An Optimized Comparative Proteomic Approach as a Tool in Neurodegenerative Disease Research
Abstract
1. Introduction
2. Proteomics as Applied to Neurodegenerative Disease Research
3. Considerations, Not Limitations
- Sample collection and preparation of in vitro cells, in vivo tissue from animal models of disease, or post mortem tissue from end-stage patients or age-matched controls.
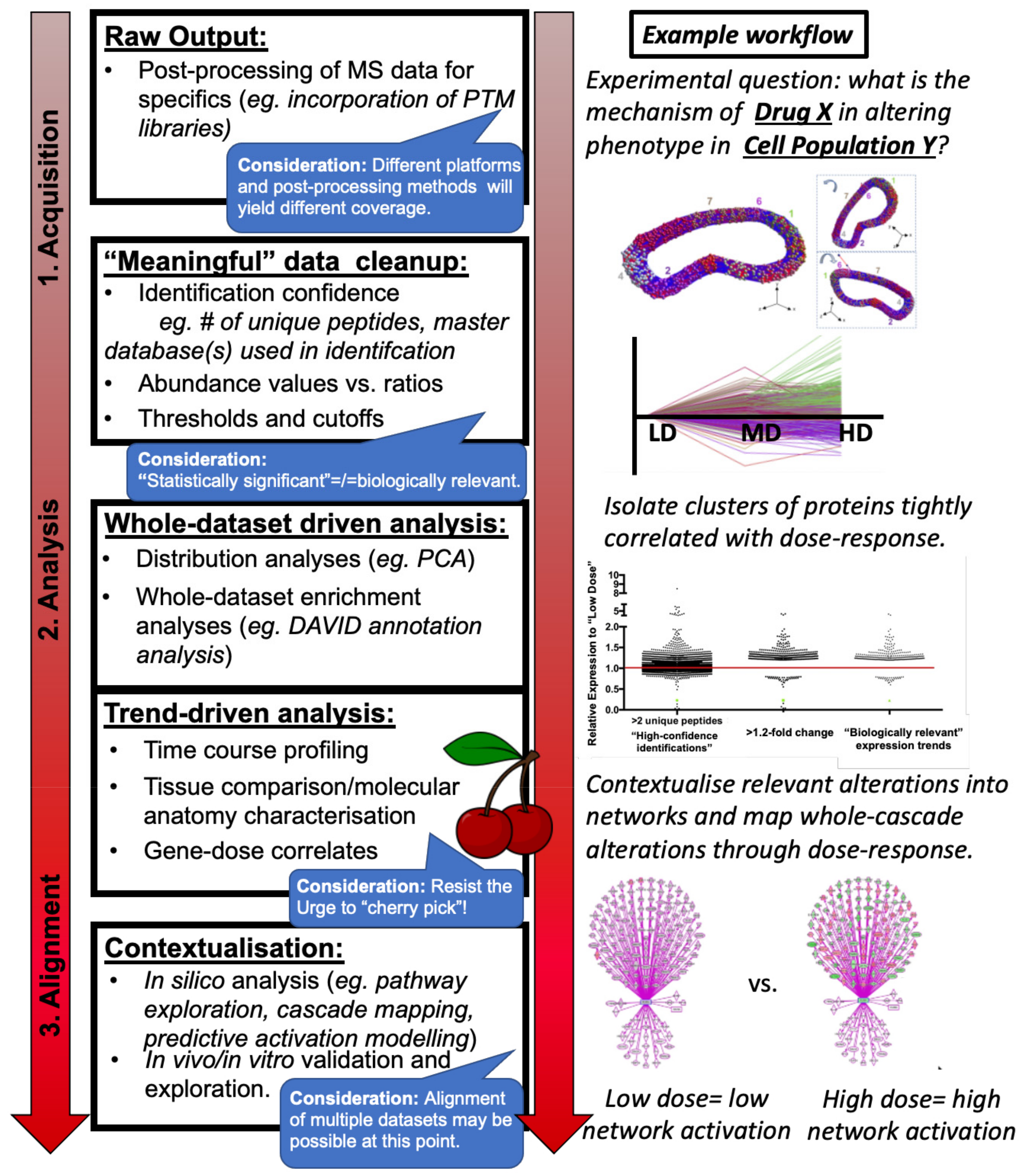
- Protein identification and absolute or relative quantitation within sample(s) including appropriate statistical analysis and multi-database searches, generally conducted within one of several commonly used proteomics software (Figure 1: Raw Output).
- Post-mass spectrometry filtering, if appropriate (Figure 1: “Meaningful Data Cleanup”) and:
3.1. Sample Collection
3.2. Mass Spectrometry Experiment and Proteomic Data Analysis
3.2.1. Quantitative Proteomic Profiling via Mass Spectrometry
3.2.2. Pre-Processing Analysis and Normalisation
3.2.3. Post-Processing Analysis
3.2.4. Data Deposition
3.3. Downstream Data Analysis
3.3.1. Enrichment Analysis: As a Whole vs. for Individual Trends
3.3.2. Network and Pathway Analysis
4. Comparative Proteomics May Uncover Common Regulators of Neuronal Stability within and between Distinct Models of Disease
4.1. Comparative Approaches for Complex or Unknown Aetiologies
4.2. Incorporation of Human Proteome Analysis
5. Concluding Commentary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
Appendix A.1. Methods
Appendix A.1.1. Source Datasets
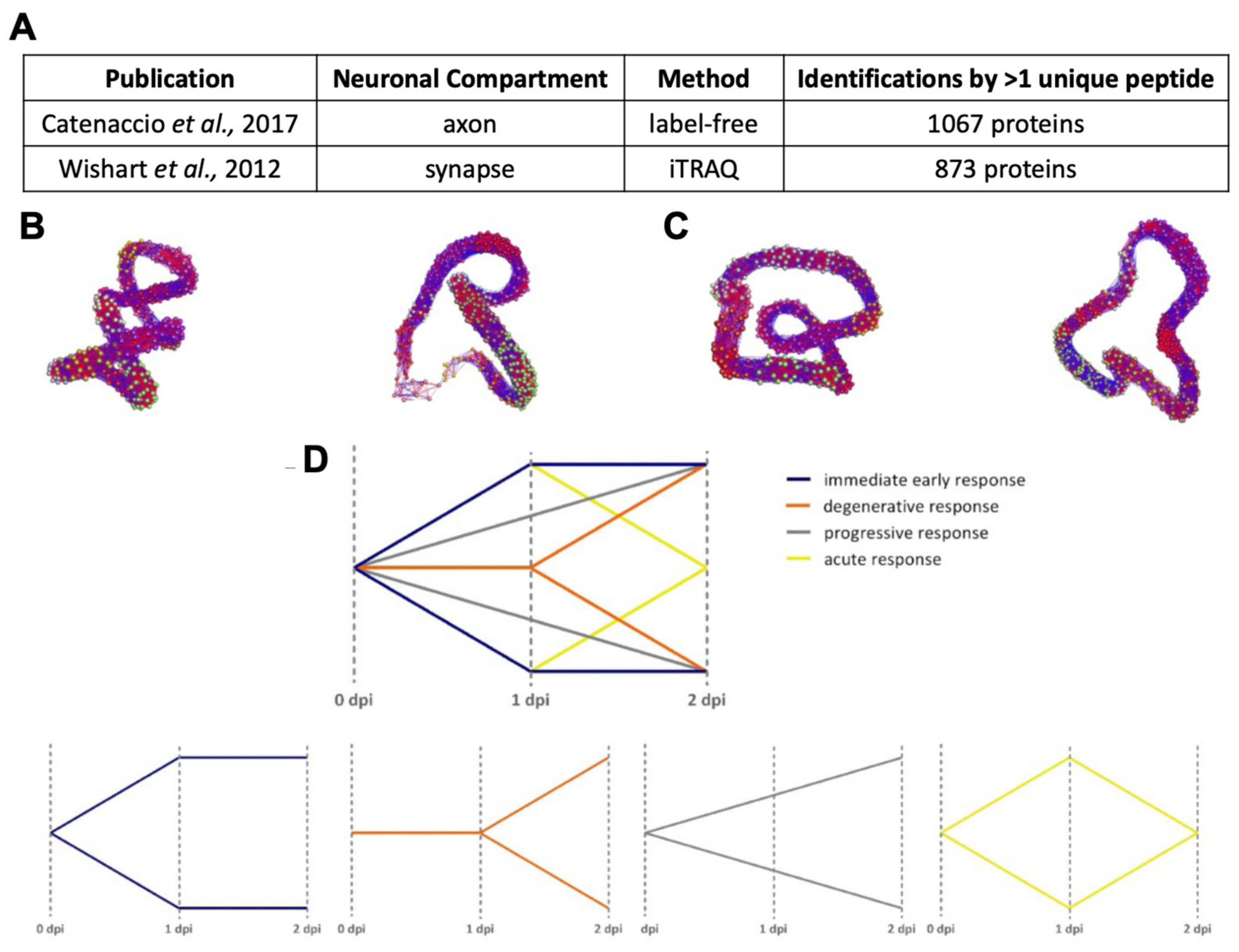
Appendix A.1.2. Standardisation for Alignment
Appendix A.1.3. BioLayout Express3D
Appendix A.1.4. DAVID
Appendix A.1.5. Ingenuity Pathway Analysis
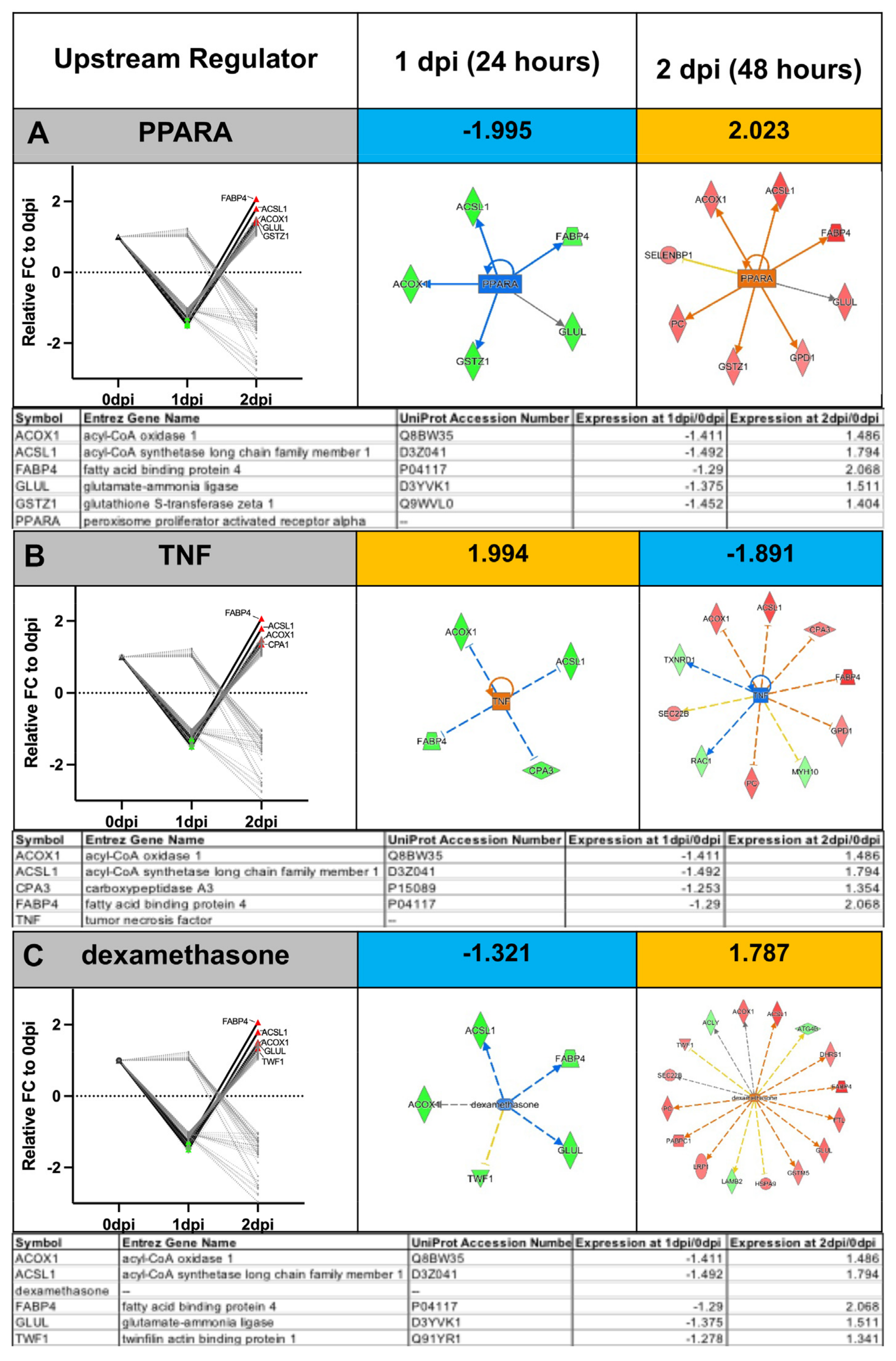

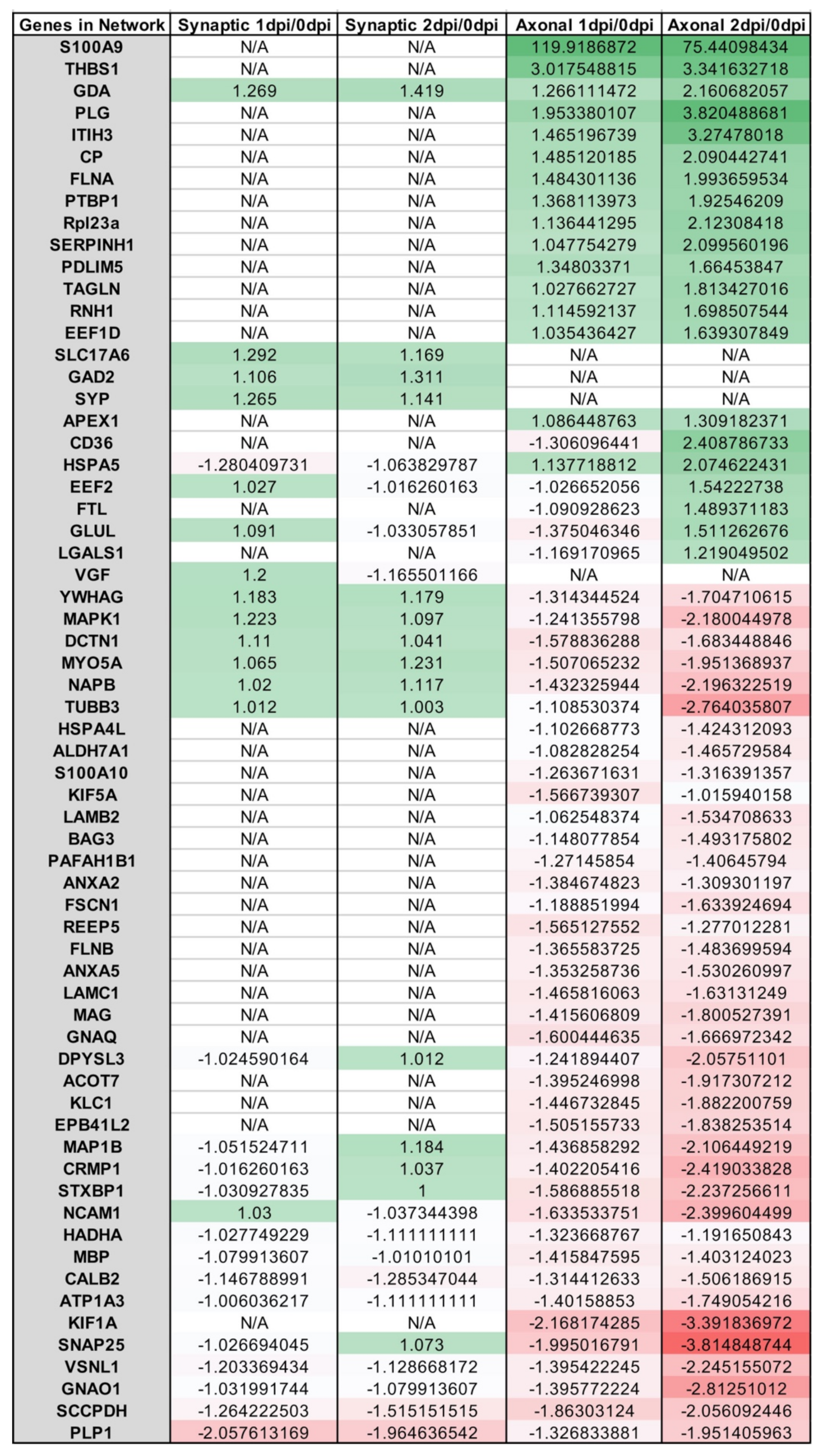
Appendix A.2. Example Results
References
- Ramdas, S.; Servais, L. New treatments in spinal muscular atrophy: An overview of currently available data. Expert Opin. Pharmacother. 2020, 21, 307–315. [Google Scholar] [CrossRef] [PubMed]
- Mendell, J.R.; Al-Zaidy, S.; Shell, R.; Arnold, W.D.; Rodino-Klapac, L.R.; Prior, T.W.; Lowes, L.; Alfano, L.; Berry, K.; Church, K.; et al. Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy. N120ew Engl. J. Med. 2017, 377, 1713–1722. [Google Scholar] [CrossRef]
- Shorrock, H.K.; Gillingwater, T.H.; Groen, E.J.N. Overview of Current Drugs and Molecules in Development for Spinal Muscular Atrophy Therapy. Drugs 2018, 78, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Vuillemenot, B.R.; Kennedy, D.; Reed, R.P.; Boyd, R.B.; Butt, M.T.; Musson, D.G.; Keve, S.; Cahayag, R.; Tsuruda, L.S.; O’Neill, C.A. Recombinant human tripeptidyl peptidase-1 infusion to the monkey CNS: Safety, pharmacokinetics, and distribution. Toxicol. Appl. Pharmacol. 2014, 277, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Markham, A. Cerliponase Alfa: First Global Approval. Drugs 2017, 77, 1247–1249. [Google Scholar] [CrossRef] [PubMed]
- Specchio, N.; Pietrafusa, N.; Trivisano, M. Changing Times for CLN2 Disease: The Era of Enzyme Replacement Therapy. Ther. Clin. Risk Manag. 2020, 16, 213–222. [Google Scholar] [CrossRef]
- Schulz, A.; Ajayi, T.; Specchio, N.; Reyes, E.D.L.; Gissen, P.; Ballon, D.; Dyke, J.P.; Cahan, H.; Slasor, P.; Jacoby, D.; et al. Study of Intraventricular Cerliponase Alfa for CLN2 Disease. N. Engl. J. Med. 2018, 378, 1898–1907. [Google Scholar] [CrossRef]
- Fridman, V.; Suriyanarayanan, S.; Novak, P.; David, W.; Macklin, E.A.; McKenna-Yasek, D.; Walsh, K.; Aziz-Bose, R.; Oaklander, A.L.; Brown, R.; et al. Randomized trial of l-serine in patients with hereditary sensory and autonomic neuropathy type 1. Neurology 2019, 92, e359–e370. [Google Scholar] [CrossRef]
- Jones, R.A.; Harrison, C.; Eaton, S.L.; Hurtado, M.L.; Graham, L.C.; Alkhammash, L.; Oladiran, O.A.; Gale, A.; Lamont, D.J.; Simpson, H.; et al. Cellular and Molecular Anatomy of the Human Neuromuscular Junction. Cell Rep. 2017, 21, 2348–2356. [Google Scholar] [CrossRef]
- Eaton, S.L.; Wishart, T.M. Bridging the gap: Large animal models in neurodegenerative research. Mamm. Genome 2017, 28, 324–337. [Google Scholar] [CrossRef]
