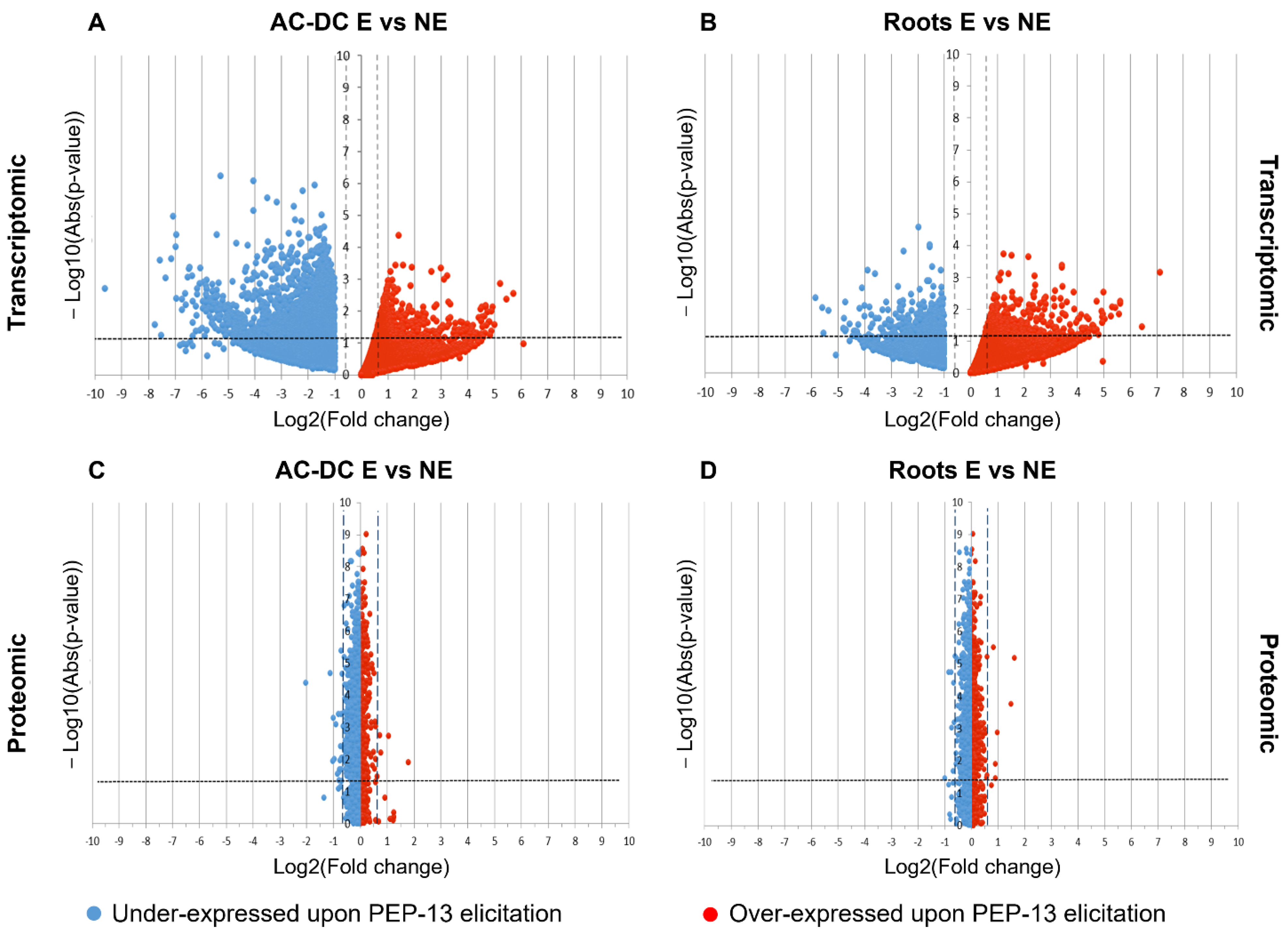
3.2. Induced Defense Mechanisms upon PEP-13 Elicitation
3.2.1. Transcriptomic and Proteomic Analysis of Induced Defense Response in the RET
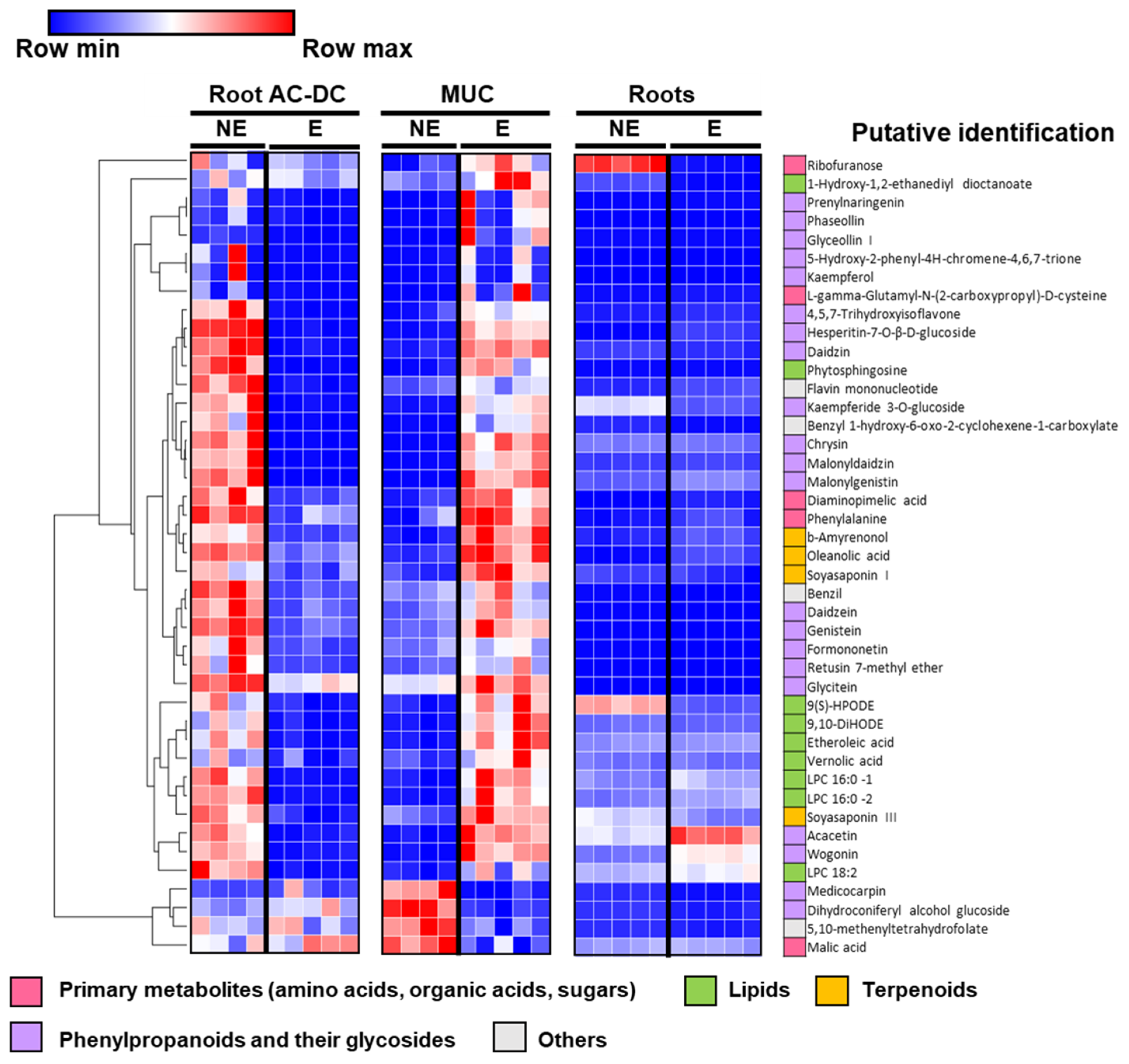
3.2.2. Metabolomic Analysis of Induced Defense Response in the RET
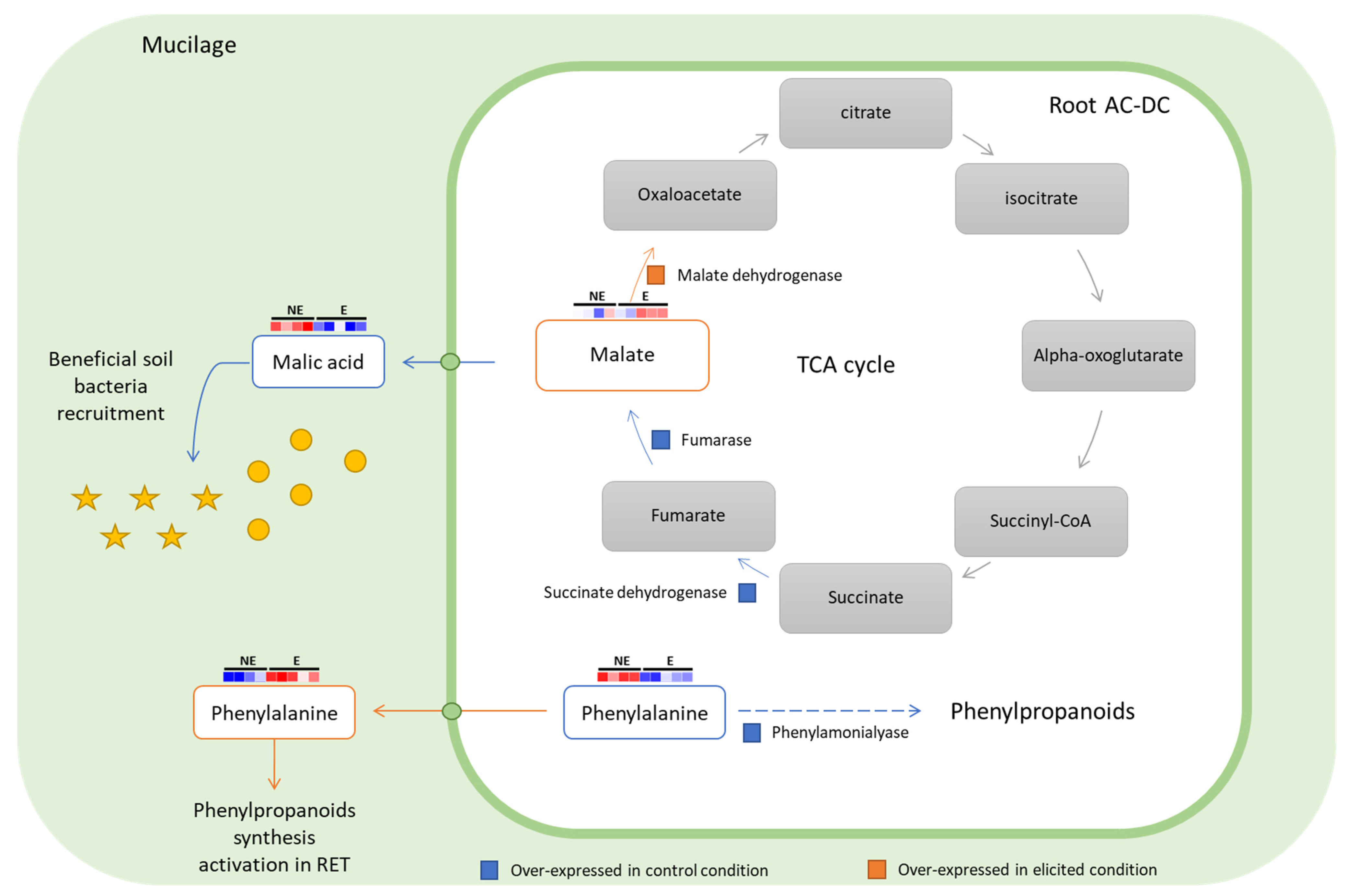
3.2.3. Focus on Primary Metabolism
3.3. Deciphering the Basal and Induced Defense Responses in the RET
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tian, H.; De Smet, I.; Ding, Z. Shaping a root system: Regulating lateral versus primary root growth. Trends Plant Sci. 2014, 19, 426–431. [Google Scholar] [CrossRef] [PubMed]
- Benfey, P.; Linstead, P.; Roberts, K.; Schiefelbein, J.; Hauser, M.-T.; Aeschbacher, R. Root development in Arabidopsis: Four mutants with dramatically altered root morphogenesis. Development 1993, 119, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Hawes, M.C.; Bengough, G.; Cassab, G.; Ponce, G. Root Caps and Rhizosphere. J. Plant Growth Regul. 2002, 21, 352–367. [Google Scholar] [CrossRef]
- Vicré, M.; Santaella, C.; Blanchet, S.; Gateau, A.; Driouich, A. Root Border-Like Cells of Arabidopsis. Microscopical Characterization and Role in the Interaction with Rhizobacteria. Plant Physiol. 2005, 138, 998–1008. [Google Scholar] [CrossRef] [Green Version]
- Driouich, A.; Follet-Gueye, M.-L.; Vicré-Gibouin, M.; Hawes, M. Root border cells and secretions as critical elements in plant host defense. Curr. Opin. Plant Biol. 2013, 16, 489–495. [Google Scholar] [CrossRef] [PubMed]
- Driouich, A.; Smith, C.; Ropitaux, M.; Chambard, M.; Boulogne, I.; Bernard, S.; Follet-Gueye, M.; Vicré, M.; Moore, J. Root extracellular traps versus neutrophil extracellular traps in host defence, a case of functional convergence? Biol. Rev. 2019, 94, 1685–1700. [Google Scholar] [CrossRef]
- Ettema, C.H.; Wardle, D.A. Spatial soil ecology. Trends Ecol. Evol. 2002, 17, 177–183. [Google Scholar] [CrossRef]
- Lampugnani, E.; Khan, G.A.; Somssich, M.; Persson, S. Building a plant cell wall at a glance. J. Cell Sci. 2018, 131, jcs207373. [Google Scholar] [CrossRef] [Green Version]
- Anderson, C.T.; Kieber, J.J. Dynamic Construction, Perception, and Remodeling of Plant Cell Walls. Annu. Rev. Plant Biol. 2020, 71, 39–69. [Google Scholar] [CrossRef] [Green Version]
- Showalter, A.M. Structure and Function of Plant Cell Wall Proteins. Plant Cell 1993, 5, 9–23. [Google Scholar]
- Nguema-Ona, E.; Vicrã-Gibouin, M.; Gottã, M.; Eplancot, B.; Elerouge, P.; Ebardor, M.; Edriouich, A. Cell wall O-glycoproteins and N-glycoproteins: Aspects of biosynthesis and function. Front. Plant Sci. 2014, 5, 499. [Google Scholar] [CrossRef] [Green Version]
- Held, M.; Tan, L.; Kamyab, A.; Hare, M.; Shpak, E.; Kieliszewski, M.J. Di-isodityrosine Is the Intermolecular Cross-link of Isodityrosine-rich Extensin Analogs Cross-linked in Vitro. J. Biol. Chem. 2004, 279, 55474–55482. [Google Scholar] [CrossRef] [Green Version]