- Nelvagal, H.R.; Lange, J.; Takahashi, K.; Tarczyluk-Wells, M.A.; Cooper, J.D. Pathomechanisms in the neuronal ceroid lipofuscinoses. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2020, 1866, 165570. [Google Scholar]
- Cooper, J.D.; Tarczyluk, M.A.; Nelvagal, H.R. Towards a new understanding of NCL pathogenesis. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2015, 1852, 2256–2261. [Google Scholar] [CrossRef]
- Dawson, T.M.; Golde, T.E.; Lagier-Tourenne, C. Animal models of neurodegenerative diseases. Nat. Neurosci. 2018, 21, 1370–1379. [Google Scholar] [CrossRef] [PubMed]
- Centa, J.L.; Jodelka, F.M.; Hinrich, A.J.; Johnson, T.B.; Ochaba, J.; Jackson, M.; Duelli, D.M.; Weimer, J.M.; Rigo, F.; Hastings, M.L. Therapeutic efficacy of antisense oligonucleotides in mouse models of CLN3 Batten disease. Nat. Med. 2020, 26, 1444–1451. [Google Scholar] [CrossRef] [PubMed]
- Goetz, C.G. The History of Parkinson’s Disease: Early Clinical Descriptions and Neurological Therapies. Cold Spring Harb. Perspect. Med. 2011, 1, a008862. [Google Scholar] [CrossRef]
- Stelzmann, R.A.; Schnitzlein, H.N.; Murtagh, F.R. An english translation of alzheimer’s 1907 paper, “über eine eigenartige erkankung der hirnrinde”. Clin. Anat. 1995, 8, 429–431. [Google Scholar]
- Kay, K.R.; Smith, C.; Wright, A.K.; Serrano-Pozo, A.; Pooler, A.M.; Koffie, R.; Bastin, M.E.; Bak, T.H.; Abrahams, S.; Kopeikina, K.J.; et al. Studying synapses in human brain with array tomography and electron microscopy. Nat. Protoc. 2013, 8, 1366–1380. [Google Scholar] [CrossRef]
- Corder, E.H.; Saunders, A.M.; Strittmatter, W.J.; Schmechel, D.E.; Gaskell, P.C.; Small, G.W.; Roses, A.D.; Haines, J.L.; Pericak-Vance, M.A. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer's disease in late onset families. Science 1993, 261, 921–923. [Google Scholar] [CrossRef]
- Rebeck, G.W.; Reiter, J.S.; Strickland, D.K.; Hyman, B.T. Apolipoprotein E in sporadic Alzheimer’s disease: Allelic variation and receptor interactions. Neuron 1993, 11, 575–580. [Google Scholar] [CrossRef]
- Chartier-Hariln, M.-C.; Parfitt, M.; Legrain, S.; Pérez-Tur, J.; Brousseau, T.; Evans, A.; Berr, C.; Vldal, O.; Roques, P.; Gourlet, V.; et al. Apolipoprotein e, ε4 allele as a major risk factor for sporadic early and late-onset forms of alzheimer’s disease: Analysis of the 19q13.2 chromosomal region. Human Mol. Genet. 1994, 3, 569–574. [Google Scholar]
- Mauricio, R.; Benn, C.; Davis, J.; Dawson, G.; Dawson, L.A.; Evans, A.; Fox, N.; Gallacher, J.; Hutton, M.; Isaac, J.; et al. Tackling gaps in developing life-changing treatments for dementia. Alzheimer’s Dement. Transl. Res. Clin. Interv. 2019, 5, 241–253. [Google Scholar]
- Jankowsky, J.L.; Zheng, H. Practical considerations for choosing a mouse model of Alzheimer’s disease. Mol. Neurodegener. 2017, 12, 1–22. [Google Scholar] [CrossRef]
- Jagmag, S.A.; Tripathi, N.; Shukla, S.D.; Maiti, S.; Khurana, S. Evaluation of Models of Parkinson's Disease. Front. Neurosci. 2016, 9, 503. [Google Scholar] [CrossRef] [PubMed]
- Hesse, R.; Hurtado, M.L.; Jackson, R.J.; Eaton, S.L.; Herrmann, A.G.; Colom-Cadena, M.; Tzioras, M.; King, D.; Rose, J.; Tulloch, J.; et al. Comparative profiling of the synaptic proteome from Alzheimer’s disease patients with focus on the APOE genotype. Acta Neuropathol. Commun. 2019, 7, 1–18. [Google Scholar] [CrossRef]
- Wingo, A.P.; Dammer, E.B.; Breen, M.S.; Logsdon, B.A.; Duong, D.M.; Troncosco, J.C.; Thambisetty, M.; Beach, T.G.; Serrano, G.E.; Reiman, E.M.; et al. Large-scale proteomic analysis of human brain identifies proteins associated with cognitive trajectory in advanced age. Nat. Commun. 2019, 10, 1619. [Google Scholar] [CrossRef]
- Sharma, K.; Schmitt, S.; Bergner, C.; Tyanovaet, S.; Kannaiyanal, N.; Manrique-Hoyos, N.; Kongi, K.; Cantuti, L.; Hanisch, U.-K.; Philips, M.-A.; et al. Cell type- and brain region-resolved mouse brain proteome. Nat. Neurosci. 2015, 18, 1819–1831. [Google Scholar]
- Carlyle, B.; Kitchen, R.R.; Kanyo, J.E.; Voss, E.Z.; Pletikos, M.; Sousa, A.M.M.; Lam, T.T.; Gerstein, M.B.; Sestan, N.; Nairn, A.C. A multiregional proteomic survey of the postnatal human brain. Nat. Neurosci. 2017, 20, 1787–1795. [Google Scholar] [CrossRef]
- Muraoka, S.; Jedrychowski, M.P.; Iwahara, N.; Abdullah, M.; Onos, K.D.; Keezer, K.J.; Hu, J.; Ikezu, S.; Howell, G.R.; Gygi, S.P.; et al. Enrichment of Neurodegenerative Microglia Signature in Brain-Derived Extracellular Vesicles Isolated from Alzheimer’s Disease Mouse Models. J. Proteome Res. 2021, 20, 1733–1743. [Google Scholar] [CrossRef]
- Grabert, K.; Michoel, T.; Karavolos, M.H.; Clohisey, S.; Baillie, J.K.; Stevens, M.P.; Freeman, T.C.; Summers, K.M.; McColl, B.W. Microglial brain region—Dependent diversity and selective regional sensitivities to aging. Nat. Neurosci. 2016, 19, 504–516. [Google Scholar]
- Lana, D.; Ugolini, F.; Nosi, D.; Wenk, G.L.; Giovannini, M.G. The Emerging Role of the Interplay Among Astrocytes, Microglia, and Neurons in the Hippocampus in Health and Disease. Front. Aging Neurosci. 2021, 13, 651973. [Google Scholar] [CrossRef]
- Koellhoffer, E.C.; McCullough, L.D.; Ritzel, R. Old Maids: Aging and Its Impact on Microglia Function. Int. J. Mol. Sci. 2017, 18, 769. [Google Scholar] [CrossRef]
- Hunter, G.; Sarvestany, A.A.; Roche, S.L.; Symes, R.C.; Gillingwater, T.H. SMN-dependent intrinsic defects in Schwann cells in mouse models of spinal muscular atrophy. Hum. Mol. Genet. 2013, 23, 2235–2250. [Google Scholar] [CrossRef] [PubMed]
- Abati, E.; Citterio, G.; Bresolin, N.; Comi, G.P.; Corti, S. Glial cells involvement in spinal muscular atrophy: Could SMA be a neuroinflammatory disease? Neurobiol. Dis. 2020, 140, 104870. [Google Scholar] [CrossRef]
- Garner, J.P. The Significance of Meaning: Why Do Over 90% of Behavioral Neuroscience Results Fail to Translate to Humans, and What Can We Do to Fix It? ILAR J. 2014, 55, 438–456. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, B.T.; Livesey, M.R.; Chandran, S. Modeling the C9ORF72 repeat expansion mutation using human induced pluripotent stem cells. Brain Pathol. 2017, 27, 518–524. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tagawa, K.; Homma, H.; Saito, A.; Fujita, K.; Chen, X.; Imoto, S.; Oka, T.; Ito, H.; Motoki, K.; Yoshida, C.; et al. Comprehensive phosphoproteome analysis unravels the core signaling network that initiates the earliest synapse pathology in preclinical Alzheimer's disease brain. Hum. Mol. Genet. 2014, 24, 540–558. [Google Scholar] [CrossRef]
- Eaton, S.L.; Hurtado, M.L.; Oldknow, K.J.; Graham, L.C.; Marchant, T.W.; Gillingwater, T.; Farquharson, C.; Wishart, T.M. A Guide to Modern Quantitative Fluorescent Western Blotting with Troubleshooting Strategies. J. Vis. Exp. 2014, e52099. [Google Scholar] [CrossRef]
- Wilson, R.S.; Nairn, A.C. Cell-Type-Specific Proteomics: A Neuroscience Perspective. Proteomes 2018, 6, 51. [Google Scholar] [CrossRef]
- Shevchenko, G.; Musunuri, S.; Wetterhall, M.; Bergquist, J. Comparison of Extraction Methods for the Comprehensive Analysis of Mouse Brain Proteome using Shotgun-based Mass Spectrometry. J. Proteome Res. 2012, 11, 2441–2451. [Google Scholar] [CrossRef]
- Gulyássy, P.; Puska, G.; Györffy, B.A.; Todorov-Völgyi, K.; Juhász, G.; Drahos, L.; Kékesi, K.A. Proteomic comparison of different synaptosome preparation procedures. Amino Acids 2020, 52, 1529–1543. [Google Scholar] [CrossRef]
- Mattei, D.; Ivanov, A.; Oostrum, M.; Pantelyushin, S.; Richetto, J.; Mueller, F.; Beffinger, M.; Schellhammer, L.; Berg, J.; Wollscheid, B.; et al. Enzymatic Dissociation Induces Transcriptional and Proteotype Bias in Brain Cell Populations. Int. J. Mol. Sci. 2020, 21, 7944. [Google Scholar] [CrossRef] [PubMed]
- Hunter, G.; Roche, S.L.; Somers, E.; Fuller, H.R.; Gillingwater, T.H. The influence of storage parameters on measurement of survival motor neuron (SMN) protein levels: Implications for pre-clinical studies and clinical trials for spinal muscular atrophy. Neuromuscul. Disord. 2014, 24, 973–977. [Google Scholar] [CrossRef] [PubMed]
- Sleat, D.E.; Tannous, A.; Sohar, I.; Wiseman, J.A.; Zheng, H.; Qian, M.; Zhao, C.; Xin, W.; Barone, R.; Sims, K.B.; et al. Proteomic Analysis of Brain and Cerebrospinal Fluid from the Three Major Forms of Neuronal Ceroid Lipofuscinosis Reveals Potential Biomarkers. J. Proteome Res. 2017, 16, 3787–3804. [Google Scholar] [CrossRef] [PubMed]
- Tannous, A.; Boonen, M.; Zheng, H.; Zhao, C.; Germain, C.J.; Moore, D.F.; Sleat, D.E.; Jadot, M.; Lobel, P. Comparative Analysis of Quantitative Mass Spectrometric Methods for Subcellular Proteomics. J. Proteome Res. 2020, 19, 1718–1730. [Google Scholar] [CrossRef] [PubMed]