- Cannon, M.C.; Terneus, K.; Hall, Q.; Tan, L.; Wang, Y.; Wegenhart, B.L.; Chen, L.; Lamport, D.T.A.; Chen, Y.; Kieliszewski, M.J. Self-assembly of the plant cell wall requires an extensin scaffold. Proc. Natl. Acad. Sci. USA 2008, 105, 2226–2231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, S.; Liu, Z.; Shen, H.; Wu, D. Damage-Associated Molecular Pattern-Triggered Immunity in Plants. Front. Plant Sci. 2019, 10, 646. [Google Scholar] [CrossRef] [PubMed]
- Lanubile, A.; Muppirala, U.K.; Severin, A.J.; Marocco, A.; Munkvold, G.P. Transcriptome profiling of soybean (Glycine max) roots challenged with pathogenic and non-pathogenic isolates of Fusarium oxysporum. BMC Genom. 2015, 16, 1089. [Google Scholar] [CrossRef] [Green Version]
- Dong, S.; Wang, W.; Jiang, Y.; Ma, Z.; Yan, C.; Liu, L.; Cui, G. Antioxidant and Proteomic Analysis of Soybean Response to Drought during Soybean Flowering. Ekoloji 2019, 28, 2041–2052. [Google Scholar]
- Bai, L.; Sun, H.-B.; Liang, R.-T.; Cai, B.-Y. iTRAQ Proteomic Analysis of Continuously Cropped Soybean Root Inoculated With Funneliformis mosseae. Front. Microbiol. 2019, 10, 61. [Google Scholar] [CrossRef] [Green Version]
- Ranjan, A.; Westrick, N.M.; Jain, S.; Piotrowski, J.S.; Ranjan, M.; Kessens, R.; Stiegman, L.; Grau, C.R.; Conley, S.P.; Smith, D.; et al. Resistance against Sclerotinia sclerotiorum in soybean involves a reprogramming of the phenylpropanoid pathway and up-regulation of antifungal activity targeting ergosterol biosynthesis. Plant Biotechnol. J. 2019, 17, 1567–1581. [Google Scholar] [CrossRef] [Green Version]
- Parker, J.E.; Hahlbrock, K.; Scheel, D. Different cell-wall components from Phytophthora megasperma f. sp. glycinea elicit phytoalexin production in soybean and parsley. Planta 1988, 176, 75–82. [Google Scholar] [CrossRef]
- Graham, M.; Weidner, J.; Wheeler, K.; Pelow, M.; Graham, T. Induced expression of pathogenesis-related protein genes in soybean by wounding and the Phytophthora sojae cell wall glucan elicitor. Physiol. Mol. Plant Pathol. 2003, 63, 141–149. [Google Scholar] [CrossRef]
- Millet, Y.A.; Danna, C.H.; Clay, N.K.; Songnuan, W.; Simon, M.D.; Werck-Reichhart, D.; Ausubel, F.M. Innate Immune Responses Activated in Arabidopsis Roots by Microbe-Associated Molecular Patterns. Plant Cell 2010, 22, 973–990. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pearce, G.; Yamaguchi, Y.; Barona, G.; Ryan, C.A. A subtilisin-like protein from soybean contains an embedded, cryptic signal that activates defense-related genes. Proc. Natl. Acad. Sci. USA 2010, 107, 14921–14925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamaguchi, Y.; Barona, G.; Ryan, C.A.; Pearce, G. GmPep914, an Eight-Amino Acid Peptide Isolated from Soybean Leaves, Activates Defense-Related Genes. Plant Physiol. 2011, 156, 932–942. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.W.; Huffaker, A.; Crippen, D.; Robbins, R.T.; Goggin, F.L. Plant elicitor peptides promote plant defences against nematodes in soybean: Plant Elicitor Peptide-Induced Nematode Resistance. Mol. Plant Pathol. 2017, 19, 858–869. [Google Scholar] [CrossRef] [Green Version]