- Perkins, D.N.; Pappin, D.J.C.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Geer, L.Y.; Markey, S.P.; Kowalak, J.A.; Wagner, L.; Xu, M.; Maynard, D.M.; Yang, X.; Shi, W.; Bryant, S.H. Open Mass Spectrometry Search Algorithm. J. Proteome Res. 2004, 3, 958–964. [Google Scholar] [CrossRef] [PubMed]
- Tabb, D.L.; Fernando, C.G.; Chambers, M.C. MyriMatch: Highly Accurate Tandem Mass Spectral Peptide Identification by Multivariate Hypergeometric Analysis. J. Proteome Res. 2007, 6, 654–661. [Google Scholar] [CrossRef]
- Cox, J.; Neuhauser, N.; Michalski, A.; Scheltema, R.A.; Olsen, J.V.; Mann, M. Andromeda: A Peptide Search Engine Integrated into the MaxQuant Environment. J. Proteome Res. 2011, 10, 1794–1805. [Google Scholar] [CrossRef]
- Eng, J.K.; Jahan, T.A.; Hoopmann, M.R. Comet: An open-source MS/MS sequence database search tool. Proteomics 2013, 13, 22–24. [Google Scholar] [CrossRef]
- Kong, A.T.; LePrevost, F.V.; Avtonomov, D.M.; Mellacheruvu, D.; Nesvizhskii, A.I. MSFragger: Ultrafast and comprehensive peptide identification in mass spectrometry–based proteomics. Nat. Methods 2017, 14, 513–520. [Google Scholar] [CrossRef]
- Elias, J.E.; Gygi, S.P. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat. Methods 2007, 4, 207–214. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Ma, B. Adepts: Advanced peptide de novo sequencing with a pair of tandem mass spectra. J. Bioinform. Comput. Biol. 2010, 8, 981–994. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.H.; Zhang, X.; Xin, L.; Shan, B.; Li, M. De novo peptide sequencing by deep learning. Proc. Natl. Acad. Sci. USA 2017, 114, 8247–8252. [Google Scholar] [CrossRef] [PubMed]
- Välikangas, T.; Suomi, T.; Elo, L.L. A systematic evaluation of normalization methods in quantitative label-free proteomics. Briefings Bioinform. 2016, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Berger, J.A.; Hautaniemi, S.; Järvinen, A.-K.; Edgren, H.; Mitra, S.K.; Astola, J. Optimized LOWESS normalization parameter selection for DNA microarray data. BMC Bioinform. 2004, 5, 194. [Google Scholar] [CrossRef]
- Bern, M.W.; Goldberg, D. De Novo Analysis of Peptide Tandem Mass Spectra by Spectral Graph Partitioning. J. Comput. Biol. 2006, 13, 364–378. [Google Scholar] [CrossRef]
- Matzke, M.M.; Brown, J.N.; Gritsenko, M.A.; Metz, T.O.; Pounds, J.G.; Rodland, K.D.; Shukla, A.K.; Smith, R.D.; Waters, K.M.; McDermott, J.E.; et al. A comparative analysis of computational approaches to relative protein quantification using peptide peak intensities in label-free LC-MS proteomics experiments. Proteomics 2012, 13, 493–503. [Google Scholar] [CrossRef]
- Branson, O.E.; Freitas, M.A. A multi-model statistical approach for proteomic spectral count quantitation. J. Proteom. 2016, 144, 23–32. [Google Scholar] [CrossRef]
- Kammers, K.; Cole, R.N.; Tiengwe, C.; Ruczinski, I. Detecting significant changes in protein abundance. EuPA Open Proteom. 2015, 7, 11–19. [Google Scholar] [CrossRef]
- Chawinga, W.D.; Zinn, S. Global perspectives of research data sharing: A systematic literature review. Libr. Inf. Sci. Res. 2019, 41, 109–122. [Google Scholar] [CrossRef]
- Šoltić, D.; Fuller, H.R. Molecular Crosstalk Between Non-SMN-Related and SMN-Related Spinal Muscular Atrophy. Neurosci. Insights 2020, 15, 2633105520914301. [Google Scholar]
- Šoltić, D.; Bowerman, M.; Stock, J.; Shorrock, H.K.; Gillingwater, T.H.; Fuller, H.R. Multi-Study Proteomic and Bioinformatic Identification of Molecular Overlap between Amyotrophic Lateral Sclerosis (ALS) and Spinal Muscular Atrophy (SMA). Brain Sci. 2018, 8, 212. [Google Scholar] [CrossRef] [PubMed]
- Vaudel, M.; Verheggen, K.; Csordas, A.; Raeder, H.; Berven, F.S.; Martens, L.; Vizcaíno, J.A.; Barsnes, H. Exploring the potential of public proteomics data. Proteomics 2015, 16, 214–225. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S.; Merchant, M.; Rai, A.; Rai, S.N. Interactive Web Tool for Standardizing Proteomics Workflow for Liquid Chromatography-Mass Spectrometry Data. J. Proteom. Bioinform. 2019, 12, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W.; Csordas, A.; Sun, Z.; Jarnuczak, A.; Perez-Riverol, Y.; Ternent, T.; Campbell, D.S.; Bernal-Llinares, M.; Okuda, S.; Kawano, S.; et al. The ProteomeXchange consortium in 2017: Supporting the cultural change in proteomics public data deposition. Nucleic Acids Res. 2017, 45, D1100–D1106. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W.; Bandeira, N.; Sharma, V.; Perez-Riverol, Y.; Carver, J.J.; Kundu, D.J.; García-Seisdedos, D.; Jarnuczak, A.F.; Hewapathirana, S.; Pullman, B.S.; et al. The ProteomeXchange consortium in 2020: Enabling ‘big data’ approaches in proteomics. Nucleic Acids Res. 2020, 48, D1145–D1152. [Google Scholar] [CrossRef] [PubMed]