- Dong, H.; Shi, S.; Zhang, C.; Zhu, S.; Li, M.; Tan, J.; Yu, Y.; Lin, L.; Jia, S.; Wang, X.; et al. Transcriptomic analysis of genes in soybean in response to Peronospora manshurica infection. BMC Genom. 2018, 19, 366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morkunas, I.; Woźniak, A.; Formela, M.; Mai, V.C.; Marczak, L.; Narożna, D.; Borowiak-Sobkowiak, B.; Kühn, C.; Grimm, B. Pea aphid infestation induces changes in flavonoids, antioxidative defence, soluble sugars and sugar transporter expression in leaves of pea seedlings. Protoplasma 2015, 253, 1063–1079. [Google Scholar] [CrossRef] [PubMed]
- Jahan, A.; Harris, B.; Lowery, M.; Infante, A.M.; Percifield, R.J.; Kovinich, N. Glyceollin Transcription Factor GmMYB29A2 Regulates Soybean Resistance to Phytophthora sojae. Plant Physiol. 2020, 183, 530–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plancot, B.; Santaella, C.; Jaber, R.; Kiefer-Meyer, M.C.; Follet-Gueye, M.-L.; Leprince, J.; Gattin, I.; Souc, C.; Driouich, A.; Vicré-Gibouin, M. Deciphering the Responses of Root Border-Like Cells of Arabidopsis and Flax to Pathogen-Derived Elicitors. Plant Physiol. 2013, 163, 1584–1597. [Google Scholar] [CrossRef] [Green Version]
- Castilleux, R.; Plancot, B.; Ropitaux, M.; Carreras, A.; Leprince, J.; Boulogne, I.; Follet-Gueye, M.-L.; Popper, Z.A.; Driouich, A.; Vicré, M. Cell wall extensins in root–microbe interactions and root secretions. J. Exp. Bot. 2018, 69, 4235–4247. [Google Scholar] [CrossRef]
- Castilleux, R.; Plancot, B.; Vicré, M.; Nguema-Ona, E.; Driouich, A. Extensin, an underestimated key component of cell wall defence? Ann. Bot. 2021, 127, 709–713. [Google Scholar] [CrossRef]
- Castilleux, R.; Plancot, B.; Gügi, B.; Attard, A.; Loutelier-Bourhis, C.; Lefranc, B.; Nguema-Ona, E.; Arkoun, M.; Yvin, J.-C.; Driouich, A.; et al. Extensin arabinosylation is involved in root response to elicitors and limits oomycete colonization. Ann. Bot. 2019, 125, 751–763. [Google Scholar] [CrossRef] [PubMed]
- Cannesan, M.A.; Durand, C.; Burel, C.; Gangneux, C.; Lerouge, P.; Ishii, T.; Laval, K.; Follet-Gueye, M.-L.; Driouich, A.; Vicré-Gibouin, M. Effect of Arabinogalactan Proteins from the Root Caps of Pea and Brassica napus on Aphanomyces euteiches Zoospore Chemotaxis and Germination. Plant Physiol. 2012, 159, 1658–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koroney, A.S.; Plasson, C.; Pawlak, B.; Sidikou, R.; Driouich, A.; Menu-Bouaouiche, L.; Vicré-Gibouin, M. Root exudate of Solanum tuberosum is enriched in galactose-containing molecules and impacts the growth of Pectobacterium atrosepticum. Ann. Bot. 2016, 118, 797–808. [Google Scholar] [CrossRef] [Green Version]
- Wen, F.; VanEtten, H.D.; Tsaprailis, G.; Hawes, M.C. Extracellular Proteins in Pea Root Tip and Border Cell Exudates. Plant Physiol. 2006, 143, 773–783. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, F.; White, G.J.; VanEtten, H.D.; Xiong, Z.; Hawes, M.C. Extracellular DNA Is Required for Root Tip Resistance to Fungal Infection. Plant Physiol. 2009, 151, 820–829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ropitaux, M.; Bernard, S.; Schapman, D.; Follet-Gueye, M.-L.; Vicré, M.; Boulogne, I.; Driouich, A. Root Border Cells and Mucilage Secretions of Soybean, Glycine Max (Merr) L.: Characterization and Role in Interactions with the Oomycete Phytophthora Parasitica. Cells 2020, 9, 2215. [Google Scholar] [CrossRef] [PubMed]