- Vizcaíno, J.A.; Deutsch, E.W.; Wang, R.; Csordas, A.; Reisinger, F.; Ríos, D.; Dianes, J.A.; Sun, Z.; Farrah, T.; Bandeira, N.; et al. ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat. Biotechnol. 2014, 32, 223–226. [Google Scholar] [CrossRef]
- Roy, S.; Coldren, C.; Karunamurthy, A.; Kip, N.S.; Klee, E.W.; Lincoln, S.E.; Leon, A.; Pullambhatla, M.; Temple-Smolkin, R.L.; Voelkerding, K.V.; et al. Standards and Guidelines for Validating Next-Generation Sequencing Bioinformatics Pipelines. J. Mol. Diagn. 2018, 20, 4–27. [Google Scholar] [CrossRef]
- Kline, R.A.; Dissanayake, K.N.; Hurtado, M.L.; Martínez, N.; Ahl, A.; Mole, A.J.; Lamont, D.J.; Court, F.A.; Ribchester, R.R.; Wishart, T.; et al. Altered mitochondrial bioenergetics are responsible for the delay in Wallerian degeneration observed in neonatal mice. Neurobiol. Dis. 2019, 130, 104496. [Google Scholar] [CrossRef]
- Hurtado, M.L.; Fuller, H.R.; Wong, A.M.S.; Eaton, S.L.; Gillingwater, T.H.; Pennetta, G.; Cooper, J.D.; Wishart, T.M. Proteomic mapping of differentially vulnerable pre-synaptic populations identifies regulators of neuronal stability in vivo. Sci. Rep. 2017, 7, 12412. [Google Scholar] [CrossRef]
- Shorrock, H.K.; Hoorn, D.; Boyd, P.J.; Hurtado, M.L.; Lamont, D.J.; Wirth, B.; Sleigh, J.N.; Schiavo, G.; Wishart, T.M.; Groen, E.J.N.; et al. UBA1/GARS-dependent pathways drive sensory-motor connectivity defects in spinal muscular atrophy. Brain 2018, 141, 2878–2894. [Google Scholar] [CrossRef] [PubMed]
- Powis, R.A.; Karyka, E.; Boyd, P.; Côme, J.; Jones, R.A.; Zheng, Y.; Szunyogova, E.; Groen, E.J.; Hunter, G.; Thomson, D.; et al. Systemic restoration of UBA1 ameliorates disease in spinal muscular atrophy. JCI Insight 2016, 1, e87908. [Google Scholar] [CrossRef] [PubMed]
- Wishart, T.; Mutsaers, C.A.; Riessland, M.; Reimer, M.M.; Hunter, G.; Hannam, M.L.; Eaton, S.L.; Fuller, H.; Roche, S.L.; Somers, E.; et al. Dysregulation of ubiquitin homeostasis and β-catenin signaling promote spinal muscular atrophy. J. Clin. Investig. 2014, 124, 1821–1834. [Google Scholar] [CrossRef] [PubMed]
- Meunier, B.; Dumas, E.; Piec, I.; Béchet, D.; Hébraud, M.; Hocquette, J.-F. Assessment of Hierarchical Clustering Methodologies for Proteomic Data Mining. J. Proteome Res. 2006, 6, 358–366. [Google Scholar] [CrossRef]
- Theocharidis, A.; Van Dongen, S.; Enright, A.; Freeman, T. Network visualization and analysis of gene expression data using BioLayout Express3D. Nat. Protoc. 2009, 4, 1535–1550. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; A Lempicki, R. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008, 4, 44–57. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Graham, L.C.; Eaton, S.L.; Brunton, P.J.; Atrih, A.; Smith, C.; Lamont, D.J.; Gillingwater, T.H.; Pennetta, G.; Skehel, P.; Wishart, T.M. Proteomic profiling of neuronal mitochondria reveals modulators of synaptic architecture. Mol. Neurodegener. 2017, 12, 1–16. [Google Scholar] [CrossRef]
- Alanis-Lobato, G.; Andrade-Navarro, M.A.; Schaefer, M.H. HIPPIE v2.0: Enhancing meaningfulness and reliability of protein–protein interaction networks. Nucleic Acids Res. 2016, 45, D408–D414. [Google Scholar] [CrossRef]
- Costa, P.R.; Acencio, M.L.; Lemke, N. A machine learning approach for genome-wide prediction of morbid and druggable human genes based on systems-level data. BMC Genom. 2010, 11, S9. [Google Scholar] [CrossRef] [PubMed]
- Kandoi, G.; Acencio, M.L.; Lemke, N. Prediction of Druggable Proteins Using Machine Learning and Systems Biology: A Mini-Review. Front. Physiol. 2015, 6, 366. [Google Scholar] [CrossRef] [PubMed]