- Cannesan, M.A.; Gangneux, C.; Lanoue, A.; Giron, D.; Laval, K.; Hawes, M.; Driouich, A.; Vicré-Gibouin, M. Association between border cell responses and localized root infection by pathogenic Aphanomyces euteiches. Ann. Bot. 2011, 108, 459–469. [Google Scholar] [CrossRef] [Green Version]
- Hawes, M.C.; Brigham, L.A.; Wen, F.; Woo, H.H.; Zhu, Y. Function of Root Border Cells in Plant Health: Pioneers in the Rhizosphere. Annu. Rev. Phytopathol. 1998, 36, 311–327. [Google Scholar] [CrossRef]
- Chuberre, C.; Plancot, B.; Driouich, A.; Moore, J.; Bardor, M.; Gügi, B.; Vicré, M. Plant Immunity Is Compartmentalized and Specialized in Roots. Front. Plant Sci. 2018, 9, 1692. [Google Scholar] [CrossRef] [Green Version]
- Driouich, A.; Gaudry, A.; Pawlak, B.; Moore, J.P. Root cap–derived cells and mucilage: A protective network at the root tip. Protoplasma 2021, 258, 1179–1185. [Google Scholar] [CrossRef]
- Chambard, M.; Plasson, C.; Derambure, C.; Coutant, S.; Tournier, I.; Lefranc, B.; Leprince, J.; Kiefer-Meyer, M.-C.; Driouich, A.; Follet-Gueye, M.-L.; et al. New Insights into Plant Extracellular DNA. A Study in Soybean Root Extracellular Trap. Cells 2021, 10, 69. [Google Scholar] [CrossRef] [PubMed]
- Afgan, E.; Baker, D.; van den Beek, M.; Blankenberg, D.; Bouvier, D.; Čech, M.; Chilton, J.; Clements, D.; Coraor, N.; Eberhard, C.; et al. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Res. 2016, 44, W3–W10. [Google Scholar] [CrossRef] [Green Version]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Kentache, T.; Ben Abdelkrim, A.; Jouenne, T.; Dé, E.; Hardouin, J. Global Dynamic Proteome Study of a Pellicle-forming Acinetobacter baumannii Strain. Mol. Cell. Proteom. 2017, 16, 100–112. [Google Scholar] [CrossRef] [Green Version]
- Obry, A.; Lequerré, T.; Hardouin, J.; Boyer, O.; Fardellone, P.; Philippe, P.; Le Loët, X.; Cosette, P.; Vittecoq, O. Identification of S100A9 as Biomarker of Responsiveness to the Methotrexate/Etanercept Combination in Rheumatoid Arthritis Using a Proteomic Approach. PLoS ONE 2014, 9, e115800. [Google Scholar] [CrossRef]
- Schläpfer, P.; Zhang, P.; Wang, C.; Kim, T.; Banf, M.; Chae, L.; Dreher, K.; Chavali, A.K.; Nilo-Poyanco, R.; Bernard, T.; et al. Genome-Wide Prediction of Metabolic Enzymes, Pathways, and Gene Clusters in Plants. Plant Physiol. 2017, 173, 2041–2059. [Google Scholar] [CrossRef] [Green Version]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-independent MS/MS deconvolution for comprehensive metabolome analysis. Nat. Methods 2015, 12, 523–526. [Google Scholar] [CrossRef]
- Eriksson, L.; Johansson, E.; Kettapeh-Wold, S.; Wold, S. Introduction to Multi- and Megavariate Data Analysis Using Projection Methods (PCA & PLS); Umetrics: Umeå, Sweden, 1999. [Google Scholar]