- Ogata, H.; Goto, S.; Sato, K.; Fujibuchi, W.; Bono, H.; Kanehisa, M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 29–34. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Cerami, E.G.; Gross, B.E.; Demir, E.; Rodchenkov, I.; Babur, O.; Anwar, N.; Schultz, N.; Bader, G.; Sander, C. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 2010, 39, D685–D690. [Google Scholar] [CrossRef]
- Ravikumar, K.E.; Wagholikar, K.B.; Li, D.; Kocher, J.-P.; Liu, H. Text mining facilitates database curation - extraction of mutation-disease associations from Bio-medical literature. BMC Bioinform. 2015, 16, 185. [Google Scholar] [CrossRef]
- Kitsak, M.; Sharma, A.; Menche, J.; Guney, E.; Ghiassian, S.D.; Loscalzo, J.; Barabási, A.-L. Tissue Specificity of Human Disease Module. Sci. Rep. 2016, 6, 35241. [Google Scholar] [CrossRef]
- Kovács, I.A.; Barabási, D.L.; Barabási, A.-L. Uncovering the genetic blueprint of the C. elegans nervous system. Proc. Natl. Acad. Sci. USA 2020, 117, 33570–33577. [Google Scholar] [CrossRef]
- Sarno, F.; The International Network Medicine Consortium; Benincasa, G.; List, M.; Barabasi, A.-L.; Baumbach, J.; Ciardiello, F.; Filetti, S.; Glass, K.; Loscalzo, J.; et al. Clinical epigenetics settings for cancer and cardiovascular diseases: Real-life applications of network medicine at the bedside. Clin. Epigenetics 2021, 13, 1–38. [Google Scholar] [CrossRef]
- Roy, M.; Sorokina, O.; McLean, C.; Tapia-González, S.; DeFelipe, J.; Armstrong, J.D.; Grant, S.G.N. Regional Diversity in the Postsynaptic Proteome of the Mouse Brain. Proteomes 2018, 6, 31. [Google Scholar] [CrossRef]
- Blokhuis, A.M.; Koppers, M.; Groen, E.J.; Heuvel, D.M.A.V.D.; Modigliani, S.D.; Anink, J.J.; Fumoto, K.; Van Diggelen, F.; Snelting, A.; Sodaar, P.; et al. Comparative interactomics analysis of different ALS-associated proteins identifies converging molecular pathways. Acta Neuropathol. 2016, 132, 175–196. [Google Scholar] [CrossRef] [PubMed]
- Henstridge, C.M.; Sideris, D.I.; Carroll, E.; Rotariu, S.; Salomonsson, S.; Tzioras, M.; McKenzie, C.-A.; Smith, C.; von Arnim, C.A.F.; Ludolph, A.C.; et al. Synapse loss in the prefrontal cortex is associated with cognitive decline in amyotrophic lateral sclerosis. Acta Neuropathol. 2017, 135, 213–226. [Google Scholar] [CrossRef] [PubMed]
- Frey, D.; Schneider, C.; Xu, L.; Borg, J.; Spooren, W.; Caroni, P. Early and Selective Loss of Neuromuscular Synapse Subtypes with Low Sprouting Competence in Motoneuron Diseases. J. Neurosci. 2000, 20, 2534–2542. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J. Alzheimer’s Disease Is a Synaptic Failure. Science 2002, 298, 789–791. [Google Scholar] [CrossRef] [PubMed]
- Cunningham, C.; Deacon, R.; Wells, H.; Boche, D.; Waters, S.; Diniz, C.P.; Scott, H.; Rawlins, J.N.P.; Perry, V.H. Synaptic changes characterize early behavioural signs in the ME7 model of murine prion disease. Eur. J. Neurosci. 2003, 17, 2147–2155. [Google Scholar] [CrossRef]
- Day, M.; Wang, Z.; Ding, J.; An, X.; Ingham, C.A.; Shering, A.F.; Wokosin, D.; Ilijic, E.; Sun, Z.; Sampson, A.R.; et al. Selective elimination of glutamatergic synapses on striatopallidal neurons in Parkinson disease models. Nat. Neurosci. 2006, 9, 251–259. [Google Scholar] [CrossRef]
- Perkins, E.M.; Clarkson, Y.L.; Sabatier, N.; Longhurst, D.M.; Millward, C.P.; Jack, J.; Toraiwa, J.; Watanabe, M.; Rothstein, J.D.; Lyndon, A.R.; et al. Loss of β-III spectrin leads to Purkinje cell dysfunction recapitulating the behavior and neuropathology of spinocerebellar ataxia type 5 in humans. J. Neurosci. 2010, 30, 4857–4867. [Google Scholar] [CrossRef]
- Trotta, N.; Orso, G.; Rossetto, M.G.; Daga, A.; Broadie, K. The Hereditary Spastic Paraplegia Gene, spastin, Regulates Microtubule Stability to Modulate Synaptic Structure and Function. Curr. Biol. 2004, 14, 1135–1147. [Google Scholar] [CrossRef]
- Li, J.Y.; Plomann, M.; Brundin, P. Huntington’s disease: A synaptopathy? Trends Mol. Med. 2003, 9, 414–420. [Google Scholar] [CrossRef]
- Kielar, C.; Wishart, T.; Palmer, A.; Dihanich, S.; Wong, A.M.; Macauley, S.; Chan, C.-H.; Sands, M.; Pearce, D.A.; Cooper, J.; et al. Molecular correlates of axonal and synaptic pathology in mouse models of Batten disease. Hum. Mol. Genet. 2009, 18, 4066–4080. [Google Scholar] [CrossRef]