- van den Berg, R.A.; Hoefsloot, H.C.J.; Westerhuis, J.A.; Smilde, A.K.; Van Der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trygg, J.; Wold, S. Orthogonal projections to latent structures (O-PLS). J. Chemom. 2002, 16, 119–128. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the gap between raw spectra and functional insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deepak, S.; Shailasree, S.; Kini, R.K.; Muck, A.; Mithöfer, A.; Shetty, S.H. Hydroxyproline-rich Glycoproteins and Plant Defence. J. Phytopathol. 2010, 158, 585–593. [Google Scholar] [CrossRef]
- Tan, L.; Mort, A. Extensins at the front line of plant defence. A commentary on: ‘Extensin arabinosylation is involved in root response to elicitors and limits oomycete colonization’. Ann. Bot. 2020, 125, vii–viii. [Google Scholar] [CrossRef]
- Nguema-Ona, E.; Vicré-Gibouin, M.; Cannesan, M.-A.; Driouich, A. Arabinogalactan proteins in root–microbe interactions. Trends Plant Sci. 2013, 18, 440–449. [Google Scholar] [CrossRef]
- Laloum, Y.; Gangneux, C.; Gügi, B.; Lanoue, A.; Munsch, T.; Blum, A.; Gauthier, A.; Trinsoutrot-Gattin, I.; Boulogne, I.; Vicré, M.; et al. Faba bean root exudates alter pea root colonization by the oomycete Aphanomyces euteiches at early stages of infection. Plant Sci. 2021, 312, 111032. [Google Scholar] [CrossRef]
- Li, A.X.; Han, Y.Y.; Wang, X.; Chen, Y.H.; Zhao, M.R.; Zhou, S.-M.; Wang, W. Root-specific expression of wheat expansin gene TaEXPB23 enhances root growth and water stress tolerance in tobacco. Environ. Exp. Bot. 2015, 110, 73–84. [Google Scholar] [CrossRef]
- Cosgrove, D.J. Plant expansins: Diversity and interactions with plant cell walls. Curr. Opin. Plant Biol. 2015, 25, 162–172. [Google Scholar] [CrossRef] [Green Version]
- Song, L.; Valliyodan, B.; Prince, S.; Wan, J.; Nguyen, H.T. Characterization of the XTH Gene Family: New Insight to the Roles in Soybean Flooding Tolerance. Int. J. Mol. Sci. 2018, 19, 2705. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ropitaux, M.; Bernard, S.; Follet-Gueye, M.-L.; Vicré, M.; Boulogne, I.; Driouich, A. Xyloglucan and cellulose form molecular cross-bridges connecting root border cells in pea (Pisum sativum). Plant Physiol. Biochem. 2019, 139, 191–196. [Google Scholar] [CrossRef] [PubMed]
- Micheli, F. Pectin methylesterases: Cell wall enzymes with important roles in plant physiology. Trends Plant Sci. 2001, 6, 414–419. [Google Scholar] [CrossRef]
- Mravec, J.; Guo, X.; Hansen, A.R.; Schückel, J.; Kračun, S.K.; Mikkelsen, M.D.; Mouille, G.; Johansen, I.E.; Ulvskov, P.; Domozych, D.S.; et al. Pea Border Cell Maturation and Release Involve Complex Cell Wall Structural Dynamics. Plant Physiol. 2017, 174, 1051–1066. [Google Scholar] [CrossRef] [Green Version]
- Pandey, S. Heterotrimeric G-Protein Signaling in Plants: Conserved and Novel Mechanisms. Annu. Rev. Plant Biol. 2019, 70, 213–238. [Google Scholar] [CrossRef] [PubMed]
- Wildermuth, M.C. Plants fight fungi using kiwellin proteins. Nature 2019, 565, 575–577. [Google Scholar] [CrossRef] [PubMed]
- Ooka, H.; Satoh, K.; Doi, K.; Nagata, T.; Otomo, Y.; Murakami, K.; Matsubara, K.; Osato, N.; Kawai, J.; Carninci, P.; et al. Comprehensive Analysis of NAC Family Genes in Oryza sativa and Arabidopsis thaliana. DNA Res. 2003, 10, 239–247. [Google Scholar] [CrossRef]
- Hao, Y.-J.; Wei, W.; Song, Q.-X.; Chen, H.-W.; Zhang, Y.-Q.; Wang, F.; Zou, H.-F.; Lei, G.; Tian, A.-G.; Zhang, W.-K.; et al. Soybean NAC transcription factors promote abiotic stress tolerance and lateral root formation in transgenic plants. Plant J. 2011, 68, 302–313. [Google Scholar] [CrossRef]
- Hussain, R.M.; Ali, M.; Feng, X.; Li, X. The essence of NAC gene family to the cultivation of drought-resistant soybean (Glycine max L. Merr.) cultivars. BMC Plant Biol. 2017, 17, 55. [Google Scholar] [CrossRef] [Green Version]
- Cândido, E.D.S.; Pinto, M.F.S.; Pelegrini, P.B.; Lima, T.B.; Silva, O.N.; Pogue, R.; Grossi-De-Sá, M.F.; Franco, O.L. Plant storage proteins with antimicrobial activity: Novel insights into plant defense mechanisms. FASEB J. 2011, 25, 3290–3305. [Google Scholar] [CrossRef]
- D’Ambrosio, J.M.; Couto, D.; Fabro, G.; Scuffi, D.; Lamattina, L.; Munnik, T.; Andersson, M.X.; Alvarez, M.E.; Zipfel, C.; Laxalt, A.M. Phospholipase C2 Affects MAMP-Triggered Immunity by Modulating ROS Production. Plant Physiol. 2017, 175, 970–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Wang, X. Phospholipase D and phosphatidic acid in plant immunity. Plant Sci. 2018, 279, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Hakim; Ullah, A.; Hussain, A.; Shaban, M.; Khan, A.H.; Alariqi, M.; Gul, S.; Jun, Z.; Lin, S.; Li, J.; et al. Osmotin: A plant defense tool against biotic and abiotic stresses. Plant Physiol. Biochem. 2018, 123, 149–159. [Google Scholar] [CrossRef] [PubMed]
- Vandana, V.V.; Bhai, R.S. Differential expression of PR genes in response to Phytophthora capsici inoculation in resistant and susceptible black pepper (Piper nigrum L.) lines. Eur. J. Plant Pathol. 2017, 150, 713–724. [Google Scholar] [CrossRef]
- Dunwell, J.; Gibbings, J.G.; Mahmood, T.; Naqvi, S.M.S. Germin and Germin-like Proteins: Evolution, Structure, and Function. Crit. Rev. Plant Sci. 2008, 27, 342–375. [Google Scholar] [CrossRef]
- Phan, L.C.H.B.T.; Van Dijck, P. Biosynthesis and Degradation of Trehalose and Its Potential to Control Plant Growth, Development, and (A) Biotic Stress Tolerance. In Osmoprotectant-Mediated Abiotic Stress Tolerance in Plants: Recent Advances and Future Perspectives; Hossain, M.A., Kumar, V., Burritt, D.J., Fujita, M., Mäkelä, P.S.A., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 175–199. ISBN 978-3-030-27423-8. [Google Scholar]
- Yan, Q.; Si, J.; Cui, X.; Peng, H.; Chen, X.; Xing, H.; Dou, D. The soybean cinnamate 4-hydroxylase gene GmC4H1 contributes positively to plant defense via increasing lignin content. Plant Growth Regul. 2019, 88, 139–149. [Google Scholar] [CrossRef]
- Wang, X.; Zhao, Z.; Guo, N.; Wang, H.; Zhao, J.; Xing, H. Comparative Proteomics Analysis Reveals That Lignin Biosynthesis Contributes to Brassinosteroid-Mediated Response to Phytophthora sojae in Soybeans. J. Agric. Food Chem. 2020, 68, 5496–5506. [Google Scholar] [CrossRef]