- Gomez-Giro, G.; Arias-Fuenzalida, J.; Jarazo, J.; Zeuschner, D.; Ali, M.; Possemis, N.; Bolognin, S.; Halder, R.; Jäger, C.; Kuper, W.F.E.; et al. Synapse alterations precede neuronal damage and storage pathology in a human cerebral organoid model of CLN3-juvenile neuronal ceroid lipofuscinosis. Acta Neuropathol. Commun. 2019, 7, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Bowerman, M.; Murray, L.M.; Scamps, F.; Schneider, B.L.; Kothary, R.; Raoul, C. Pathogenic commonalities between spinal muscular atrophy and amyotrophic lateral sclerosis: Converging roads to therapeutic development. Eur. J. Med Genet. 2018, 61, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Zou, T.; Ilangovan, R.; Yu, F.; Xu, Z.; Zhou, J. SMN protects cells against mutant SOD1 toxicity by increasing chaperone activity. Biochem. Biophys. Res. Commun. 2007, 364, 850–855. [Google Scholar] [CrossRef] [PubMed]
- Turner, B.J.; Alfazema, N.; Sheean, R.K.; Sleigh, J.N.; Davies, K.E.; Horne, M.K.; Talbot, K. Overexpression of survival motor neuron improves neuromuscular function and motor neuron survival in mutant SOD1 mice. Neurobiol. Aging 2013, 35, 906–915. [Google Scholar] [CrossRef]
- Turner, B.J.; Bäumer, D.; Parkinson, N.J.; Scaber, J.; Ansorge, O.; Talbot, K. TDP-43 expression in mouse models of amyotrophic lateral sclerosis and spinal muscular atrophy. BMC Neurosci. 2008, 9, 104. [Google Scholar] [CrossRef]
- Turner, B.; Parkinson, N.J.; Davies, K.E.; Talbot, K. Survival motor neuron deficiency enhances progression in an amyotrophic lateral sclerosis mouse model. Neurobiol. Dis. 2009, 34, 511–517. [Google Scholar] [CrossRef]
- Gertz, B.; Wong, M.; Martin, L.J. Nuclear Localization of Human SOD1 and Mutant SOD1-Specific Disruption of Survival Motor Neuron Protein Complex in Transgenic Amyotrophic Lateral Sclerosis Mice. J. Neuropathol. Exp. Neurol. 2012, 71, 162–177. [Google Scholar] [CrossRef]
- Kariya, S.; Re, D.B.; Jacquier, A.; Nelson, K.; Przedborski, S.; Monani, U.R. Mutant superoxide dismutase 1 (SOD1), a cause of amyotrophic lateral sclerosis, disrupts the recruitment of SMN, the spinal muscular atrophy protein to nuclear Cajal bodies. Hum. Mol. Genet. 2012, 21, 3421–3434. [Google Scholar] [CrossRef]
- Mirra, A.; Rossi, S.; Scaricamazza, S.; Di Salvio, M.; Salvatori, I.; Valle, C.; Rusmini, P.; Poletti, A.; Cestra, G.; Carri, M.T.; et al. Functional interaction between FUS and SMN underlies SMA-like splicing changes in wild-type hFUS mice. Sci. Rep. 2017, 7, 1–14. [Google Scholar] [CrossRef]
- Groen, E.J.; Fumoto, K.; Blokhuis, A.M.; Engelen-Lee, J.; Zhou, Y.; van den Heuvel, D.M.; Koppers, M.; van Diggelen, F.; van Heest, J.; Demmers, J.A.; et al. ALS-associated mutations in FUS disrupt the axonal distribution and function of SMN. Hum. Mol. Genet. 2013, 22, 3690–3704. [Google Scholar] [CrossRef]
- Kline, R.A.; Kaifer, K.A.; Osman, E.; Carella, F.; Tiberi, A.; Ross, J.; Pennetta, G.; Lorson, C.L.; Murray, L.M. Comparison of independent screens on differentially vulnerable motor neurons reveals alpha-synuclein as a common modifier in motor neuron diseases. PLoS Genet. 2017, 13, e1006680. [Google Scholar] [CrossRef] [PubMed]
- Boyd, P.J.; Tu, W.-Y.; Shorrock, H.K.; Groen, E.J.N.; Carter, R.N.; Powis, R.A.; Thomson, S.R.; Thomson, D.; Graham, L.C.; Motyl, A.A.L.; et al. Bioenergetic status modulates motor neuron vulnerability and pathogenesis in a zebrafish model of spinal muscular atrophy. PLoS Genet. 2017, 13, e1006744. [Google Scholar] [CrossRef] [PubMed]
- Gonçalves, I.D.; Rehorst, W.A.; Kye, M.J. Dysregulation of RNA Mediated Gene Expression in Motor Neuron Diseases. CNS Neurol. Disord. Drug Targets 2016, 15, 887–895. [Google Scholar] [CrossRef] [PubMed]
- Gama-Carvalho, M.; Garcia-Vaquero, M.L.; Pinto, F.R.; Besse, F.; Weis, J.; Voigt, A.; Schulz, J.B.; Rivas, J.D.L. Linking amyotrophic lateral sclerosis and spinal muscular atrophy through RNA-transcriptome homeostasis: A genomics perspective. J. Neurochem. 2017, 141, 12–30. [Google Scholar] [CrossRef] [PubMed]