- Tiwari, S.; Shweta, S.; Prasad, M.; Lata, C. Genome-wide investigation of GRAM-domain containing genes in rice reveals their role in plant-rhizobacteria interactions and abiotic stress responses. Int. J. Biol. Macromol. 2019, 156, 1243–1257. [Google Scholar] [CrossRef]
- Ramongolalaina, C. Dynamics of symbiotic relationship of soybean with Bradyrhizobium diazoefficiens and involvements of root-secreted daidzein behind the continuous cropping. Eur. J. Soil Biol. 2019, 93, 103098. [Google Scholar] [CrossRef]
- Morris, P.; Ward, E. Chemoattraction of zoospores of the soybean pathogen, Phytophthora sojae, by isoflavones. Physiol. Mol. Plant Pathol. 1992, 40, 17–22. [Google Scholar] [CrossRef]
- Farrell, K.; Jahan, A.; Kovinich, N. Distinct Mechanisms of Biotic and Chemical Elicitors Enable Additive Elicitation of the Anticancer Phytoalexin Glyceollin I. Molecules 2017, 22, 1261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Faizal, A.; Geelen, D. Saponins and their role in biological processes in plants. Phytochem. Rev. 2013, 12, 877–893. [Google Scholar] [CrossRef]
- Tsuno, Y.; Fujimatsu, T.; Endo, K.; Sugiyama, A.; Yazaki, K. Soyasaponins: A New Class of Root Exudates in Soybean (Glycine max). Plant Cell Physiol. 2017, 59, 366–375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kruger, N.J.; von Schaewen, A. The oxidative pentose phosphate pathway: Structure and organisation. Curr. Opin. Plant Biol. 2003, 6, 236–246. [Google Scholar] [CrossRef]
- Choque, B.; Catheline, D.; Rioux, V.; Legrand, P. Linoleic acid: Between doubts and certainties. Biochimie 2014, 96, 14–21. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Wang, X.; Zhang, F.; Dong, L.; Wu, J.; Cheng, Q.; Qi, D.; Yan, X.; Jiang, L.; Fan, S.; et al. Phenylalanine ammonia-lyase2.1 contributes to the soybean response towards Phytophthora sojae infection. Sci. Rep. 2017, 7, 7242. [Google Scholar] [CrossRef] [Green Version]
- González, B.; Vera, P. Folate Metabolism Interferes with Plant Immunity through 1C Methionine Synthase-Directed Genome-wide DNA Methylation Enhancement. Mol. Plant 2019, 12, 1227–1242. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Beard, K.F.; Nunes-Nesi, A.; Fernie, A.R.; Ratcliffe, R.G. Not just a circle: Flux modes in the plant TCA cycle. Trends Plant Sci. 2010, 15, 462–470. [Google Scholar] [CrossRef]
- Rudrappa, T.; Czymmek, K.J.; Paré, P.W.; Bais, H.P. Root-Secreted Malic Acid Recruits Beneficial Soil Bacteria. Plant Physiol. 2008, 148, 1547–1556. [Google Scholar] [CrossRef] [Green Version]
- Oliva, M.; Hatan, E.; Kumar, V.; Galsurker, O.; Nisim-Levi, A.; Ovadia, R.; Galili, G.; Lewinsohn, E.; Elad, Y.; Alkan, N.; et al. Increased phenylalanine levels in plant leaves reduces susceptibility to Botrytis cinerea. Plant Sci. 2019, 290, 110289. [Google Scholar] [CrossRef] [PubMed]
- Patel, M.K.; Maurer, D.; Feygenberg, O.; Ovadia, A.; Elad, Y.; Oren-Shamir, M.; Alkan, N. Phenylalanine: A Promising Inducer of Fruit Resistance to Postharvest Pathogens. Foods 2020, 9, 646. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, R.P.; Chen, L.; Schwier, M.; Koprivova, A.; Kopriva, S. Recent advances in the role of plant metabolites in shaping the root microbiome. F1000Research 2020, 9, 151. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.; Muthreich, N.; Liao, C.; Franz-Wachtel, M.; Schütz, W.; Zhang, F.; Hochholdinger, F.; Li, C. The Mucilage Proteome of Maize (Zea mays L.) Primary Roots. J. Proteome Res. 2010, 9, 2968–2976. [Google Scholar] [CrossRef]
- Weiller, F.; Moore, J.P.; Young, P.; Driouich, A.; Vivier, M.A. The Brassicaceae species Heliophila coronopifolia Produces root border-like cells that protect the root tip and secrete defensin peptides. Ann. Bot. 2016, 119, 803–813. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Lu, S.; Liu, K.; Wang, S.; Huang, L.; Guo, L. Proteomics: A powerful tool to study plant responses to biotic stress. Plant Methods 2019, 15, 135. [Google Scholar] [CrossRef] [PubMed]
- Chivasa, S.; Simon, W.J.; Yu, X.-L.; Yalpani, N.; Slabas, A.R. Pathogen elicitor-induced changes in the maize extracellular matrix proteome. Proteomics 2005, 5, 4894–4904. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J. Flavonoid transport mechanisms: How to go, and with whom. Trends Plant Sci. 2015, 20, 576–585. [Google Scholar] [CrossRef]
- Hawes, M.C.; Pueppke, S.G. Sloughed Peripheral Root Cap Cells: Yield from Different Species and Callus Formation from Single Cells. Am. J. Bot. 1986, 73, 1466–1473. [Google Scholar] [CrossRef]
- Hawes, M.; Allen, C.; Turgeon, B.G.; Curlango-Rivera, G.; Tran, T.M.; Huskey, D.A.; Xiong, Z. Root Border Cells and Their Role in Plant Defense. Annu. Rev. Phytopathol. 2016, 54, 143–161. [Google Scholar] [CrossRef]
- Wen, F.; Curlango-Rivera, G.; Huskey, D.A.; Xiong, Z.; Hawes, M.C. Visualization of extracellular DNA released during border cell separation from the root cap. Am. J. Bot. 2017, 104, 970–978. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chambard, M.; Ben Mlouka, M.A.; Jing, L.; Plasson, C.; Cosette, P.; Leprince, J.; Follet-Gueye, M.-L.; Driouich, A.; Nguema-Ona, E.; Boulogne, I. Elicitation of Roots and AC-DC with PEP-13 Peptide Shows Differential Defense Responses in Multi-Omics. Cells 2022, 11, 2605. https://doi.org/10.3390/cells11162605
Chambard M, Ben Mlouka MA, Jing L, Plasson C, Cosette P, Leprince J, Follet-Gueye M-L, Driouich A, Nguema-Ona E, Boulogne I. Elicitation of Roots and AC-DC with PEP-13 Peptide Shows Differential Defense Responses in Multi-Omics. Cells. 2022; 11(16):2605. https://doi.org/10.3390/cells11162605
Chicago/Turabian StyleChambard, Marie, Mohamed Amine Ben Mlouka, Lun Jing, Carole Plasson, Pascal Cosette, Jérôme Leprince, Marie-Laure Follet-Gueye, Azeddine Driouich, Eric Nguema-Ona, and Isabelle Boulogne. 2022. "Elicitation of Roots and AC-DC with PEP-13 Peptide Shows Differential Defense Responses in Multi-Omics" Cells 11, no. 16: 2605. https://doi.org/10.3390/cells11162605
APA StyleChambard, M., Ben Mlouka, M. A., Jing, L., Plasson, C., Cosette, P., Leprince, J., Follet-Gueye, M. -L., Driouich, A., Nguema-Ona, E., & Boulogne, I. (2022). Elicitation of Roots and AC-DC with PEP-13 Peptide Shows Differential Defense Responses in Multi-Omics. Cells, 11(16), 2605. https://doi.org/10.3390/cells11162605