- Fuller, H.R.; Gillingwater, T.; Wishart, T.M. Commonality amid diversity: Multi-study proteomic identification of conserved disease mechanisms in spinal muscular atrophy. Neuromuscul. Disord. 2016, 26, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Kline, R.A.; Wishart, T.M.; Mills, K.; Heywood, W.E. Applying modern Omic technologies to the Neuronal Ceroid Lipofuscinoses. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2019, 1866, 165498. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, N.; Akiyama, T.; Warita, H.; Aoki, M. Omics Approach to Axonal Dysfunction of Motor Neurons in Amyotrophic Lateral Sclerosis (ALS). Front. Neurosci. 2020, 14, 194. [Google Scholar] [CrossRef]
- Sowell, R.A.; Owen, J.B.; Butterfield, D.A. Proteomics in animal models of Alzheimer's and Parkinson’s diseases. Ageing Res. Rev. 2009, 8, 1–17. [Google Scholar] [CrossRef]
- Ubaida-Mohien, C.; Lyashkov, A.; Gonzalez-Freire, M.; Tharakan, R.; Shardell, M.; Moaddel, R.; Semba, R.D.; Chia, C.W.; Gorospe, M.; Sen, R.; et al. Discovery proteomics in aging human skeletal muscle finds change in spliceosome, immunity, proteostasis and mitochondria. eLife 2019, 8, e49874. [Google Scholar] [CrossRef]
- Bai, B.; Wang, X.; Li, Y.; Chen, P.-C.; Yu, K.; Dey, K.K.; Yarbro, J.M.; Han, X.; Lutz, B.M.; Rao, S.; et al. Deep Multilayer Brain Proteomics Identifies Molecular Networks in Alzheimer’s Disease Progression. Neuron 2020, 105, 975–991.e7. [Google Scholar] [CrossRef]
- Higginbotham, L.; Ping, L.; Dammer, E.B.; Duong, D.M.; Zhou, M.; Gearing, M.; Hurst, C.; Glass, J.D.; Factor, S.A.; Johnson, E.C.B.; et al. Integrated proteomics reveals brain-based cerebrospinal fluid biomarkers in asymptomatic and symptomatic Alzheimer’s disease. Sci. Adv. 2020, 6, eaaz9360. [Google Scholar] [CrossRef] [PubMed]
- Johnson, E.C.B.; Dammer, E.B.; Duong, D.; Ping, L.; Zhou, M.; Yin, L.; Higginbotham, L.A.; Guajardo, A.; White, B.; Troncoso, J.C.; et al. Large-scale proteomic analysis of Alzheimer’s disease brain and cerebrospinal fluid reveals early changes in energy metabolism associated with microglia and astrocyte activation. Nat. Med. 2020, 26, 769–780. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Huang, Z.; Feng, C.; Guan, C.; Li, R.; Xie, H.; Chen, J.; Li, M.; Que, R.; Deng, B.; et al. Multimodal analysis of gene expression from postmortem brains and blood identifies synaptic vesicle trafficking genes to be associated with Parkinson’s disease. Brief. Bioinform. 2021, 22, bbaa244. [Google Scholar] [CrossRef] [PubMed]
- Catenaccio, A.; Hurtado, M.L.; Diaz, P.; Lamont, D.J.; Wishart, T.M.; Court, F.A. Molecular analysis of axonal-intrinsic and glial-associated co-regulation of axon degeneration. Cell Death Dis. 2017, 8, e3166. [Google Scholar] [CrossRef]
- Wishart, T.; Rooney, T.M.; Lamont, D.J.; Wright, A.K.; Morton, A.J.; Jackson, M.; Freeman, M.; Gillingwater, T.H. Combining Comparative Proteomics and Molecular Genetics Uncovers Regulators of Synaptic and Axonal Stability and Degeneration In Vivo. PLoS Genet. 2012, 8, e1002936. [Google Scholar] [CrossRef]
- Wishart, T.; Parson, S.H.; Gillingwater, T.H. Synaptic Vulnerability in Neurodegenerative Disease. J. Neuropathol. Exp. Neurol. 2006, 65, 733–739. [Google Scholar] [CrossRef]
- Coleman, M.P.; Freeman, M.R. Wallerian degeneration, wld(s), and nmnat. Annu. Rev. Neurosci. 2010, 33, 245–267. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kline, R.A.; Lößlein, L.; Kurian, D.; Aguilar Martí, J.; Eaton, S.L.; Court, F.A.; Gillingwater, T.H.; Wishart, T.M. An Optimized Comparative Proteomic Approach as a Tool in Neurodegenerative Disease Research. Cells 2022, 11, 2653. https://doi.org/10.3390/cells11172653
Kline RA, Lößlein L, Kurian D, Aguilar Martí J, Eaton SL, Court FA, Gillingwater TH, Wishart TM. An Optimized Comparative Proteomic Approach as a Tool in Neurodegenerative Disease Research. Cells. 2022; 11(17):2653. https://doi.org/10.3390/cells11172653
Chicago/Turabian StyleKline, Rachel A., Lena Lößlein, Dominic Kurian, Judit Aguilar Martí, Samantha L. Eaton, Felipe A. Court, Thomas H. Gillingwater, and Thomas M. Wishart. 2022. "An Optimized Comparative Proteomic Approach as a Tool in Neurodegenerative Disease Research" Cells 11, no. 17: 2653. https://doi.org/10.3390/cells11172653
APA StyleKline, R. A., Lößlein, L., Kurian, D., Aguilar Martí, J., Eaton, S. L., Court, F. A., Gillingwater, T. H., & Wishart, T. M. (2022). An Optimized Comparative Proteomic Approach as a Tool in Neurodegenerative Disease Research. Cells, 11(17), 2653. https://doi.org/10.3390/cells11172653








